Open Access Article
Open Access ArticleImproved bovine embryo production in an oviduct-on-a-chip system: prevention of poly-spermic fertilization and parthenogenic activation†
Marcia A. M. M.
Ferraz
a,
Heiko H. W.
Henning
b,
Pedro F.
Costa
cd,
Jos
Malda
bcd,
Ferry P.
Melchels
cd,
R.
Wubbolts
e,
Tom A. E.
Stout
ab,
Peter L. A. M.
Vos
a and
Bart M.
Gadella
 *ae
*ae
aDepartment of Farm Animal Health, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands. E-mail: b.m.gadella@uu.nl; Tel: +31 302535386
bDepartment of Equine Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands
cDepartment of Orthopedics, Utrecht Medical Center, Utrecht, The Netherlands
dUtrecht Biofabrication Facility, Utrecht Medical Center, Utrecht, The Netherlands
eDepartment of Biochemistry and Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands
First published on 7th February 2017
Abstract
The oviduct provides the natural micro-environment for gamete interaction, fertilization and early embryo development in mammals, such as the cow. In conventional culture systems, bovine oviduct epithelial cells (BOEC) undergo a rapid loss of essential differentiated cell properties; we aimed to develop a more physiological in vitro oviduct culture system capable of supporting fertilization. U-shaped chambers were produced using stereo-lithography and mounted with polycarbonate membranes, which were used as culture inserts for primary BOECs. Cells were grown to confluence and cultured at an air–liquid interface for 4 to 6 weeks and subsequently either fixed for immune staining, incubated with sperm cells for live-cell imaging, or used in an oocyte penetration study. Confluent BOEC cultures maintained polarization and differentiation status for at least 6 weeks. When sperm and oocytes were introduced into the system, the BOECs supported oocyte penetration in the absence of artificial sperm capacitation factors while also preventing polyspermy and parthenogenic activation, both of which occur in classical in vitro fertilization systems. Moreover, this “oviduct-on-a-chip” allowed live imaging of sperm-oviduct epithelium binding and release. Taken together, we describe for the first time the use of 3D-printing as a step further on bio-mimicking the oviduct, with polarized and differentiated BOECs in a tubular shape that can be perfused or manipulated, which is suitable for live imaging and supports in vitro fertilization.
Introduction
In mammals, the oviducts are paired organs that connect the uterus to the respective ovaries. The oviduct also forms the specific niche in which mammalian fertilization takes place. Its lumen provides the physiological microenvironment required for gamete interaction and early embryo development.1–5 Sperm enter the oviduct from the isthmic end, which is connected to the uterus by the utero-tubal junction. Close contact of sperm with the epithelium of the oviductal isthmus has been proven to be important for extending sperm survival, in a so-called ‘sperm reservoir’. It also serves to trigger subsequent activation (i.e. capacitation) around the time of ovulation. This allows sperm to detach from the isthmus and to ascend into the ampulla where fertilization will take place.5–9 The ampulla of the oviduct is connected to the funnel-shaped infundibulum, which catches the freshly ovulated cumulus oocyte complex (COC) and directs it further into the ampulla. Final modifications of the COC takes place in the ampulla which will ensure that the oocyte is ready to become fertilized by a sperm cell.10,11 After fertilization, the first embryonic divisions and further development take place in the oviduct and, once the morula stage is achieved, the bovine embryo will leave the isthmic part of the oviduct to enter the uterus.Conditions for supporting fertilization and early embryo development in vitro have been developed for a wide range of species. However, despite advances in reproductive biotechnology and embryo culture media, it is clear that in vitro produced embryos differ markedly from those that develop in vivo.12–15 Despite common belief that the oviduct is more than a simple tube allowing the transport of gametes and early stage embryos, the findings that in vitro embryos are of reduced developmental competence convincingly demonstrates the importance of the oviduct environment for optimal embryo development. Both the gametes and the early embryo are in close contact with the epithelial lining the oviduct. This epithelium is composed of a mixture of ciliated and non-ciliated, i.e. secretory, cells. The oviduct tubular morphology with its intricately folded morphology16 influences the flux of fluids. Fluid movements are created with muscular contractions and ciliary beating which both actively support the transport of the sperm and oocyte to the ampulla, where fertilization takes place. In this respect, the critical contribution of the oviduct to the complex regulated processes of fertilization of the oocyte and optimal early embryo development remains to be elucidated in detail.17
One of the reasons why oviduct physiology and function has poorly been studied, due to the location of the organ being deep within the abdominal cavity. This makes it difficult to perform in vivo observational studies in mammals. Consequently, various in vitro models have been designed to study the role of oviduct epithelial cells in gamete interaction and fertilization. The most commonly used models are based on monolayer cultures of oviduct epithelial cells,7,8,18,19 or on explant cultures of oviduct tissue that forms cellular vesicles with ciliary beating activity.20–22 Standard in vitro oviduct monolayer cultures (OMs, 2D culture) are typically hampered by a rapid transformation of the differentiated, cuboidal–columnar oviduct epithelial cells (OECs) into flattened cells with a complete loss of cilia and with a reduced secretory ability.18,23–25 Recently, the use of porous membrane inserts to allow oviduct epithelial cells to be cultured at an air–liquid interface, has been shown to allow the formation of epithelial monolayers that preserve their epithelial secretory and ciliary beating activity.8,23,26–28 Although, this has been a break-through in terms of cell culture, commercial insert systems do have a number of limitations for some experimental purposes. For example, it is not possible to perform live cell imaging within most inserts, and perfusion is difficult because the inserts are flat circular discs rather than mimicking the tubular structure of the oviduct.
Ideally, an in vitro model of the oviduct would be compartmentalized with a basolateral perfusion compartment mimicking the blood circulation, and an independently apical perfusion compartment mimicking the luminal fluid movements of the oviduct. Such a system would allow mimicking the endocrine changes that do occur during a natural estrous cycle at the basal side and facilitate the apical addition and removal of gametes, embryos, and medium or cell secretions. Indeed, it was recently demonstrated that specific tissue morphology and functions can be preserved better in customized three-dimensional (3D) culture systems than in conventional 2D systems.24,29–33
Three-dimensional (3D) printing technology can generate prototypes rapidly, allowing researchers to design and print devices within a short period of time.34 Combined with microfluidic technology, 3D printing has led to the creation of “organs-on-a-chip” to study human and animal physiology in an organ-specific context and, thereby, create models for researching specific aspects of health, disease and toxicology.31
The advances of 3D printing and cell insert culture systems and the lack of a physiological in vitro model to study oviduct function, led us to design and print a tube-like chamber in which BOECs can be cultured at an air–liquid interface that supports further epithelial polarization and differentiation during long-term culture period. We tested the designed chamber for its suitability for live imaging the interaction between sperm and oviduct cells. Furthermore, the functionality of the epithelial cells cultured in a 3D chamber for supporting fertilization is demonstrated in an oocyte penetration approach. Using this oviduct-on-a-chip design, we aim to better understand the interactive role of the oviduct environment supporting gamete interaction, early embryonic development, and ultimately to be able to produce in vitro embryos more similar to in vivo embryos than is currently possible.
Materials and methods
Chemicals
Unless stated otherwise, all chemicals used were obtained from Sigma Chemical Co. (St. Louis, MO) and were of the highest purity available.Three-dimensional chamber design and printing
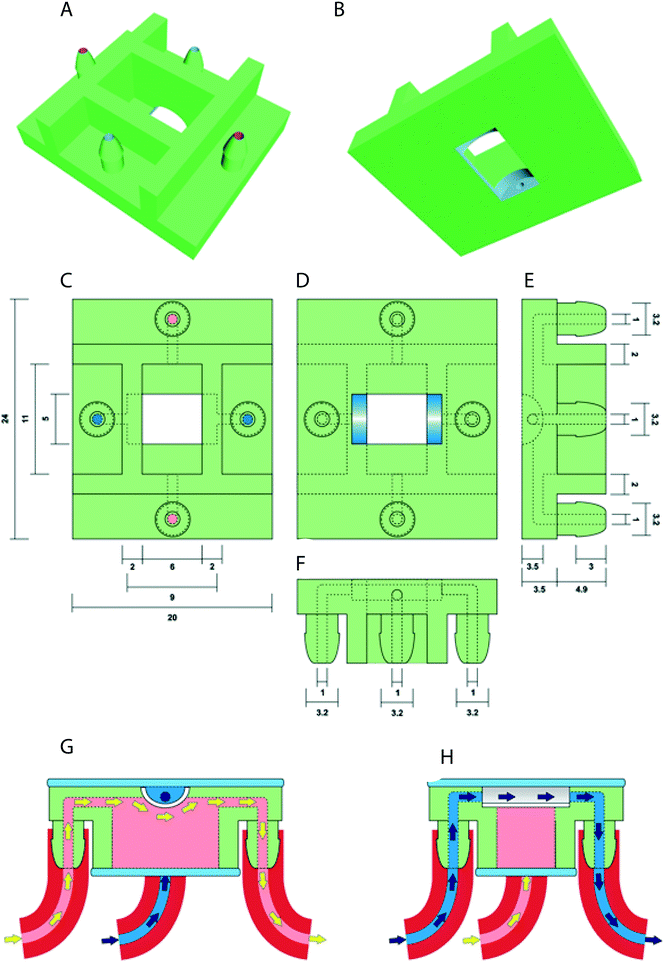
The prototype design of the oviduct-on-a-chip was created using Tinkercad (Autodesk Inc., San Francisco, CA, USA). The design included flat upper and lower surfaces to allow attachment to a glass slide for future perfusion and imaging. A curved inner chamber was created to better mimic the tubular surface of the oviduct, while remaining shallow enough not to interfere with imaging. Inlets and outlets in both the apical and basolateral compartments were included, with the size and shape of the inlets designed to permit easy attachment of tubing and to allow adequate fluid flow. An outer cuboid chamber shape was used to facilitate later up-scaling by printing multiple conjoined parallel chambers. The design was exported from Tinkercad as an STL file, and then imported into Mimics software (Materialise NV, Leuven, Belgium) to verify and repair any mesh errors and generate printing support structures. The screenshot of the prototype including its inlets and outlets is shown in Fig. 1.Three-dimensional printing of the device was performed using a photo-cured resin, PIC100 (Envisiontec GmbH, Gladbeck, Germany), via a Perfactory 3 Mini 3D printer (Envisiontec GmbH, Gladbeck, Germany) at a resolution of 50 μm, which exploits the photo-polymerization technique for 3D printing.
Post-curing, mounting a porous membrane and sterilization of the 3D chamber
To avoid the leakage of compounds from the printed material that might interfere with cell viability, removal of excess resin was performed by a 15 minute immersion in ethanol. After complete air drying, the chambers were immersed 3 times for 2 hours each in isopropanol solution. After repeated air drying, the chambers were light-cured using 4000 flashes in an Otoflash G171 (Envisiontec GmbH, Gladbeck, Germany).The polycarbonate membrane (0.4 μm pores; SABEU GmbH & Co. KG, Germany) was attached to the chamber using the silicone elastomer Kwik-Sil (World Precision Instruments Inc., Florida, USA) and cured for 5 minutes at room temperature. Before incubation with cells, the chambers were sterilized by immersion for 1 hour in 70% ethanol, washed three-times for 30 minutes each in phosphate-buffered saline solution (PBS; 163.9 mM Na+, 140.3 mM Cl−, 8.7 mM HPO43−, 1.8 mM H2PO4−, pH 7.4; Braun, Melsungen, Germany) and washed for 1 hour in HEPES buffered Medium 199 (Gibco BRL, Paisley, U.K.) supplemented with 100 U mL−1 penicillin and 100 μg mL−1 streptomycin (Gibco BRL, Paisley, U.K.).
Isolation of oviduct cells and long term oviduct cell culture
Cow oviducts were collected from a local abattoir immediately after slaughter and transported to the laboratory on ice, within two hours. The oviducts were dissected free of surrounding tissue and washed three times in cold PBS supplemented with 100 U mL−1 of penicillin and 100 μg mL−1 of streptomycin. BOECs were isolated by squeezing the total oviduct contents out of the ampullary end of the oviducts, and collected in HEPES buffered Medium 199 supplemented with 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. The cells were washed twice by centrifuging for 500 × g for 10 minutes at 25 °C in HEPES buffered Medium 199 supplemented with 100 U mL−1 of penicillin and 100 μg mL−1 of streptomycin. The cells were then cultured for 24 hours in HEPES buffered Medium 199 supplemented with 100 U mL−1 penicillin, 100 μg mL−1 streptomycin and 10% fetal calf serum (FCS; Bovogen Biologicals, Melbourne, Australia). During these 24 hours, the cells arranged themselves into floating vesicles with outward facing actively beating cilia; these vesicles were collected, centrifuged at 500 × g for 10 minutes at 25 °C, resuspended in DMEM/Ham's F12 medium (DMEM/F-12 Glutamax I, Gibco BRL, Paisley, U.K.) supplemented with 1.4 mM hydrocortisone, 5 mg mL−1 insulin, 10 mg mL−1 transferrin, 2.7 mM epinephrine, 9.7 nM tri-iodothyronine, 0.5 ng mL−1 epidermal growth factor, 50 nM trans-retinoic acid, 2% bovine pituitary extract (containing 14 mg mL−1 protein), 1.5 mg mL−1 BSA, 100 mg mL−1 gentamycin, and 2.5 mg mL−1 amphotericin B (3D culture medium, adapted from24), and pipetted up and down several times to mechanically separate the cells. Next, cells were seeded either into: (i) the oviduct-on-a-chip (3D culture; 0.6 × 106 cells per cm2) or (ii) into 24 well culture dishes with glass coverslips in the bottom of the wells (2D culture; 0.3 × 106 cells per cm2). Cells in both systems were cultured in 3D culture medium in a humidified atmosphere of 5% CO2-in-air at 38.5 °C until they reached confluence (5–7 days). Once the cells had reached confluence, an air–liquid interface was established in the 3D culture by removing the medium in the apical compartment. Cells in the 3D chambers were cultured at an air–liquid interface for up to 42 days in a humidified atmosphere of 5% CO2-in-air at 38.5 °C. The culture medium was completely refreshed twice a week in both systems.Oocyte collection and in vitro maturation
Oocyte collection and maturation was performed as described somewhere else.35 Briefly, bovine ovaries were collected from a local abattoir and transported to the laboratory within 2 hours after dissection. The ovaries were washed in physiological saline (0.9% w/v NaCl) and held in physiological saline containing 100 U mL−1 penicillin and 100 μg mL−1 streptomycin at a temperature of 30 °C. The fluid and cumulus oocyte complexes (COCs) were aspirated from follicles with a diameter ranging from 2 to 8 mm and were collected into a 50 ml conical tube using a 19-gauge needle and a vacuum pump. COCs with a minimum of three layers of intact cumulus cells were selected and first washed in HEPES-buffered M199 (Gibco BRL, Paisley, U.K.) before being washed and cultured in maturation medium (M199 supplemented with 0.02 IU mL−1 follicle-stimulating hormone [Sioux Biochemical Inc., Sioux Center, IA]), 0.02 IU mL−1 luteinizing hormone (Sioux Biochemical Inc.), 7.71 μg mL−1 cysteamine, 10 ng mL−1 epidermal growth factor in 0.1% w/v fatty acid-free bovine serum albumin (BSA) and 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. Selected COCs were cultured in four-well culture plates (Nunc A/S, Roskilde, Denmark) containing maturation medium. The oocytes were matured in groups of 50 COCs in 500 μl maturation medium and incubated in a humidified atmosphere of 5% CO2-in-air for 24 hours at 38.5 °C.Sperm washing and staining with mitotracker
Frozen sperm, from 3 different bulls, were thawed at 37 °C for 30 seconds and washed by centrifugation at 100 × g for 10 minutes through a BoviPure discontinuous gradient, following manufacture instructions (Nidacon International AB, Gothenburg, Sweden) at room temperature. The supernatant was removed, the pellet resuspended in 3 mL of BoviPure wash solution, and centrifuged again at 100 × g for 5 minutes. Spermatozoa from the 3 pellets were pooled and then incubated for 30 minutes with 200 nM mitotracker green FM® or mitotracker red FM® (MTG and MTR respectively; Molecular Probes Inc., Eugene, USA) in fertilization medium (modified Tyrode's medium supplemented with 25 mM sodium bicarbonate, 22 mM lactate, 1 mM pyruvate, 6 mg mL−1 fatty acid-free BSA) containing 100 U mL−1 penicillin and 100 μg mL−1 streptomycin instead of gentamycin and without glucose or activation factors (heparin, D-penicillamine, hypotaurine and epinephrine). The mitotracker stained spermatozoa were then washed three times in fertilization medium without activation factors by centrifuging at 100 × g for 5 minutes and used for in vitro fertilization.In vitro fertilization
MTG stained sperm were added to the fertilization medium at a final concentration of 1 × 106 sperm cells per mL in the presence (control IVF, 500 μL volume) or absence (3D culture, 2D culture and no activation factors control IVF; 80, 500 and 500 μL volume, respectively) of 10 μg mL−1 heparin, 20 μM D-penicillamine, 10 μM hypotaurine, and 1 μM epinephrine (activation factors). For the 3D culture IVF, the sperm suspension (80 μL) was manually perfused to the apical compartment and – after 2 hours – unattached sperm were perfused out of the system by flushing 240 μL of PBS over the apical side of the BOEC and immediately thereafter, a total of 25 COCs were perfused to the apical compartment in 80 μL of fertilization medium without activation factors of each of the 3D culture chambers (n = 8 chambers; 25 COCs per chamber). In 2D cultures 50 COCs were added in 500 μL of fertilization medium without activating factors. A standard IVF protocol,35 with or without activation factors, was performed as a control on 300 COCs. After 24 h of co-incubation under a humidified atmosphere of 5% CO2-in-air at 38.5 °C, cumulus cells were removed by pipetting and the presumptive zygotes were fixed and stained with the membrane permeable DNA stain Hoechst 33342 (5 μg mL−1 in PBS) to distinguish parthenotes and poly-spermic from mono-spermic fertilized oocytes. All experiments were performed in 4 replicates, using 2 different animals per replicate for the 2D and 3D cultures groups.The sperm cells perfused out of the 3D culture chambers were centrifuged at 100 × g for 5 minutes, resuspended in 50 μL of fertilization medium, and the number of recovered sperm cells were calculated in order to determine the number of spermatozoa that remained bound to the epithelial cells. A routine IVF with the same number of sperm cells as the ones that remained attached to the epithelial cells in the 3D culture (69 × 103 sperm cells per well) and a routine IVF with same proportion of sperm cells that remained attached to the 3D culture were performed (0.431 × 106 sperm cells per well, 86.25% of sperm cells used for control IVF). After 24 hours of co-incubation under a humidified atmosphere of 5% CO2-in-air at 38.5 °C, presumptive zygotes were fixed and stained as described above.
Ciliation of cells and cell morphology
At weeks 3, 4, 5 and 6 of air–liquid interface culture, two oviduct-on-a-chip chambers and 1 coverslip from the 2D culture was sacrificed per animal (n = 4) for assessment of cilia formation on epithelial cells. The membranes were dismounted from the chamber for immune fluorescent staining. The membranes or cover slips were washed in PBS, fixed in 4% paraformaldehyde dissolved in PBS, and permeabilized for 30 minutes using 0.5% Triton-X100 in PBS. Non-specific binding was blocked by incubation for 1 hour in PBS containing 5% normal goat serum, at room temperature. The cells were then incubated overnight at 4 °C with a mouse anti-acetylated α-tubulin primary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution with PBS Santa Cruz Biotechnology, Santa Cruz, CA). The next morning the cells were washed three times in PBS (5 minutes per wash) and incubated with an Alexa 488 conjugated goat anti-mouse antibody (1
100 dilution with PBS Santa Cruz Biotechnology, Santa Cruz, CA). The next morning the cells were washed three times in PBS (5 minutes per wash) and incubated with an Alexa 488 conjugated goat anti-mouse antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution with PBS, Santa Cruz Biotechnology, Santa Cruz, CA) at room temperature for 1 hour. Hoechst 33342 (5 μg mL−1) was used to stain cell nuclei and phalloidin conjugated to Alexa 568 (1
100 dilution with PBS, Santa Cruz Biotechnology, Santa Cruz, CA) at room temperature for 1 hour. Hoechst 33342 (5 μg mL−1) was used to stain cell nuclei and phalloidin conjugated to Alexa 568 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution with PBS) was used to stain actin filaments. Negative controls were performed by omitting incubation with the primary antibody. Analysis was performed by laser scanning confocal microscopy using a TCS SPE-II system (Leica Microsystems GmbH, Wetzlar, Germany) attached to an inverted semi-automated DMI4000 microscope (Leica) with a 40× NA 1.25 magnification objective. Five random field of views in the center of the membrane and coverslip were imaged for each animal and group and, at least, 350 cells per animal and per group were classified; the percentage of ciliated cells was determined. Moreover, Z-stacks of 0.2 μm were obtained by laser scanning confocal microscopy at 100× NA 1.40 magnification objective. 3D constructs of the cells were performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA) to demonstrate cell morphology.
100 dilution with PBS) was used to stain actin filaments. Negative controls were performed by omitting incubation with the primary antibody. Analysis was performed by laser scanning confocal microscopy using a TCS SPE-II system (Leica Microsystems GmbH, Wetzlar, Germany) attached to an inverted semi-automated DMI4000 microscope (Leica) with a 40× NA 1.25 magnification objective. Five random field of views in the center of the membrane and coverslip were imaged for each animal and group and, at least, 350 cells per animal and per group were classified; the percentage of ciliated cells was determined. Moreover, Z-stacks of 0.2 μm were obtained by laser scanning confocal microscopy at 100× NA 1.40 magnification objective. 3D constructs of the cells were performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA) to demonstrate cell morphology.
Pieces of 5 mm from ampullary and isthmic regions of the oviduct ipsilateral to the ovary with an active corpus luteum were fixed for 24 hours in 4% w/v paraformaldehyde, paraffin embedded and sections of 4 μm were stained as described above.
Live cell imaging
After one week of the air–liquid interface culture, the oviduct-on-a-chip was incubated with MTR labeled sperm and stained with Hoechst 33342 (5 μg mL−1) in the 3D culture medium for 30 minutes. Live cell imaging was done on a Nikon Eclipse TE2000 equipped with the perfect focus system with a two-channel simultaneous imaging system by exciting with the lasers Vortran 405 nm and Cobolt Jive 561 nm, using the filters ET-DAPI (490/00) and ET-DSRed (490/05). Images from both channels were detected with a 20× magnification long distance objective (Plan Apo 20×/NA 0.75 dry) with a speed of 60 frames per second.Detection of oocyte penetration
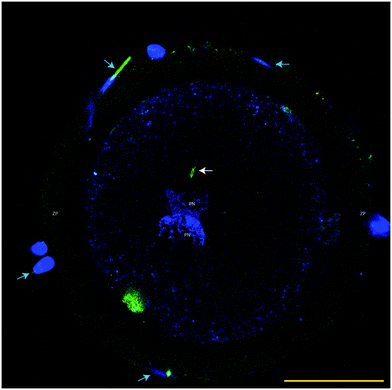
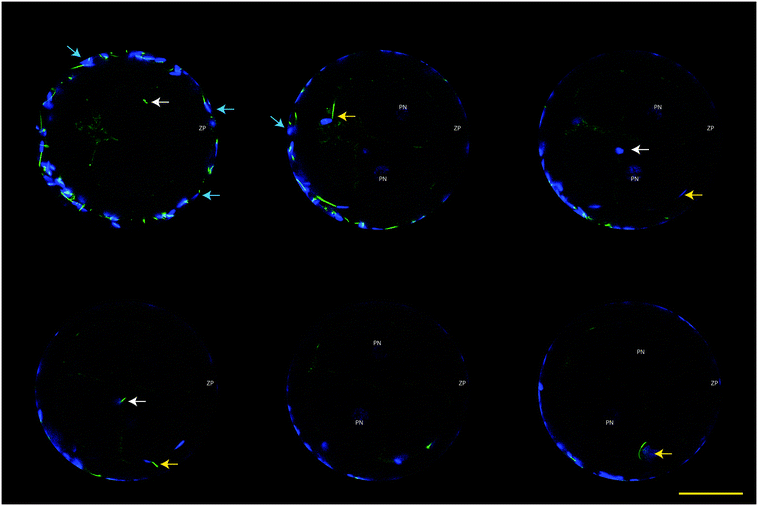
Fixed presumptive zygotes were stained with Hoechst 33342 (5 μg mL−1 in PBS) for 30 min, washed three times in PBS containing 3 mg mL−1 polyvinyl pyrrolidone (PVP) and then mounted into a 0.12 mm eight-well Secure-Seal Spacer (Molecular Probes) on a glass slide (Superfrost Plus; Menzel, Braunschweig, Germany), covered with Vectashield antifade (Vector Laboratories, Burlingame, CA), and sealed with a coverslip. Slides were analyzed by Laser scanning confocal microscopy using a TCS SPE-II system (Leica Microsystems GmbH, Wetzlar, Germany) attached to an inverted semi-automated DMI4000 microscope (Leica) with a 40× NA 1.25 magnification objective. The number of oocytes with the presence of labeled sperm mid-piece(s) within the ooplasm was determined (i.e. sperm-penetrated oocytes). Poly-spermy was identified by the detection of 2 or more sperm mid-pieces within the ooplasm, while parthenotes were identified in the case when 2 or more nuclei were detected without the presence of a sperm mid-piece.Data analysis
The data were analyzed using IBM SPSS Statistics (version 24). A Shapiro–Wilk test was performed and all groups were normally distributed. Mean ± standard deviation is provided and differences between groups were examined by one-way ANOVA, followed by a Tukey's post hoc analysis (p < 0.05).Results and discussion
In this study, we designed and successfully 3D printed an oviduct-on-a-chip model using a stereo-lithographic technique (Fig. 1). The chamber was designed such that its dimensions were compatible with live-cell imaging. Nowadays, 3D printing generates fast prototyping process technology, allowing researchers to design and print devices in a short period of time.34 On the other hand, research employing technologies, such as 3D printing and microfluidics, to bio-mimic 3D cultures for reproductive events is scarce and only a handful of papers on 3D microfluidics and gamete research have been published.36–40The oviductal lumen has a complex morphology due to folding of the mucosa of the oviduct wall. This folding varies in the different anatomical parts of the oviduct.41 Exactly mimicking those folding in vitro is difficult and does not allow accurate live imaging. Therefore, we had to compromise our bio-mimicked model and decided to create an U-shape topology as this construct, at least, would allow live imaging and perfusion of the system, also providing a niche where more cell contact area is offered for introduced COCs. The oviduct-on-a-chip was designed in such a way that the distance between cells adhered to the porous filter and the glass coverslip was less than 2 mm, to meet the working distance of objectives available and permit live-cell imaging using an inverted epifluorescence microscope after incubation of the cells with MTR pre-labeled sperm (Movie S1†). To our knowledge, this is the first published 3D printed device with a half-pipe shaped porous filter for BOEC culture. The potential benefits of this system for BOEC culture extend beyond the accessibility for live-cell imaging, since both the apical and basolateral compartments can be independently perfused or otherwise manipulated. The possibility of live imaging within the device is a significant advantage over currently available commercial porous membrane systems, and should allow a greater range of in vitro experiments, in particular those focusing on the changes within sperm cells during their incubation with oviduct epithelial cells just prior to fertilization and further embryonic development under different conditions.
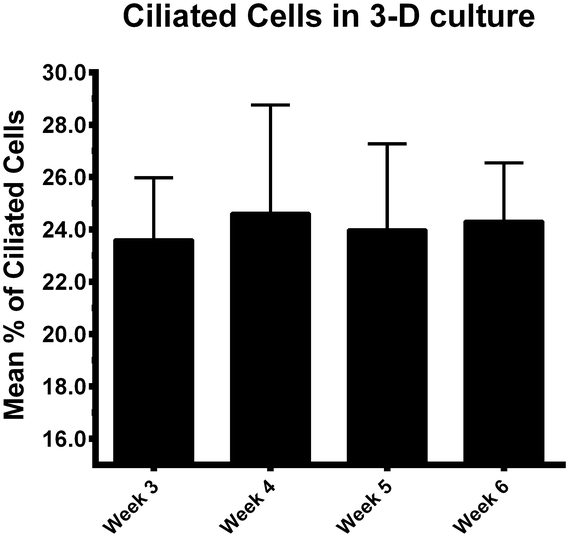
When BOECs were cultured under 2D conditions the cells did not become ciliated but lost their columnar epithelium shape instead and became flat. This is in line with previous reports that describe this process known as de-differentiation.25,27,42–44 In contrast, BOECs cultured in the 3D printed device regained and maintained their ciliated and cuboidal to columnar pseudostratified epithelium for a period of at least 6 weeks, with a mixed population of ciliated and non-ciliated secretory cells (Fig. 2A–C). This morphology was comparable to the in vivo oviduct epithelium (Fig. 2D and E). It was also possible to observe the formation of actin rich protrusions in non-ciliated cells, the secretory bulbs. Cilia emerged at the air–liquid interface side of the cells (apical) within about 2 weeks of culture, was complete within 3 weeks and remained stable during weeks 3–6 of culture (P < 0.05; Fig. 3). Similar results have been described, previously, using porous membranes cultured at an air–liquid interface system for OECs derived from different species including mouse, cow, pig, monkey and man.8,24,26–28,42,45 However, such systems do not allow live-cell analysis and monitoring in contrast to the BOEC system described in our current study.
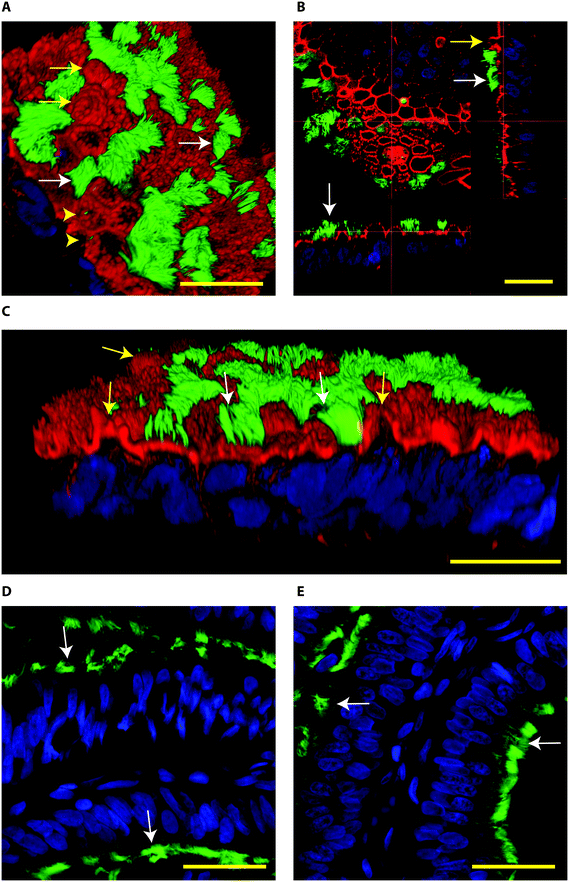 | ||
| Fig. 2 Confocal immune fluorescent images of bovine oviduct epithelial cells (BOECs) in 3D culture at an air–liquid interface for 28 days (A–C) and from paraffin sections of oviductal isthmus and ampulla (D and E, respectively). Acetylated α-tubulin was used to stain secondary cilia (green), phalloidin to label actin filaments (red in A–C) and Hoechst 33342 to stain nuclei (blue). A–C: Note the presence of ciliated cells (green, white arrows), actin rich secretory protrusions (red, yellow arrows) and primary cilia (yellow arrow heads). In B, note the cuboid to columnar pseudostratified epithelium. D and E: Note columnar pseudostratified morphology of oviduct paraffin sections, similar to the one encountered in the 3D cultured BOEC. In paraffin embedded sections the phalloidin staining was not observed. The Z-stacks from top to bottom of the cells cultured on the 3D system can also be observed in the Movie S2.† Bars = 25 μm. | ||
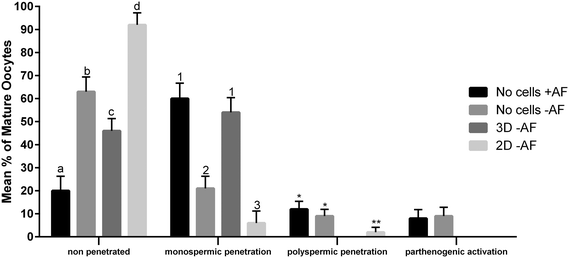
The functionality of our oviduct-on-a-chip system was tested using a bio-monitoring assay in which sperm penetration of the oocyte was scored (this is indicative for fertilization). Although a lower percentage of oocytes was penetrated, compared to a standard bovine IVF system (Fig. 4), the oviduct-on-a-chip system resulted in a similar proportion of oocytes that were mono-spermic fertilized (Fig. 4 and 5). Remarkably, and in contrast to standard IVF, no parthenogenic oocyte activation nor poly-spermic fertilization was observed in our developed oviduct-on-a-chip fertilization system (Fig. 4 and 6). In contrast, parthenogenesis and poly-spermy both occurred with an incidence of approximately 10% in the routine IVF system (Fig. 4). Thus the developed oviduct-on-a-chip allowed similar normal fertilization of oocytes and completely reduced the incidence of abnormal fertilization/activation of oocytes when compared to routine IVF. Further to this, it should be noted that in our developed oviduct-on-a-chip system we have not added factors to the incubation media to stimulate sperm activation and capacitation in the model, in contrast to conventional bovine IVF where such additions are a routine requirement. Thus the apical fluid compartment must have been conditioned by the secretions of the polarized BOECs allowing similar mono-spermic fertilization rates to those achieved via conventional IVF.
The reduced rates in poly-spermy and parthenogenic activation of oocytes in the 3D BOEC system was not due to a reduction of the amount sperm (non-bound sperm were perfused away) when compared to conventional IVF: the majority of sperm (86.3 ± 2.9%) remained attached to the 3D BOEC (representing 69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 2300 sperm per 3D-BOEC). When a similar proportional reduction of sperm was used in conventional IVF (i.e. only 0.43 × 106 instead of 0.5 × 106 sperm per well) no differences on mono-spermic penetration, poly-spermic penetration and parthenogenic activation was observed (for control 60.1%, 12.4% and 8.9%; for reduced number of sperm 55.63%, 12.23% and 10.11%, respectively; p > 0.05). In another control we compared the fertilization results using 69
000 ± 2300 sperm per 3D-BOEC). When a similar proportional reduction of sperm was used in conventional IVF (i.e. only 0.43 × 106 instead of 0.5 × 106 sperm per well) no differences on mono-spermic penetration, poly-spermic penetration and parthenogenic activation was observed (for control 60.1%, 12.4% and 8.9%; for reduced number of sperm 55.63%, 12.23% and 10.11%, respectively; p > 0.05). In another control we compared the fertilization results using 69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells (the same number of sperm that remained bound in the 3D-BOEC) with 500
000 cells (the same number of sperm that remained bound in the 3D-BOEC) with 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells (normally used in conventional IVF). The large reduction of sperm resulted in severely reduced mono-spermic fertilization (20.5% versus 60.1%; p < 0.05) and a concomitant reduction of poly-spermic fertilization (5.1% versus 12.5%; p < 0.05) while the parthenogenic activation rates remained the same (10.3% and 8.9%, p > 0.05). Note that both poly-spermic fertilization and parthenogenic activation of oocytes was not observed in the 3D-BOEC system: (i) the binding and activation of sperm to the 3D-BOEC and the absence of the activation factors are required to achieve high mono-spermic fertilization rates in combination with complete abolishment of poly-spermic fertilization and parthenogenic activation. (ii) In the 3D-BOEC system a strong reduction of number of sperm (13.8%) leads to similar mono-spermic fertilization rates when compared to conventional IVF while such a reduction of sperm in conventional IVF leads to a severe reduction of mono-spermic fertilization rates. (iii) Reducing the amount of sperm in conventional IVF, in presence of activation factors, does not reduce poly-spermic fertilization/mono-spermic fertilization ratio (both are reduced to >60%) and does not reduce the incidence of parthenogenic activation. Altogether these data confirm that the higher efficiency of mono-spermic fertilization (at low sperm dose) and the abolition of poly-spermic fertilization as well as parthenogenic activation in the 3D BOEC culture IVF is due to an interaction between the sperm and/or the oocyte with the oviduct cells and/or secretions rather than to the severe reduction in number of sperm when compared to conventional IVF.
000 cells (normally used in conventional IVF). The large reduction of sperm resulted in severely reduced mono-spermic fertilization (20.5% versus 60.1%; p < 0.05) and a concomitant reduction of poly-spermic fertilization (5.1% versus 12.5%; p < 0.05) while the parthenogenic activation rates remained the same (10.3% and 8.9%, p > 0.05). Note that both poly-spermic fertilization and parthenogenic activation of oocytes was not observed in the 3D-BOEC system: (i) the binding and activation of sperm to the 3D-BOEC and the absence of the activation factors are required to achieve high mono-spermic fertilization rates in combination with complete abolishment of poly-spermic fertilization and parthenogenic activation. (ii) In the 3D-BOEC system a strong reduction of number of sperm (13.8%) leads to similar mono-spermic fertilization rates when compared to conventional IVF while such a reduction of sperm in conventional IVF leads to a severe reduction of mono-spermic fertilization rates. (iii) Reducing the amount of sperm in conventional IVF, in presence of activation factors, does not reduce poly-spermic fertilization/mono-spermic fertilization ratio (both are reduced to >60%) and does not reduce the incidence of parthenogenic activation. Altogether these data confirm that the higher efficiency of mono-spermic fertilization (at low sperm dose) and the abolition of poly-spermic fertilization as well as parthenogenic activation in the 3D BOEC culture IVF is due to an interaction between the sperm and/or the oocyte with the oviduct cells and/or secretions rather than to the severe reduction in number of sperm when compared to conventional IVF.
Our results may indicate that routine IVF misses the optimal conditioning factors that are secreted by the oviduct epithelium and this absence caused the noted increase in rates of oocytes that are abnormally parthenogenic activated or poly-spermic fertilized. Note that the activating factors used in conventional IVF are not responsible for the incidence of poly-spermic fertilization and/or parthenogenic activation of the oocytes (Fig. 4). They only served to increase the mono-spermic fertilization rate to similar levels when compared to the 3D-BOEC, albeit approximately 7.2 times more sperm were needed in the conventional IVF when compared to the 3D-BOEC. This result further supports the notion that the oviduct-on-a-chip have conditioned and optimized the apical environment for mono-spermic fertilization. Altogether our data indicate that when OECs are cultured into a polarized and differentiated state within a 3D topology, they appear to condition apical medium as they exclusively support mono-spermic fertilization. In the 2D culture system, where the majority of OECs were flat and non-ciliated, the conditioning of the apical medium was insufficient and did not inhibit poly-spermic fertilization (even though the total penetration rate was reduced).
A reduction of poly-spermy with even higher (74–84%) fertilization rates was demonstrated previously in an IVF system using BOECs cultured on porous membrane inserts.8 The major difference of that study and our approach is that we did not add any sperm activating components to the 3D BOEC culture medium. Moreover, in our oviduct-on-a-chip system we not only showed reduced rates but even a complete absence of both poly-spermic fertilization and parthenogenic activation. With regards to the higher fertilization rates in the former BOEC study,8 the addition of estrous cow serum may have stimulated changes in the BOECs secretory activity. Studies to examine the influence of factors such as endocrine stimulation, as well to investigate the influence of different segments of the oviduct (ampulla vs. isthmus) on sperm activation and embryo development are planned for the oviduct-on-a-chip system.
The concept that conditioning of the apical medium by the BOECs in the 3D system is responsible for preventing poly-spermy, is in line with previously described, inhibitory, effects of oviduct fluid on poly-spermic fertilization in cows and pigs.46,47 Moreover, studies have also reported beneficial effects of oviduct fluid and/or oviduct proteins on sperm motility, acrosome reaction, bull fertility3,8,48–54 and on oocyte and embryo development and quality.1,47,55–61 Despite of this, the oviduct has remained a largely neglected organ when designing IVF procedures in man and domestic animals.17
Note that epigenetic modulation of the maturing oocyte and the early developing embryo can also be of concern while producing embryos in vitro. In vivo these epigenetic events take place in the oviduct and are thought to allow reprogramming of the embryonic genome. For instance, the methylation of sperm DNA is erased in the paternal pronucleus after fertilization. Amongst other functions this process allows specific pluripotency genes to be expressed. Failure of, or disturbances to, this process leads to impaired embryo development.62 Interestingly, bovine blastocysts developed after culturing embryos partially in vitro and partially in vivo have been shown to differ in DNA methylation patterns when compared to blastocysts developed completely in vivo and to those completely developed in vitro.13
A classic example of the possible epigenetic effects of in vitro embryo production conditions on embryo development is the large offspring syndrome (LOS), which is characterized by increased size and weight at birth, breathing difficulties, reluctance to suckle, and perinatal death of the born calves.63 The LOS was described in cattle and sheep derived from in vitro cultured embryos in the presence of elevated serum concentrations.63 The pathogenesis of the syndrome is not completely clear, but there is evidence that a loss of gene-imprinting and overexpression of insulin growth factor 2 (IGF2) receptor may be an important contributor.64,65 The epigenetic changes that may be induced during embryo culture emphasize the need for improved in vitro embryo production systems. Not only the quantitative production of blastocysts, but also the quality and genetic normality of such embryos produced are highly relevant. We believe that the oviduct-on-a-chip approach will be an ideal starting point to better mimic the physiological environment for mammalian fertilization and embryo production. This more physiological environment likely serves to reduce metabolic and genetic programming abnormalities caused by in vitro embryo production conditions.
Conclusions
In conclusion, a 3D oviduct-on-a-chip model with a U-shaped porous membrane allowed BOEC polarization that could be maintained during long-term culture (over 6 weeks). In this system the oviduct cells under culture must have conditioned the apical medium, as this allowed proper sperm and oocyte interactions, fertilization and completely abolished poly-spermic fertilization and parthenogenic activation of oocytes in the absence of added sperm activating factors. The oviduct-on-a-chip system is easy to manipulate, can be used for introduction, manipulation and live microscopic visualization of sperm cells, oocytes and early embryos and study their cellular processes around fertilization. The fact that fertilization is exclusively mono-spermic may become relevant for assisted reproductive technologies for both bovine and other mammalian species.Acknowledgements
This research is part of the research focus theme Life Sciences of Utrecht University, experiments were performed in the research group Fertility and Reproduction and confocal imaging was performed at the Centre for Cell Imaging, both on the Faculty of Veterinary Medicine of the same university.References
- R. E. Lloyd, R. Romar, C. Matás, A. Gutiérrez-Adán, W. V. Holt and P. Coy, Reproduction, 2009, 137, 679–687 CrossRef CAS PubMed.
- P. Coy, F. A. García-Vázquez, P. E. Visconti and M. Avilés, Reproduction, 2012, 144, 649–660 CrossRef CAS PubMed.
- G. Killian, J. Anim. Sci., 2011, 89, 1315–1322 CrossRef CAS PubMed.
- B. R. Zhang, B. Larsson, N. Lundeheim and H. Rodriguez-Martinez, Int. J. Androl., 1998, 21, 207–216 CrossRef CAS PubMed.
- S. S. Suarez, Cell Tissue Res., 2016, 363, 185–194 CrossRef PubMed.
- R. Talevi and R. Gualtieri, Theriogenology, 2010, 73, 796–801 CrossRef CAS PubMed.
- R. Gualtieri, V. Mollo, V. Barbato and R. Talevi, Theriogenology, 2010, 73, 1037–1043 CrossRef CAS PubMed.
- J. W. Pollard, C. Plante, W. A. King, P. J. Hansen, K. J. Betteridge and S. S. Suarez, Biol. Reprod., 1991, 44, 102–107 CrossRef CAS PubMed.
- T. Mcnutt and G. Killian, J. Androl., 1991, 12, 244–252 CAS.
- C. Gabler, S. Odau, K. Muller, J. Schon, A. Bondzio and R. Einspanier, J. Physiol. Pharmacol., 2008, 59, 29–42 Search PubMed.
- R. F. Gonçalves, A. L. Staros and G. J. Killian, Reprod. Domest. Anim., 2008, 43, 720–729 CrossRef PubMed.
- A. Gad, M. Hoelker, U. Besenfelder, V. Havlicek, U. Cinar, F. Rings, E. Held, I. Dufort, M.-A. Sirard, K. Schellander and D. Tesfaye, Biol. Reprod., 2012, 87, 100 CrossRef PubMed.
- D. Salilew-Wondim, E. Fournier, M. Hoelker, M. Saeed-Zidane, E. Tholen, C. Looft, C. Neuhoff, U. Besenfelder, V. Havlicek, F. Rings, D. Gagné, M. A. Sirard, C. Robert, H. A. Shojaei Saadi, A. Gad, K. Schellander and D. Tesfaye, PLoS One, 2015, 10, 1–31 Search PubMed.
- S. Betsha, M. Hoelker, D. Salilew-Wondim, E. Held, F. Rings, C. Große-Brinkhause, M. U. Cinar, V. Havlicek, U. Besenfelder, E. Tholen, C. Looft, K. Schellander and D. Tesfaye, Mol. Reprod. Dev., 2013, 80, 315–333 CrossRef CAS PubMed.
- D. Barrera, E. V. García, F. Sinowatz, G. A. Palma, M. A. Jiménez-Díaz and D. C. Miceli, Anat., Histol., Embryol., 2013, 42, 312–315 CrossRef CAS PubMed.
- K. Miki and D. E. Clapham, Curr. Biol., 2013, 23, 443–452 CrossRef CAS PubMed.
- Y. Ménézo, P. Guérin and K. Elder, Reprod. BioMed. Online, 2015, 30, 233–240 CrossRef PubMed.
- S. E. Ulbrich, K. Zitta, S. Hiendleder and E. Wolf, Theriogenology, 2010, 73, 802–816 CrossRef CAS PubMed.
- P. Morales, V. Palma, A. M. Salgado and M. Villalon, Hum. Reprod., 1996, 11, 1504–1509 CrossRef CAS PubMed.
- H. Nelis, K. D'Herde, K. Goossens, L. Vandenberghe, B. Leemans, K. Forier, K. Smits, K. Braeckmans, L. Peelman and A. Van Soom, Reprod., Fertil. Dev., 2014, 26, 954–966 CrossRef PubMed.
- B. Leemans, B. M. Gadella, T. A. E. Stout, E. Sostaric, C. De Schauwer, H. M. Nelis, M. Hoogewijs and A. Van Soom, Reproduction, 2016, 151, 313–330 CrossRef CAS PubMed.
- V. Maillo, R. Lopera-Vasquez, M. Hamdi, A. Gutierrez-Adan, P. Lonergan and D. Rizos, Theriogenology, 2016, 86, 443–450 CrossRef PubMed.
- R. Gualtieri, V. Mollo, S. Braun, V. Barbato, I. Fiorentino and R. Talevi, Theriogenology, 2013, 79, 429–435 CrossRef CAS PubMed.
- R. Gualtieri, V. Mollo, S. Braun, V. Barbato, I. Fiorentino and R. Talevi, Theriogenology, 2012, 78, 1456–1464 CrossRef CAS PubMed.
- E. Sostaric, S. J. Dieleman, C. H. A. van de Lest, B. Colenbrander, P. L. A. M. Vos, N. Garcia-Gil and B. M. Gadella, Mol. Reprod. Dev., 2008, 75, 60–74 CrossRef PubMed.
- S. Chen, R. Einspanier and J. Schoen, Theriogenology, 2013, 80, 862–869 CrossRef CAS PubMed.
- M. Rajagopal, T. L. Tollner, W. E. Finkbeiner, G. N. Cherr and J. H. Widdicombe, In Vitro Cell. Dev. Biol., 2006, 42, 248–254 CAS.
- S. Fotheringham, K. Levanon and R. Drapkin, J. Visualized Exp., 2011, 3–7 Search PubMed.
- F. Pampaloni, E. G. Reynaud and E. H. K. Stelzer, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS PubMed.
- J. Yu, S. Peng, D. Luo and J. C. March, Biotechnol. Bioeng., 2012, 109, 2173–2178 CrossRef CAS PubMed.
- D. Huh, G. A. Hamilton and D. E. Ingber, Trends Cell Biol., 2011, 21, 745–754 CrossRef CAS PubMed.
- M. Kessler, K. Hoffmann, V. Brinkmann, O. Thieck, S. Jackisch, B. Toelle, H. Berger, H.-J. Mollenkopf, M. Mangler, J. Sehouli, C. Fotopoulou and T. F. Meyer, Nat. Commun., 2015, 6, 8989 CrossRef CAS PubMed.
- C. M. Costello, J. Hongpeng, S. Shaffiey, J. Yu, N. K. Jain, D. Hackam and J. C. March, Biotechnol. Bioeng., 2014, 111, 1222–1232 CrossRef CAS PubMed.
- N. P. Macdonald, F. Zhu, C. J. Hall, J. Reboud, P. S. Crosier, E. E. Patton, D. Wlodkowic and J. M. Cooper, Lab Chip, 2016, 16, 291–297 RSC.
- H. T. A. A. Van Tol, F. J. C. M. C. M. Van Eerdenburg, B. Colenbrander and B. A. J. J. Roelen, Mol. Reprod. Dev., 2008, 75, 578–587 CrossRef CAS PubMed.
- T. Scherr, G. L. Knapp, A. Guitreau, D. S.-W. Park, T. Tiersch, K. Nandakumar and W. T. Monroe, Biomed. Microdevices, 2015, 17, 1–10 CrossRef CAS PubMed.
- S. L. Angione, N. Oulhen, L. M. Brayboy, A. Tripathi and G. M. Wessel, Fertil. Steril., 2015, 103, 281–290 CrossRef PubMed.
- S. M. Knowlton, M. Sadasivam and S. Tasoglu, Trends Biotechnol., 2016, 33, 221–229 CrossRef PubMed.
- T. M. El-Sherry, M. Elsayed, H. K. Abdelhafez and M. Abdelgawad, Integr. Biol., 2014, 6, 1111–1121 RSC.
- M. S. Kim, C. Y. Bae, G. Wee, Y. M. Han and J. K. Park, Electrophoresis, 2009, 30, 3276–3282 CrossRef CAS PubMed.
- H. Abe, Histol. Histopathol., 1996, 11, 743–768 CAS.
- J. Reischl, K. Prelle, H. Schöl, C. Neumüller, R. Einspanier, F. Sinowatz and E. Wolf, Cell Tissue Res., 1999, 296, 371–383 CrossRef CAS PubMed.
- M. S. Joshi, Microsc. Res. Tech., 1995, 31, 507–518 CrossRef CAS PubMed.
- A. M. Karst and R. Drapkin, Nat. Protoc., 2012, 7, 1755–1764 CrossRef CAS PubMed.
- M. T. Comer, H. J. Leese and J. Southgate, Hum. Reprod., 1998, 13, 3114–3120 CrossRef CAS PubMed.
- P. Coy and M. Avilés, Biol. Rev. Cambridge Philos. Soc., 2010, 85, 593–605 Search PubMed.
- I. Mondéjar, I. Martínez-Martínez, M. Avilés and P. Coy, Biol. Reprod., 2013, 89, 67 CrossRef PubMed.
- E. K. Topper, G. J. Killian, A. Way, B. Engel and H. Woelders, J. Reprod. Fertil., 1999, 115, 175–183 CrossRef CAS.
- C. Rodríguez and G. Killian, Anim. Reprod. Sci., 1998, 54, 1–12 CrossRef.
- G. J. Killian, Anim. Reprod. Sci., 2004, 82–83, 141–153 CrossRef CAS PubMed.
- D. W. Erikson, A. L. Way, R. P. Bertolla, D. A. Chapman and G. J. Killian, Anim. Reprod., 2007, 4, 103–112 Search PubMed.
- I. A. Taitzoglou, A. N. Kokoli and G. J. Killian, Int. J. Androl., 2007, 30, 108–114 CrossRef CAS PubMed.
- C. J. de Jonge, C. L. R. Barratt, E. W. A. Radwanska and I. A. N. D. Cooke, J. Androl., 1993, 14, 359–365 CAS.
- R. S. King, S. H. Anderson and G. J. Killian, J. Androl., 1994, 15, 468–478 CAS.
- U. Besenfelder, V. Havlicek and G. Brem, Reprod. Domest. Anim., 2012, 47, 156–163 CrossRef PubMed.
- E. Monaco, B. Gasparrini, L. Boccia, A. De Rosa, L. Attanasio, L. Zicarelli and G. Killian, Theriogenology, 2009, 71, 450–457 CrossRef CAS PubMed.
- S. Kölle, S. Reese and W. Kummer, Theriogenology, 2010, 73, 786–795 CrossRef PubMed.
- G. Zaccagnini, B. Maione, R. Lorenzini and C. Spadafora, Biol. Reprod., 1998, 59, 1549–1553 CrossRef CAS PubMed.
- R. Lopera-Vásquez, M. Hamdi, B. Fernandez-Fuertes, V. Maillo, P. Beltrán-Breña, A. Calle, A. Redruello, S. López-Martín, A. Gutierrez-Adán, M. Yañez-Mó, M. Á. Ramirez and D. Rizos, PLoS One, 2016, 11, e0148083 Search PubMed.
- V. Maillo, P. O. Gaora, N. Forde, U. Besenfelder, V. Havlicek, G. W. Burns, T. E. Spencer, A. Gutierrez-Adan, P. Lonergan and D. Rizos, Biol. Reprod., 2015, 92, 144 CrossRef PubMed.
- B. V. Sanches, J. H. F. Pontes, A. C. Basso, C. R. Ferreira, F. Perecin and M. M. Seneda, Reprod. Domest. Anim., 2013, 48, 7–9 CrossRef PubMed.
- A. Bakhtari and P. J. Ross, Epigenetics, 2014, 9, 1271–1279 CrossRef PubMed.
- L. E. Young, K. D. Sinclair and I. Wilmut, Rev. Reprod., 1998, 3, 155–163 CrossRef CAS PubMed.
- L. E. Young, K. Fernandes, T. G. McEvoy, S. C. Butterwith, C. G. Gutierrez, C. Carolan, P. J. Broadbent, J. J. Robinson, I. Wilmut and K. D. Sinclair, Nat. Genet., 2001, 27, 153–154 CrossRef CAS PubMed.
- L. E. Young, A. E. Schnieke, K. J. McCreath, S. Wieckowski, G. Konfortova, K. Fernandes, G. Ptak, A. J. Kind, I. Wilmut, P. Loi and R. Feil, Mech. Dev., 2003, 120, 1433–1442 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: ESI is available as two supplementary movies. See DOI: 10.1039/c6lc01566b |
| This journal is © The Royal Society of Chemistry 2017 |