Open Access Article
Open Access ArticleDNA condensation in live E. coli provides evidence for transertion†
Anil K.
Gorle
a,
Amy L.
Bottomley
b,
Elizabeth J.
Harry
b,
J. Grant
Collins
*a,
F. Richard
Keene
 *cd and
Clifford E.
Woodward
*a
*cd and
Clifford E.
Woodward
*a
aSchool of Physical, Environmental and Mathematical Sciences, University of New South Wales, Australian Defence Force Academy, Canberra, ACT 2600, Australia. E-mail: g.collins@adfa.edu.au; c.woodward@adfa.edu.au
bThe ithree institute, University of Technology Sydney, PO Box 123, Broadway, NSW 2007, Australia
cCentre for Biodiscovery & Molecular Development of Therapeutics, James Cook University, Townsville, QLD 4811, Australia
dSchool of Physical Sciences, University of Adelaide, Adelaide, SA 5066, Australia. E-mail: richard.keene@adelaide.edu.au
First published on 20th February 2017
Abstract
Condensation studies of chromosomal DNA in E. coli with a tetranuclear ruthenium complex are carried out and images obtained with wide-field fluorescence microscopy. Remarkably different condensate morphologies resulted, depending upon the treatment protocol. The occurrence of condensed nucleoid spirals in live bacteria provides evidence for the transertion hypothesis.
In Escherichia coli (E. coli), chromosomal DNA has a contour length approximately 1000-fold larger than the ∼1 μm cell width, forcing it to adopt a compact super-coiled structure within the cell cavity.1–3 However, the mechanisms by which this is achieved are still not well understood.1–3 It is conjectured that depletion forces in the presence of other cell components (protein and ribosomes) are sufficient to stabilise a nucleoid phase that remains centrally located due to entropic repulsion from the cell wall.4,5 These compressive forces are hypothesised to be opposed by an expansive mechanical force due to transertion, when proteins are inserted directly into the membrane while being expressed by the nucleoid. While a number of studies support the transertion hypothesis, further work is required to establish its veracity.1,6,7
We have recently demonstrated that a series of linear multinuclear ruthenium(II) complexes with a flexible bbn bridging ligand {bbn = bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane} exhibit excellent antimicrobial properties against both Gram positive and Gram negative bacteria.8–11 In E. coli, wide-field fluorescence microscopy revealed that the dinuclear (+4) Rubb16 complex was able to condense both ribosomal material as well as chromosomal DNA.11 Highly-charged cations induce condensation by an attractive charge correlation mechanism, which can dominate the entropic repulsion due to ion crowding and favour a compact macro-ionic structure.12
Studies (predominately in vitro) have revealed that the structure of condensed DNA is strongly affected by the size and intrinsic curvature of the DNA molecule.13,14 Furthermore, magnetic tweezers have been used to show that applied forces on single DNA molecules can also affect the condensed morphology.15 With this in mind, we sought to use DNA condensation in live E. coli by ruthenium(II) complexes as a means to probe the nature of the forces acting on chromosomal DNA within the cell cavity and, in particular, the role played by putative transertion links.
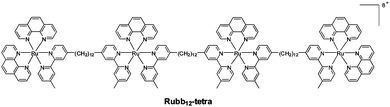
In this study, we used a tetranuclear ruthenium complex (Rubb12-tetra; Fig. 1) as the condensing agent. The greater charge of this complex (+8) should enhance DNA condensation, compared to the (+4) complex used in our previous experiments.11 Using wide-field fluorescence microscopy, we investigated the morphology of the condensed chromosomal DNA in native E. coli, and also where the cells were pre-treated with the bacterial agents rifampicin and chloramphenicol, which halt transcription and translation respectively.16
The minimum inhibitory concentration (MIC) of Rubb12-tetra against E. coli MG1665 was determined to be 9.7 μM. E. coli cells were incubated with Rubb12-tetra at concentrations ranging from of 0.5× MIC to 2.0× MIC for either 30 or 60 minutes at 37 °C. After the cells were washed with phosphate buffer, they were incubated with the DNA-selective dye DAPI at room temperature for 20 minutes before processing them for microscopic imaging.
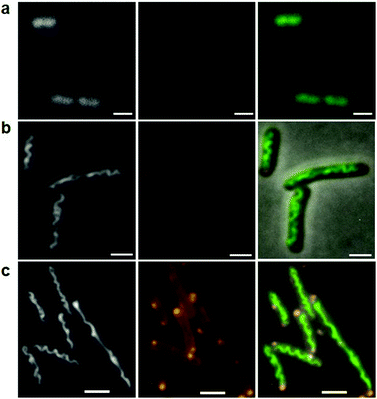
Fig. 2 compares typical images of the general population of both viable (dividing) and non-viable cells treated with different concentrations of ruthenium complex. We observed relatively high emission of the ruthenium complex, at both the central and polar regions of treated cells (coloured orange in Fig. 2). This pattern was also observed in earlier studies with Rubb16.11 We proposed there that the bright regions were due to aggregation of polysomes following association of the ruthenium complex with the constituent ribosomal RNA. Polysomes consist of 30S and 50S ribosomal subunits assembled on messenger RNA, to form necklaces of (negatively-charged) 70S ribosomes.17 The metal complex can aggregate polysomes via strong electrostatic correlations.12 These aggregates more likely form mid-cell and at the poles, where the polysomal concentration is relatively high. The similarity of the emission patterns for both Rubb16 and the bulkier Rubb12-tetra complex strongly supports such a mechanism, rather than an explanation based on specific binding of the metal complex.
In Fig. 2 we also show the fluorescence of the DNA-selective dye DAPI at both 1× and 2× MIC, Fig. 2(b) and (c). Comparison with untreated cells, Fig. 2(a), demonstrates that the Rubb12-tetra complex caused the nucleoid to become axially condensed into a rope that adopts a spiral shape, aligned with the long axis of the cell. While the ruthenium complex is likely to associate strongly with the DNA, we have shown that its emission is significantly quenched in the nucleoid environment (ESI:† Fig. S1). The degree of DNA condensation increased with both the incubation time and the concentration of the metal complex. The condensed DNA spirals were observed in essentially all the incubated cells with a similar number of turns, albeit with some fluctuation in pitch along their length. Furthermore, the spirals were stable over the longest observation time (at least 60 minutes) which is large compared to typical relaxation times for DNA condensation (in vitro).13 Importantly, we also observed several highly elongated cells wherein the condensed DNA spiral appears to stretch, so as to fill the intracellular space (e.g., see the elongated cell in the bottom panel in Fig. 2). Elongation occurs in (presumably) non-viable cells due to the continued uniform growth of the cell wall without division. In the elongated cell seen here, the condensed DNA rope appears to display a similar number of turns as in the shorter cells, but with a greater degree of twist (number of rotations around the central axis) and reduced writhe (degree of coiling). This indicates that the condensed DNA stretches upon cell elongation and that replication has been halted (or slowed), most likely due to the effects of condensation. Stretching, coupled with increasing twist, indicates that the nucleoid is attached to the inner membrane close to both poles and possibly more spots.
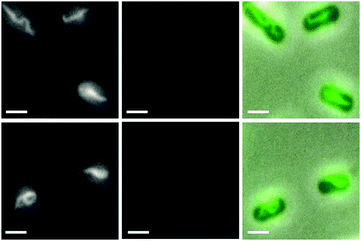
To further probe this system, we also carried out condensation experiments on cells pre-treated with anti-bacterial agents. E. coli cells were first treated with rifampicin (RIF) for 30 minutes. RIF causes the nucleoid to expand to fill the intracellular volume (see ESI,† Fig. S2, panel a), as is well known.16 RIF blocks transcription, causing almost all ribosomes to be converted to their 30S and 50S subunits. In the presence of these smaller particles, de-mixing of the nucleoid is suppressed causing the nucleoid to expand into the cell volume.4,5,11,16 Subsequent incubation of RIF-treated cells with the Rubb12-tetra complex (15 minutes at 1× MIC) caused the dispersed nucleoid to condense (examples shown in Fig. 3, and ESI,† Fig. S3).
Most of the cells (>90%) displayed linear nucleoid strands, which had quite a different morphology to the spirals seen in Fig. 2. As shown in Fig. 3, some of the linear condensates appeared to be forming loops, suggesting they were transforming into a more condensed structure, most likely toroids.18 Indeed, a few cells (∼1%) contained a globular condensed nucleoid at the centre of the cell, although it was not possible to ascertain if these were toroidal. These morphologies are similar to those observed for DNA condensation in vitro, where linear and toroidal morphologies are the preferred shape of condensed DNA.13 We did not observe significant ruthenium complex emission, indicating that the ribosomal subunits are not easily condensed.
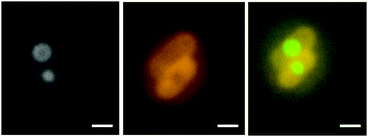
E. coli cells were also pre-treated with chloramphenicol (CAM). CAM halts translation giving rise to stalled polysomes and a diminished population of 30S and 50S ribosomal sub-units. As shown in ESI,† Fig. S2 (panel b), CAM causes the nucleoid to condense into a compact globular shape (probably toroids),19 presumably driven by the osmotic pressure of the stalled polysomes and other cellular components, which remain de-mixed from the nucleoid phase. Following CAM treatment and subsequent incubation with the Rubb12-tetra complex (15 minutes at 1× MIC), we observed that the shape of the nucleoid condensates were more clearly toroidal (Fig. 4) – with yet again no evidence of the spiral morphology seen earlier. On the other hand, we did see emission from the metal complex, which is presumably associated with the polysomes.
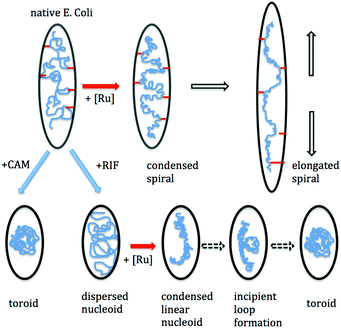
Our observations suggest that two different (stable) nucleoid morphologies are possible in live bacteria in the presence of a sufficient amount of metal complex – toroidal (or globular) or an extended spiral. The former occurs after pre-treatment with either RIF or CAM, while the spiral morphology is exhibited in previously untreated cells. CAM treatment shows that a toroidal or globular morphology for the nucleoid may also occur in the presence of a large number of polysomes. While such conditions also prevail in rapidly growing cells, in that case the nucleoid maintains a relatively expanded morphology. The transertion hypothesis proposes that in rapidly growing cells, one end of the message (mRNA) for membrane proteins is being transcribed by the chromosome, while translated proteins are being inserted into the inner membrane at the other end. Thus the nucleoid is transiently attached to the membrane via a transcription-translation-insertion chain.20 It has been proposed that approximately ten transertion connections may occur along the length of the nucleoid at any one time,1 and it is this tethering which stabilises the nucleoid against further osmotic compaction.
Treatment with either RIF and CAM would break transertion chains due to halting of either transcription (RIF) or translation (CAM).5 Consistent with this, is our finding that subsequent incubation with the tetranuclear complex causes the nucleoid to condense in a manner which mimics that of free DNA in vitro. Condensation of the nucleoid with Rubb12-tetra can be explained by strong charge fluctuations, which give rise to attractions between DNA segments.12 These attractions cause the nucleoid to readily form an axially-condensed rope-like structure. In native cells with the ruthenium complex, the rope adopts a well-defined spiral superstructure with many turns (Fig. 2). However, if pre-treatment with RIF occurs prior to metal complex addition, a much more linear rope appears to condense, from the initially dispersed nucleoid. Furthermore, this appears to be a meta-stable state with incipient loop formation indicating a transition to stable toroids (Fig. 3). Indeed, a small percentage of cells displayed a globular nucleoid, and it is reasonable to expect that longer incubation times (>15 minutes) with the metal complex would increase the occurrence of these. On the other hand, if cells are pre-treated with CAM, addition of the metal complex preserves the globular (toroidal) condensate. A summary of this behaviour is depicted in Fig. 5.
The spiral morphology observed for the condensed DNA in native cells is surprising, as in vitro studies suggest that a compact structure should be the most stable form.13 This leads us to propose that the nucleoid is attached to the cell wall and, were it not for the frustration caused by this attachment, electrostatic correlations would favour a compact morphology (probably toroids). If this attachment is achieved through the putative transertion links, then these must be preserved upon addition of the metal complex. This can be achieved via condensation of the polysome chain involved in transertion, thus halting translation of partially imbedded membrane proteins. Previous work by us provided evidence for polysomal condensation in the presence of the metal complex,11 which is further supported in this study. At the other end of the transertion chain, transcription can be stalled via DNA condensation. One explanation for the spiral shape is that it results from supercoiling due to torsional stress which is pinned at the membrane by transertion links upon condensation. The torsional stress is relieved by increasing the degree of writhe in the nucleoid.21 Alternatively, supercoiling may be caused by electrostatic correlations between DNA segments. In this case, the helices can be formed via attracting loops of condensed DNA, frustrated by transertion links. Further, strong evidence of membrane attachment is provided by the nucleoid behaviour in elongated cells. Here the degree of writhe in the nucleoid appears to decrease, leading to greater twist in the condensed rope. That is, the nucleoid appears to extend with cell elongation without unwinding, indicating the extended nucleoid is under tension, rather than compression. Clearly, this can only occur if the DNA is attached to the membrane.
In conclusion, there are very few studies of DNA condensation in live bacteria. This is the first time that a spiral-shaped condensate has been observed in live bacteria. Our findings support the existence of transertion in E. coli cells, which appears to be an important fundamental mechanism in the cellular biology.
References
- C. L. Woldringh, Mol. Microbiol., 2002, 45, 17 CrossRef CAS PubMed.
- T. Cremer and C. Cremer, Chromosome territories, nuclear architecture and gene regulation in mammalian cells, Nat. Rev. Genet., 2001, 2, 292–301 CrossRef CAS PubMed.
- A. S. Nouwens, F. G. Hopwood, M. Traini, K. L. Williams and B. J. Walsh, Organization of the Prokaryotic Genome, ed. R. L. Charlebois, American Society for Microbiology Press, Washington, DC, 1999, ch. 10, pp. 331–346 Search PubMed.
- T. Odijk, Biophys. Chem., 1998, 73, 23–30 CrossRef CAS PubMed.
- J. Mondal, B. P. Bratton, Y. Li, A. Yethiraj and J. C. Weisshaar, Biophys. J., 2011, 100, 2605 CrossRef CAS PubMed.
- E. A. Libby, M. Roggiani and M. Goulian, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7445 CrossRef CAS PubMed.
- M. Roggiani and M. Goulian, Annu. Rev. Genet., 2015, 49, 115 CrossRef CAS PubMed.
- F. Li, M. Feterl, Y. Mulyana, J. M. Warner, J. G. Collins and F. R. Keene, J. Antimicrob. Chemother., 2012, 67, 2686 CrossRef CAS PubMed.
- A. K. Gorle, M. Feterl, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2014, 43, 16713 RSC.
- F. Li, Y. Mulyana, M. Feterl, J. M. Warner, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 5032 RSC.
- F. Li, E. J. Harry, A. L. Bottomley, M. D. Edstein, G. W. Birrell, C. E. Woodward, F. R. Keene and J. G. Collins, Chem. Sci., 2014, 5, 685 RSC.
- L. Guldbrand, B. G. Jonsson., H. Wennerstrom and P. Linse, J. Chem. Phys., 1984, 80, 2221 CrossRef CAS.
- I. D. Vilfan, C. C. Conwell, T. Sarkar and N. V. Hud, Biochemistry, 2006, 45, 8174 CrossRef CAS PubMed.
- V. B. Teif and K. Bohinc, Prog. Biophys. Mol. Biol., 2011, 105, 208 CrossRef CAS PubMed.
- K. Besteman, S. Hage, N. H. Dekker and S. G. Lemay, Phys. Rev. Lett., 2007, 98, 058103 CrossRef CAS PubMed.
- S. Bakshi, H. Choi, J. Mondal and J. C. Weisshaar, Mol. Microbiol., 2014, 94, 871 CrossRef CAS PubMed.
- F. C. Neidhardt, Escherichia coli and salmonella: cellular and molecular biology, American society for microbiology, Washington, DC, 2nd edn, 1996 Search PubMed.
- C. C. Conwell, I. D. Vilfan and N. V. Hud, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9296 CrossRef CAS PubMed.
- S. B. Zimmerman, J. Struct. Biol., 2002, 138, 199 CrossRef CAS PubMed.
- C. L. Woldringh, P. R. Jensen and H. V. Westerhoff, FEMS Microbiol. Lett., 1995, 131, 235 CrossRef CAS PubMed.
- N. Gilbert and J. Allan, Curr. Opin. Genet. Dev., 2014, 25, 15 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Quenching of Ru complex emission upon DNA binding (Fig. S1); fluorescence images of RFI-treated and CAM-treated E. coli (Fig. S2): and Experimental methods. See DOI: 10.1039/c6mb00753h |
| This journal is © The Royal Society of Chemistry 2017 |