Patterning of sharp cellular interfaces with a reconfigurable elastic substrate†
Allison
Curtis
a,
David J.
Li
 a,
Brian
DeVeale
b,
Kento
Onishi
c,
Monica Y.
Kim
a,
Robert
Blelloch
b,
Diana J.
Laird
b and
Elliot E.
Hui
a,
Brian
DeVeale
b,
Kento
Onishi
c,
Monica Y.
Kim
a,
Robert
Blelloch
b,
Diana J.
Laird
b and
Elliot E.
Hui
 *a
*a
aDepartment of Biomedical Engineering, University of California, Irvine, California 92697-2715, USA. E-mail: eehui@uci.edu
bEli and Edythe Broad Center of Regeneration Medicine and Stem Cell Research, University of California, San Francisco, California, USA
cDepartment of Bioengineering, University of California, Berkeley, California, USA
First published on 21st December 2016
Abstract
Micropatterned cocultures are a useful experimental tool for the study of cell–cell interactions. Patterning methods often rely on sequential seeding of different cell types or removal of a barrier separating two populations, but it is difficult to pattern sharp interfaces between pure populations with low cross-contamination when using these approaches. Patterning by the use of reconfigurable substrates can overcome these limitations, but such methods can be costly and challenging to employ in a typical biology laboratory. Here, we describe a low-cost and simple-to-use reconfigurable substrate comprised of a transparent elastic material that is partially cut to form a slit that opens when the device is stretched. The slit seals back up when released, allowing two initially separate, adherent cell populations to be brought together to form a contact interface. Fluorescent imaging of patterned cocultures demonstrates the early establishment of a sharp cellular interface. As a proof of principle, we demonstrate the use of this device to study competition at the interface of two stem cell populations.
Insight, innovation, integrationThe ability to arrange multiple cell types with a defined spatial organization in culture has been useful for a variety of studies to understand how cells communicate and interact with one another in tissues. This work was specifically focused on defining a sharp border between two adjoining cell populations, which many previous approaches have struggled to achieve. This problem was solved by the development of a new microengineered device, which was designed for use in conventional cell biology laboratories. As a demonstration, the system was employed to study competition between DNA-damaged and undamaged stem cells. Due to the sharp initial border, it was possible to observe and quantify cell invasion distances as small as tens of micrometers. |
Introduction
Biological tissues possess a complex architecture that relies on the precise positioning of different cell types within their microenvironment. Spatial proximity of cells and their interactions with one another are fundamental to tissue and organ function. Cell micropatterning has become a useful technique in the study of such interactions, providing precise control over the spatial organization of cell populations in vitro. In particular, methods for patterning two cell populations have enabled the investigation of phenotypic optimization,1 the role of direct cell–cell contact,2–4 gradients in paracrine signaling,5,6 and migration and invasion between populations.7 Boundaries between cellular compartments are a fundamental feature of complex tissues. The ability to pattern a sharp interface between two cell populations could enable in vitro studies of phenomena such as the establishment of morphogen gradients, the specification of specialized border cells, and the maintenance of cell compartment boundaries, or their disruption.8,9Typically, patterned cocultures are achieved by sequential seeding of two different cell types. First, one cell population is confined via masking, selective adhesion, or microfluidic flow, followed by the addition of the second cell type into the remaining areas where the first cells were not placed.10 While these techniques can be successful in generating a fairly organized tissue construct, cross-contamination can be significant as the second population is seeded directly over the first group of patterned cells (Fig. 4A). Another approach is to place a removable barrier between the two cell populations. Pure cell populations may be seeded on either side of the barrier, and once the barrier is removed, the cells can migrate towards each other to close the gap, as in a wound-healing assay. This method can be implemented by a number of commercial products including removable stencils from Ibidi (Martinsried, Germany), Cell Biolabs (San Diego, USA), and Nunc Lab-Tek (Thermo Fisher Scientific). A significant drawback, however, is that the cells typically do not maintain a clean front as they advance, resulting in a ragged interface between the two populations (Fig. 4B), which may interfere with accurate measurement of cell invasion or signaling gradients.
An alternative approach is to employ a reconfigurable substrate.4,11 Here, different populations of cells are adhered onto plates that can then be repositioned to establish patterned cultures. Since each cell type can be seeded in isolation, cross contamination can be avoided. Cells may be grown to confluence prior to patterning, resulting in very well-defined boundaries between populations. Our previously reported comb device4 consists of microfabricated silicon parts that are independently seeded with different cell types and then locked together to form patterned cocultures. This device has been successfully employed to dissect cell–cell interactions in the liver,3,4 modulate coculture interactions to drive the differentiation of stem cells,2,12 and interrogate tumor-stromal signaling.5 However, while reconfigurable micromechanical substrates can pattern sharp cellular interfaces, the devices are costly, require a significant amount of training to operate, and are optically opaque, complicating their use with the inverted microscopes commonly employed by biologists.
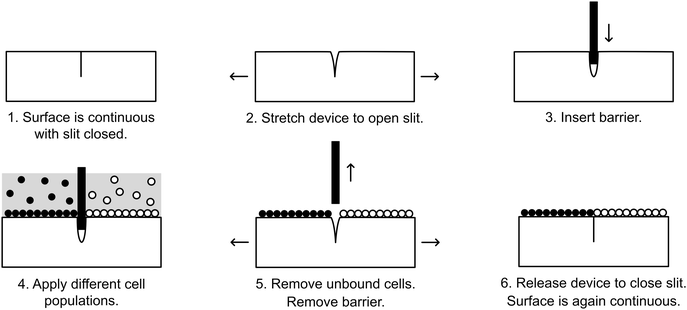
Here, we present a simple-to-use and inexpensive reconfigurable substrate for cellular interface patterning. The substrate is comprised of a transparent elastic material, polydimethylsiloxane (PDMS), that is partially cut to form a slit that opens when the device is stretched, allowing insertion of a thin barrier. Cells may then be seeded simultaneously on opposite sides of the barrier. When the barrier is removed, the slit seals back up, allowing two initially separate, adherent cell populations to be brought together to form a contact interface (Fig. 1). We demonstrate that this device can pattern cellular interfaces that are sharper and cleaner than can be achieved by alternative patterning methods. Through fluorescence microscopy and automated image processing, the position of the interface can be quantitatively mapped over time. As an example of the utility our system, we demonstrate a cell invasion assay and employ this assay to study competition at the interface of two stem cell populations.
Materials and methods
Device fabrication
Molds were created by attaching laser-cut acrylic parts (20 mm diameter circles × 6 mm) to the bottom of a square 100 mm polystyrene dish (Fisher Scientific, USA) by using cyanoacrylate glue (Gorilla Glue Company, USA). Polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning, USA) was prepared per manufacturer recommendation, mixed at a ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
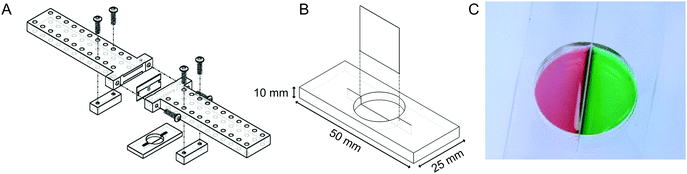
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 base to curing agent, and degassed. 95 g of PDMS was then poured into the mold and cured for 4 hours at 65 °C. The molded PDMS was removed and cut into individual devices (50 mm × 25 mm × 8.25 mm). A shallow slit was cut across the bottom of each molded well by using a razor blade. The apparatus illustrated in Fig. 2A was employed to ensure that the depth of cut was repeatable and did not penetrate all of the way through the bottom of the well. Angled cuts were performed by hand without using the apparatus.
1 base to curing agent, and degassed. 95 g of PDMS was then poured into the mold and cured for 4 hours at 65 °C. The molded PDMS was removed and cut into individual devices (50 mm × 25 mm × 8.25 mm). A shallow slit was cut across the bottom of each molded well by using a razor blade. The apparatus illustrated in Fig. 2A was employed to ensure that the depth of cut was repeatable and did not penetrate all of the way through the bottom of the well. Angled cuts were performed by hand without using the apparatus.
Cell culture
NIH 3T3 cells (ATCC, USA) expressing H2B-mTurquoise and H2B-citrine were kindly provided by Dr. Michael Elowitz (California Institute of Technology, USA). 3T3 cells were maintained in DMEM media (GIBCO, USA) supplemented with 10% BCS (GIBCO, USA), 1% L-glutamine (Caisson Laboratories, Inc., USA), and 1% penicillin/streptomycin (Caisson Laboratories, Inc., USA) at 37 °C and 5% CO2. NMUMG cells (ATCC, USA) were maintained in DMEM media supplemented with 10% FBS (GIBCO, USA), 1% L-glutamine, and 1% penicillin/streptomycin at 37 °C and 5% CO2. Prior to device seeding, cells were trypsinized and re-suspended in 500 μL of fresh media.Cell seeding into elastic devices
Devices were UV-sterilized overnight and treated for 60 seconds with a plasma formed from room air. Device wells were incubated in 200 μL of 5 μg mL−1 human fibronectin (Sigma-Aldrich, USA) at room temperature (RT) for 3 hours. The wells were then washed once with sterile PBS. Fig. 1 outlines the strategy for seeding cells into the device. The device was bent to stretch open the slit, and a glass coverslip (#1, FisherScientific, USA) was inserted. With the coverslip barrier in place, 3T3-H2b-mTurquoise (0.5 × 106 in 500 mL media) and 3T3-H2b-citrine (0.5 × 106 cells in 500 mL media) were seeded into the two isolated chambers and incubated for 3–5 hours to allow cell adhesion. The wells were then gently aspirated and washed three times with warm PBS. Following aspiration, the device was stretched to open the slit and the coverslip barrier was removed. The cells were washed another three times with warm PBS in order to remove unattached cells, and finally 1 mL of media was added.Cell seeding into Ibidi inserts
A 2-well culture insert (Ibidi, Martinsried, Germany) was placed in the center of a well of a 12-well tissue culture plate (Fisher Scientific, USA). For patterning by sequential seeding, 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 3T3-citrine cells in 70 μL of media were pipetted into one of the insert wells, while the other insert well (70 μL) and the remainder of the culture well (1 mL) were filled with plain media. Cells were grown to confluence overnight. Unattached cells were then aspirated and the insert well was rinsed once with warm media. All media was aspirated and the insert was removed from the well bottom using sterile tweezers. The well was then filled with 1 mL of warm media and incubated for 15 minutes. After media aspiration, 250
000 3T3-citrine cells in 70 μL of media were pipetted into one of the insert wells, while the other insert well (70 μL) and the remainder of the culture well (1 mL) were filled with plain media. Cells were grown to confluence overnight. Unattached cells were then aspirated and the insert well was rinsed once with warm media. All media was aspirated and the insert was removed from the well bottom using sterile tweezers. The well was then filled with 1 mL of warm media and incubated for 15 minutes. After media aspiration, 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 3T3-mTurquoise cells were seeded into the well in 1 mL of media. The culture plates were shaken vigorously every 30 minutes for the first 3 hours to ensure uniform cell distribution, then incubated overnight. The next day, the wells were rinsed once with warm media and replaced with 1 mL of fresh media.
000 3T3-mTurquoise cells were seeded into the well in 1 mL of media. The culture plates were shaken vigorously every 30 minutes for the first 3 hours to ensure uniform cell distribution, then incubated overnight. The next day, the wells were rinsed once with warm media and replaced with 1 mL of fresh media.
For patterning interfaces by cell migration, 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 3T3-citrine cells and 35
000 3T3-citrine cells and 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 3T3-mTurquoise cells, each in 70 μL of media, were seeded into neighboring insert wells and the remainder of the culture well was filled with 1 mL of plain media. Cells were incubated and grown to confluence overnight, followed by a rinse with warm media and insert removal. The well was then replenished with 1 mL of media and incubated for 48 hours in order to allow the two cell populations to migrate towards each other and form an interface, as in a wound healing assay.
000 3T3-mTurquoise cells, each in 70 μL of media, were seeded into neighboring insert wells and the remainder of the culture well was filled with 1 mL of plain media. Cells were incubated and grown to confluence overnight, followed by a rinse with warm media and insert removal. The well was then replenished with 1 mL of media and incubated for 48 hours in order to allow the two cell populations to migrate towards each other and form an interface, as in a wound healing assay.
Cell imaging
Imaging was performed using an inverted epifluorescence microscope (Eclipse, Nikon, USA) equipped with the appropriate CFP and YFP filter cubes for visualization of nuclear expression of mTurquoise and citrine, respectively. Images were taken with the field of view centered on the middle of the well. Brightfield, CFP, and YFP images were taken of the same field of view without adjusting the focus.Characterizing slit opening and closing dynamics
Devices were sterilized overnight under UV light, plasma treated for 60 seconds and coated with 200 μL of human fibronectin at 5 μg mL−1 for 3 hours at RT. Wells were washed one time with warm PBS, aspirated, and the device was bent apart for insertion of a coverslip barrier. 500 μL of media was then added to each half of the well, and the device was kept at 37 °C and 5% CO2 for 3 hours. Devices were removed from the incubator, and washed once with warm PBS. The liquid was then aspirated, followed by removal of the coverslip barrier. 1 mL warm media was then added to the well. Devices were either imaged immediately, or placed back into the incubator and imaged at later time points. Imaging was performed with a 20× objective on an upright microscope (Olympus, U-TVO.63XC), allowing the size of the slit opening to be measured. Image acquisition was performed with QCapture software (QImaging, BC, Canada). For dynamic imaging, the software was set to acquire images automatically every 10 seconds over the course of 20 minutes. Focus adjustments were made by hand as needed in between acquisitions.Image processing
ImageJ (NIH, USA) was used for all image processing. Linear filters were applied to improve image clarity. Fluorescence images were also corrected using the background subtraction plugin, which employs a rolling ball radius algorithm. A single background subtraction was performed on all fluorescence images with the rolling ball radius set to 50 μm.Cell invasion assay
Devices were sterilized overnight (O/N) under UV light then pre-treated O/N with 1.25% fibronectin (Sigma, F1141) and 0.02% gelatin to promote growth as a monolayer. Immediately prior to cell seeding, 22 × 30 coverslips (Fisherbrand, USA) were inserted into the slit of each device.ROSAmT/mG mouse embryonic stem cells (mESCs) maintained in 15% FBS and LIF13 were trypsinized and each side of the device was seeded with a volume of 250 μL. 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 control mESCs were seeded on either side of the device. Alternatively, 200
000 control mESCs were seeded on either side of the device. Alternatively, 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 damaged mESCs were seeded. Cells were damaged by treatment with 100 nM doxorubicin for four hours (Cell Signaling, 5927S). Coverslips were removed 3.5 hours after seeding. Once seeded on the device, cells were refed daily, cultured for a total of 60 hours and then imaged on a Leica DMI 4000.
000 damaged mESCs were seeded. Cells were damaged by treatment with 100 nM doxorubicin for four hours (Cell Signaling, 5927S). Coverslips were removed 3.5 hours after seeding. Once seeded on the device, cells were refed daily, cultured for a total of 60 hours and then imaged on a Leica DMI 4000.
Border identification algorithm
To identify the border of the two PDMS halves and the invasion distance across the border of cells from either side, an algorithm was written and run in MATLAB®. The code for this script is available as part of the ESI.† The inputs of the algorithm are the directories in which the images from three channels (brightfield and two fluorescent channels) are stored. The outputs are plots showing the junction of the PDMS halves and leading edges of the two cell populations on either side of the junction, all superimposed onto contrast corrected images of their respective channels (Fig. S2, ESI†). Additionally, a table of values is generated, measuring average area and distance of ingression where positive values represent travel of the leading edge across to the other side of the junction.To perform these tasks, the algorithm first identified the junction of the PDMS halves by analyzing the brightfield (BF) channel. Briefly, pixel intensity values were measured and the difference between each pixel and its horizontally adjacent neighbor was calculated. At the junction, there is a vertical strip of consecutive positive values. Long vertical strips of positive values were identified, binarized, and fit to a line to determine slope and intersection with the x-axis. Images from all channels were rotated if the slope exceeded a defined threshold and this whole process was repeated once. The border was identified as the intersection of this fit line with the x-axis.
Then, using Sobel edge detection, edges were identified in the two fluorescence channels. Sobel values were generated by applying a transformation matrix on all pixels of the fluorescent images. These Sobel values were then binarized according to a threshold (Sobel threshold or ST) that was determined automatically. First, a threshold (pixel intensity threshold or PT) was identified for the pixel intensity values of each fluorescence channels using the ISODATA method.14 This PT was used to segregate Sobel values to those associated with “bright” and “dim”. Next, the ST was determined by taking the mean less the standard deviation of the “bright” Sobel values. After binarization using the ST, the boundary was drawn using the boundaries function in the MATLAB® image processing toolbox. Additionally, regions opposite of the leading edge were computationally filled in to ensure gaps in cells due to under confluence did not affect analysis. The total area of the PDMS half was subtracted from the total area of the drawn boundary of cells to determine area of invasion (or ingression). This area was then divided by the vertical junction length to determine average distance of invasion (or ingression).
Results and discussion
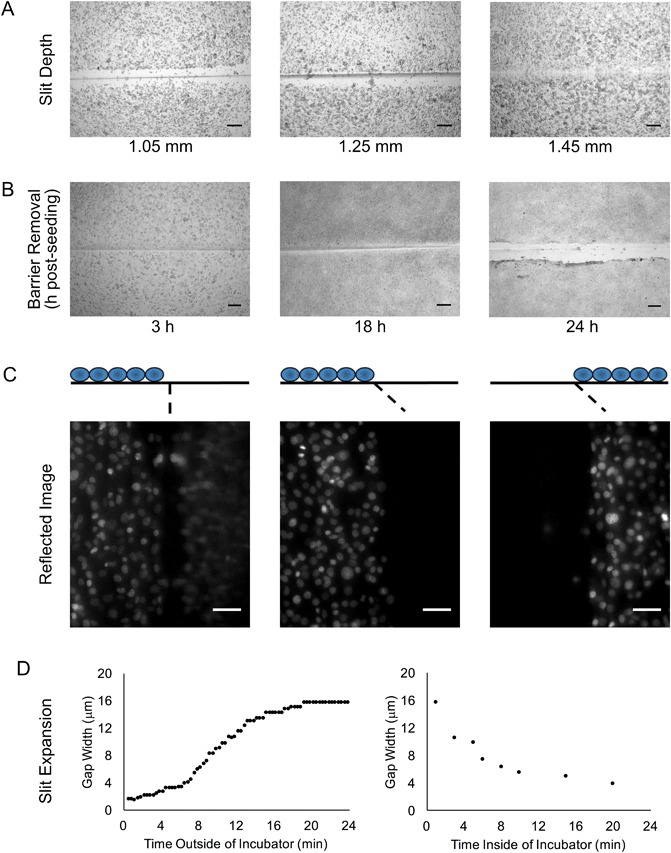
We sought to develop a device to pattern a sharp border between two adjoining cell populations. Our strategy is illustrated in Fig. 1. An elastic substrate contains a slit, which opens upon stretching of the device to allow insertion of a thin barrier. The barrier separates the substrate into two regions that may be separately seeded with cells. After the barrier is removed, the slit reseals, bringing the two cell populations directly together. To fabricate this device, we employed PDMS, an optically clear elastomer that is suitable for culturing a wide range of mammalian cell types.15 Importantly, the optical properties of PDMS allows the device to be compatible with fluorescence imaging on inverted biological microscopes. Wells were molded out of PDMS and then cut with a razor blade to form a shallow slit across the bottom of the well. The razor blade was mounted on an apparatus to ensure a reproducible depth of cut (Fig. 2A). The cut was designed to extend horizontally into the well walls (Fig. 2B) such that a cover slip inserted into the slit would divide the well into isolated compartments (Fig. 2C). The overall device dimensions were cut to conform to the size of a standard microscope slide in order to facilitate compatibility with microscope slide mounts.We found that if the slit was cut too shallow, cell adhesion was poor in the vicinity of the slit (Fig. 3A), perhaps due to compressive deformation of the PDMS around the barrier. This effectively determined the minimum thickness of the well bottom. When using a #1 coverslip as the barrier, a slit depth of at least 1.45 mm was required to ensure good cell adhesion up to the edge of the slit. Similarly, we found that leaving the barrier in place for an extended period of time following cell seeding could result in the sheeting off of cells when the barrier was finally removed, perhaps due to the formation of a continuous cell sheet that becomes attached to the coverslip barrier (Fig. 3B). Consequently, we adopted a standard incubation time of 3–5 hours between cell seeding and barrier removal. The use of a nonadhesive coating on the barrier surface could perhaps be helpful in reducing cell sheeting, but this was not investigated.
During fluorescence microscopy at higher magnifications, we often observed ghost images in the close vicinity of the slit (Fig. 3C). These appeared to be reflected images of cells from the opposite side of the slit. Interestingly, these ghost images could be eliminated by cutting the slit at an angle of at least 20° off vertical (Fig. 3C). The mirroring effect likely arises from an incompletely closed slit that is filled with media; light can be reflected from the PDMS–media interface due to the index mismatch. Our rationale in angling the slit was to direct the reflected light away from the microscope objective, but it may also be that the angled slit is better able to seal closed.
We noted that even with no barrier in the device, the slit did not close up perfectly, but rather a gap remained. Furthermore, the width of the gap seemed to be inconsistent, even on a single device. Upon further examination, we observed that after removal of the device from the cell culture incubator, the gap width expanded from less than 4 μm to about 16 μm over the course of about 20 minutes (Fig. 3D). When the device was placed back into the incubator, the gap width decreased back to 4 μm over a similar time course. This change in gap width could adversely affect experiments that are dependent on cell contact or cell migration across the gap. If frequent imaging is required, a microscope stage top incubator is the best solution for mitigating this concern. Even if this is not available, the gap width does remain relatively stable over the first few minutes out of the incubator (Fig. 3D), and so brief periods of removal from the incubator may not be detrimental. The observed gap width dynamics are likely related to the temperature change encountered when the device is removed from the incubator. Assuming a PDMS coefficient of thermal expansion of 310 × 10−6 per °C, a temperature differential of 12 °C should produce a 37 μm change in length for a 1 cm object, which is on the same order of magnitude as the changes in gap width that were observed.
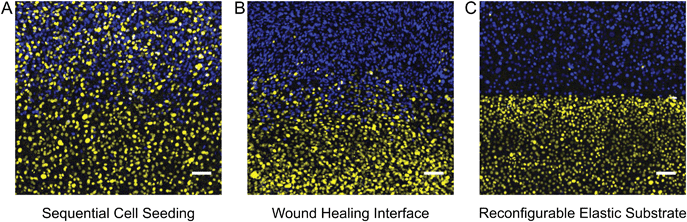
We sought to compare the quality of the coculture interface patterned with our device against the most common alternative patterning methods. A common method for creating micropatterned cocultures involves first patterning one cell population and then seeding a second population around the first pattern.1,7 It has been reported that with this sequential seeding approach, it is difficult to avoid cross-contamination between the two cell populations.16 We demonstrated patterning by sequential seeding by employing Ibidi stencils to pattern an initial population of mTurquoise-labeled 3T3 fibroblasts, then removing the stencil after overnight incubation and seeding a second population of citrine-labeled 3T3 fibroblasts. Substantial adhesion of citrine cells in mTurquoise regions was observed (Fig. 4A). Another common method for patterning cocultures is to seed two cell populations in neighboring wells of a stencil and then to remove the stencil to allow the cells to migrate together as in a wound-healing assay.7 We demonstrated this approach by employing Ibidi stencils to pattern mTurquoise-labeled 3T3 fibroblasts and citrine-labeled 3T3 fibroblasts simultaneously in separate wells, then removing the stencil after overnight incubation and allowing the two populations to migrate together over the course of 48 hours (Fig. S1, ESI†). Cross-contamination between the two populations was much improved over the sequential seeding approach, but the border between the two populations was not very sharp since the cells did not advance with a uniform front (Fig. 4B). Finally, we patterned the same 3T3 fibroblasts by using our reconfigurable elastic substrate. Citrine and mTurquoise cells were seeded simultaneously on opposite sides of the coverslip barrier and incubated overnight; the barrier was then removed to establish a contact interface almost immediately. Cross-contamination between the two populations was minimal, and the border between the two populations was much sharper than with either of the other two methods (Fig. 4C). Use of a reconfigurable substrate is thus the better approach for patterning a sharp interface between two cell populations.
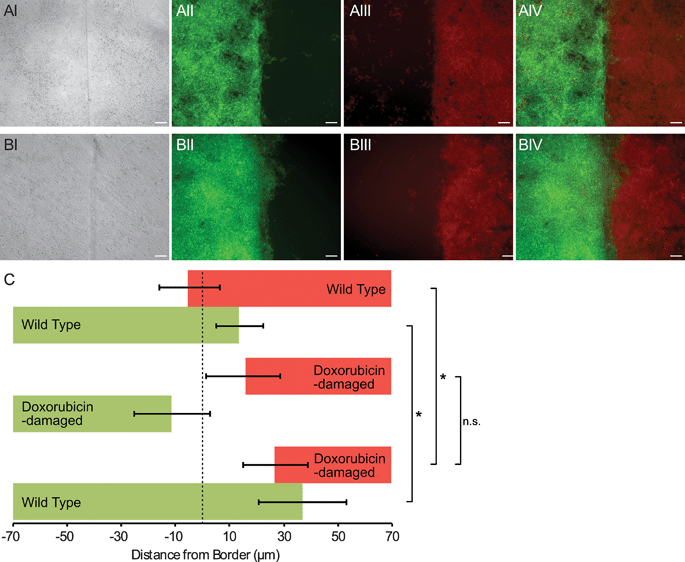
The sharp border possible with our device was useful for quantifying invasion of one cell population into another. To demonstrate this method, we patterned wild-type mouse embryonic stem cells (mES) bordering DNA-damaged mES cells. After 60 hours of culture, each cell population was imaged under epi-fluorescence, and a border identification algorithm was applied (Fig. S2, ESI†). We observed that the undamaged mES cells invaded the damaged mES cell region, penetrating an average of about 30 μm (Fig. 5). When two undamaged mES populations were patterned, the border did not undergo any net displacement. These data are consistent with the hypothesis that competition between cells occurs in the developing embryo, and in this competition DNA-damaged cells are disadvantaged in comparison to their undamaged counterparts.17,18 However, a control experiment with two populations of DNA-damaged mES cells showed that both populations receded away from the original border (Fig. 5). Thus, it was unclear whether undamaged mES cells actively induced cell death in damaged cells, or damaged cells receded autonomously and undamaged cells simply filled the vacated area. Nevertheless, these experiments demonstrated that our method was capable of quantifying cell invasion at a coculture interface with as little as tens of microns of net border displacement.
Conclusion
We have demonstrated a patterning method that establishes very sharp borders between two cell populations. Our method achieved cocultures with sharper borders and less cross-contamination than common alternative patterning strategies. Furthermore, the sharp interface proved useful in a coculture cell invasion assay. The device can be scaled to use larger or smaller wells, or to employ multiple wells controlled by a single barrier (Fig. S3, ESI†). Multiple slits within a single well can also be employed, for example to pattern a triculture. It may also be possible to adapt the system to establish non-contacting cocultures in addition to contacting cocultures, or even to switch between the two states dynamically.4 Unlike previously reported reconfigurable culture systems, this device is easy to operate, cheap to manufacture, and compatible with inverted biological microscopes, which may increase the number of researchers who are able to access this technology to study cell–cell interactions.Acknowledgements
We thank Joe Markson and Pulin Li from the Elowitz lab for their helpful feedback on the device design as well as for providing cell lines. This work was supported by the Defense Advanced Research Projects Agency (D13AP00044 to EEH), the National Institutes of Health (DP2OD007420 to DJL), and the University of California Cancer Research Coordinating Committee funds. AC was supported by the Biophotonics across Energy, Space, and Time (BEST) Interdisciplinary Graduate Education and Research Traineeship program through the National Science Foundation.References
- S. N. Bhatia, U. J. Balis, M. L. Yarmush and M. Toner, FASEB J., 1999, 13, 1883–1900 CAS.
- N. Rao, G. N. Grover, L. G. Vincent, S. C. Evans, Y. S. Choi, K. H. Spencer, E. E. Hui, A. J. Engler and K. L. Christman, Integr. Biol., 2013, 5, 1344–1354 RSC.
- S. March, E. E. Hui, G. H. Underhill, S. Khetani and S. N. Bhatia, Hepatology, 2009, 50, 920–928 CrossRef CAS PubMed.
- E. E. Hui and S. N. Bhatia, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 5722–5726 CrossRef CAS PubMed.
- K. H. Spencer, M. Y. Kim, C. C. Hughes and E. E. Hui, Integr. Biol., 2014, 6, 382–387 RSC.
- V. V. Abhyankar, M. A. Lokuta, A. Huttenlocher and D. J. Beebe, Lab Chip, 2006, 6, 389–393 RSC.
- M. J. Stine, C. J. Wang, W. F. Moriarty, B. Ryu, R. Cheong, W. H. Westra, A. Levchenko and R. M. Alani, Cancer Res., 2011, 71, 2433–2444 CrossRef CAS PubMed.
- K. D. Irvine and C. Rauskolb, Annu. Rev. Cell Dev. Biol., 2001, 17, 189–214 CrossRef CAS PubMed.
- D. J. Montell, Nat. Rev. Mol. Cell Biol., 2003, 4, 13–24 CrossRef CAS PubMed.
- D. Falconnet, G. Csucs, H. M. Grandin and M. Textor, Biomaterials, 2006, 27, 3044–3063 CrossRef CAS PubMed.
- H. Kaji, T. Yokoi, T. Kawashima and M. Nishizawa, Lab Chip, 2009, 9, 427–432 RSC.
- N. Rao, S. Evans, D. Stewart, K. H. Spencer, F. Sheikh, E. E. Hui and K. L. Christman, Biomed. Microdevices, 2013, 15, 161–169 CrossRef PubMed.
- M. D. Muzumdar, B. Tasic, K. Miyamichi, L. Li and L. Luo, Genesis, 2007, 45, 593–605 CrossRef CAS PubMed.
- T. Ridler and S. Calvard, IEEE Trans. Syst. Man Cybern., 1978, 8, 630–632 CrossRef.
- J. N. Lee, X. Jiang, D. Ryan and G. M. Whitesides, Langmuir, 2004, 20, 11684–11691 CrossRef CAS PubMed.
- E. E. Hui and S. N. Bhatia, Langmuir, 2007, 23, 4103–4107 CrossRef CAS PubMed.
- C. Claveria, G. Giovinazzo, R. Sierra and M. Torres, Nature, 2013, 500, 39–44 CrossRef CAS PubMed.
- M. Sancho, A. Di-Gregorio, N. George, S. Pozzi, J. M. Sánchez, B. Pernaute and T. A. Rodríguez, Dev. Cell, 2013, 26, 19–30 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ib00203j |
| This journal is © The Royal Society of Chemistry 2017 |