Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Virus inactivation in stored human urine, sludge and animal manure under typical conditions of storage or mesophilic anaerobic digestion†
Loïc
Decrey
 and
Tamar
Kohn
*
and
Tamar
Kohn
*
Laboratory of Environmental Chemistry, School of Architecture, Civil and Environmental Engineering (ENAC), École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland. E-mail: tamar.kohn@epfl.ch; Fax: +41 (0)21 693 8070; Tel: +41 (0)21 693 0891
First published on 17th March 2017
Abstract
Viruses represent major disease transmitting agents carried by human excreta and animal manure. Understanding virus inactivation is therefore essential in preventing microbial spread due to inadequate treatment of these materials. Here, we investigated the inactivation kinetics of the single-stranded (ss) RNA phage MS2, DNA phages T4 and ΦX174, and the double-stranded DNA human adenovirus in stored human urine, sludge, and animal manure, at temperatures and pH values typical of storage under naturally occurring conditions or mesophilic anaerobic digestion (<40 °C). The ssRNA phage MS2 was most readily inactivated in all samples compared to the other viruses tested. This is consistent with previous findings in well-controlled buffer solutions of similar composition, where inactivation was found to be governed by bases (NH3, carbonate, hydroxide) that catalyze the transesterification and cleavage of the ssRNA. Correspondingly, MS2 inactivation kinetics in real matrices could be adequately modelled by only taking into account the effects of temperature, pH, carbonate and ammonia on the integrity of ssRNA. DNA viruses were more persistent compared to MS2; however, inactivation in selected sludge and manure samples proceeded at faster rates compared to well-controlled buffer solutions of similar composition. This indicates a contribution of microbial or enzymatic activity to inactivation of DNA viruses. Overall, this study identifies the most important factors contributing to inactivation of viruses in human excreta and manure, and highlights the differences in inactivation kinetics and mechanisms between ssRNA and DNA viruses.
Water impactViruses are among the most resistant pathogens in human excreta and manure (HEAM). A better understanding of the kinetics and mechanisms driving virus inactivation in HEAM is instrumental to design and optimize HEAM treatment and reduce pathogen dissemination into the environment. In particular, it enables to evaluate virus stability, identify resistant viruses, and assess the public health risks posed by virus transmitted by HEAM. |
1 Introduction
Prior to reuse or discharge, human excreta and animal manure (HEAM) need to be correctly managed to avoid the transmission of pollutants (macro- and micro-pollutants) and pathogens to the environment.1–4 If adequately treated, however, environmental pollution and public health issues resulting from the disposal of HEAM can be minimized.5–8 Organized excreta containment, collection and treatment is one of the pillars in breaking the transmission cycle of fecal-orally transmitted pathogens.9 Currently only a small percentage of human excreta collected is safely managed, in particular in cities in lower-income countries, though on-site storage or treatment offer an opportunity to reduce the pathogen load before disposal.10,11 On-site treatment should in particular focus on viruses and helminths, since they were shown to be the most persistent pathogens in excreta, in particular during storage at ambient temperature.12–14 Their inactivation kinetics, and the factors that promote inactivation in HEAM, however, remain poorly understood.In previous work, we characterized virus inactivation in well-controlled, homogenous laboratory solutions under conditions of pH, temperature and chemical composition typically encountered during storage or mesophilic anaerobic digestion of HEAM.15,16 Single-stranded (ss)RNA viruses, such as enteroviruses or MS2 coliphage, were shown to be rapidly inactivated. Inactivation resulted from base-catalyzed transesterification of the ssRNA, which causes the genome to cleave.15 The efficiency of a base to induce inactivation depends on the pKa of its conjugated acid and its concentration. Under the solution conditions considered in Decrey et al.,15 the most important bases promoting inactivation were hydroxide and ammonia, though other bases such as (bi-)carbonate also contributed. Based on this mechanistic insight, a model to estimate the inactivation rate constant under typical urine, sludge and manure storage conditions was established for MS2 coliphage, which was shown to be a conservative surrogate of other ssRNA viruses in such conditions.16 Using the solution composition as the input, this model was able to accurately estimate the MS2 inactivation rate constant at 35 °C and over a pH range of 7.5–9.5. In contrast to ssRNA viruses, double-stranded (ds) RNA and both ssDNA and dsDNA viruses, which are not amenable to genome transesterification, exhibited low inactivation rates under the same conditions. More extreme conditions of pH or temperature, such as those encountered in thermophilic digestion or alkaline treatment, were required to achieve appreciable inactivation rates.16
Compared to laboratory solutions, real HEAM matrices exhibit a higher level of complexity: they contain particles, additional chemical constituents (e.g., metals or organic acids), as well as live communities of microorganisms that may contribute to virus inactivation. The current study extends our understanding of virus inactivation from laboratory solutions to real matrices associated with on-site HEAM storage (sludge, stored human urine, animal manure). Specifically, we aim to determine if the principles of virus inactivation established in well-controlled solution also apply to HEAM matrices. Our objectives were i) to validate the previous approach to estimate MS2 inactivation for the more complex conditions encountered in HEAM; and ii) to establish if the inactivation trends observed in laboratory solutions for viruses with different genome types correspond to those in real matrices. To attain these objectives, the pH, temperature and ion composition of different (diluted) stored urine, sludge and manure solutions was determined, and MS2 inactivation in these solutions was monitored and compared to the predicted inactivation. Furthermore, the inactivation kinetics of the base-sensitive ssRNA phage MS2 as well as the more resistant dsDNA phage T4, ssDNA phage ΦX174 and the dsDNA human adenovirus (HAdV) were determined in stored urine, sludge and manure, and were compared to results from controlled laboratory studies. The phages were chosen to span a range of susceptibilities to solution conditions typical of HEAM, and to represent different genome types.16 HAdV was chosen as a representative of a base-resistant human virus. Finally, experiments were conducted to determine the effect of HEAM-associated environmental parameters, such as microbial activity or the presence of metals, on inactivation.
2 Materials and methods
2.1 Virus and cells
HAdV type 2 was provided by Rosina Gironès (Laboratory of Virus Contaminants of Water and Food, University of Barcelona). HAdV was propagated on A549 human lung carcinoma epithelial cells, provided by the University Hospital of Lausanne. A549 cells were cultivated in high-glucose, pyruvate Dulbecco's modified Eagle's medium (DMEM; Invitrogen). The media was supplemented with penicillin (20 U mL−1), streptomycin (20 μg mL−1) (Invitrogen), and 2 or 10% fetal bovine serum (FBS; Invitrogen) and cells were incubated at 37 °C in 5% CO2 and 95% humidity. Viruses were propagated by spiking 10 μl of HAdV (1010–1011 most probable number of cytopathogenic units (MPNCU) mL−1) into 160 cm2 flasks (TPP Techno Plastic Products, Trasadingen, Switzerland) containing 95% confluent cells, and were purified as described in Bosshard et al.17 From each flask, 1 ml of samples containing 1010–1011 MPNCU mL−1 of HAdV was collected and stored at 4 °C as virus stocks for the experiments. New stocks were produced before each set of experiments. Virus titers were determined as MPNCU mL−1 from 5 × 100 μl of samples on 96-wells plates (Greiner Bio-One, Frickenhausen, Germany) as described in Bosshard et al.17 Briefly, the DMEM containing 10% FBS on a 95% confluent cell monolayer was replaced by 100 μl of virus solution and augmented with 200 μl of DMEM containing 2% FBS. Cytopathogenic units could be discerned after an incubation time of 14 days. The detection limit was 102–103 MPNCU mL−1.2.2 Phages and bacteria
Coliphages MS2 (DSMZ 13767) and ΦX174 (DSMZ 4497) and their host Escherichia coli (DSMZ 5695 and DSMZ 13127, respectively) were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). Coliphage T4 and its E. coli host (B1) were provided by Petr Leiman (Laboratory of Structural Biology and Biophysics, EPFL). Media used to grow E. coli B1 were free of any antibiotics. All phages were propagated and purified as described previously.18 Stock solutions were stored in the fridge, and the same stocks were used for all the experiments. Infectivity was assessed using the double agar layer method as described elsewhere.19 The detection limit for all phages was 300 PFU mL−1.2.3 Stored urine, sludge and manure
Undiluted stored human urine was obtained from the Swiss Federal Institute of Aquatic Sciences (Eawag) in Dübendorf, Switzerland (CH), where urine is collected from waterless urinals and men's NoMix toilets and women's NoMix toilets respectively. Three batches of male stored urine taken in 3 different years (2012, 2013, 2014) and one batch of female stored urine (2014) were obtained and used for experimentation. An additional batch of stored urine was collected from urine diverting dry toilets in Durban, South Africa (SA) in 2014. Diluted stored urine was obtained by mixing undiluted stored urine (male, CH, 2012) and MilliQ water at urine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water ratios of 1
water ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9. In total, the various stored urine batches and dilutions yielded 15 urine samples (U1–U15; Table 1). For experiments with sludge, three batches of sludge were used: two batches (S1, S3) consisted of a synthetic fecal sludge. This sludge was made of walnuts, straw flour, kaolinite, sodium phosphate, ammonium chloride and potassium nitrate, and was digested by an inoculum of bacteria obtained from a thermophilic anaerobic digester, as described by Gallandat et al.20 An additional batch (S2) consisted of fecal sludge collected from septic tanks in Switzerland. Two batches of pig (M1, M2) and cow manure (M3, M4) were collected in the Swiss country-side.
9. In total, the various stored urine batches and dilutions yielded 15 urine samples (U1–U15; Table 1). For experiments with sludge, three batches of sludge were used: two batches (S1, S3) consisted of a synthetic fecal sludge. This sludge was made of walnuts, straw flour, kaolinite, sodium phosphate, ammonium chloride and potassium nitrate, and was digested by an inoculum of bacteria obtained from a thermophilic anaerobic digester, as described by Gallandat et al.20 An additional batch (S2) consisted of fecal sludge collected from septic tanks in Switzerland. Two batches of pig (M1, M2) and cow manure (M3, M4) were collected in the Swiss country-side.
| ID | Sample description | T [°C] | pH | EC [mS cm−1] | Dilutiona | Total solids% | {NH3} [mmol L−1] | Virus tested | ||
|---|---|---|---|---|---|---|---|---|---|---|
a Urine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water ratio. water ratio.
|
||||||||||
| U1 | Urine | CH | Male (2012) | 20 | 8.47 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
15.8 | MS2 | ||
| U2 | 8.49 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
9.6 | MS2 | ||||||
| U3 | 8.45 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 9 |
1.9 | MS2 | ||||||
| U4 | 35 | 8.15 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
19.1 | MS2, HAdV | |||||
| U5 | 8.19 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
24.6 | MS2 | ||||||
| U6 | 8.19 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
24.4 | MS2 | ||||||
| U7 | 8.22 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
13.6 | MS2 | ||||||
| U8 | 8.15 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
12.2 | MS2 | ||||||
| U9 | 8.19 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
8.4 | MS2 | ||||||
| U10 | 8.13 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 9 |
2.5 | MS2 | ||||||
| U11 | 8.13 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 9 |
2.7 | MS2 | ||||||
| U12 | CH | Male (2013) | 35 | 8.72 | 33.6 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
81.0 | MS2, HAdV, ϕX174, T4 | ||
| U13 | CH | Male (2014) | 35 | 8.79 | 33.0 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
106.0 | MS2 | ||
| U14 | CH | Female | 35 | 8.49 | 16.0 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
28.2 | MS2 | ||
| U15 | SA | Mix | 35 | 8.48 | 33.6 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
71.1 | MS2 | ||
| S1 | Sludge | CH | Synthetic | 35 | 8.24 | 3.3 | 27.7 | MS2 | ||
| S2 | CH | Septic tank | 35 | 7.76 | 6.7 | 1.3 | MS2, HAdV, ϕX174, T4 | |||
| S3 | CH | Synthetic | 35 | 7.39 | 2.5 | 0.2 | MS2 | |||
| M1 | Manure | CH | Pig | 35 | 7.79 | 2.3 | 8.4 | MS2 | ||
| M2 | CH | 35 | 8.08 | 2.0 | 8.5 | MS2 | ||||
| M3 | CH | Cow | 35 | 8.05 | 4.6 | 35.7 | MS2 | |||
| M4 | CH | 35 | 8.23 | 1.7 | 14.2 | MS2, HAdV, ϕX174, T4 | ||||
Upon arrival in the lab, the stored urine, sludge and manure were stored at 4 °C from weeks to months until use. Prior to characterization, all urine samples were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 minutes. For sludge and manure characterization, 5–10 ml of MilliQ water were added, and the sample was shaken for 10–15 minutes and centrifuged at 4000 × g for 15 minutes. Stored urine, sludge and manure physical and chemical characteristics (summarized in Tables 1 and S1†) were determined as follows: pH was measured at experimental temperature (780 pH Meter with primatrode with NTC no. 6.0228.010, Metrohm, Herisau, Switzerland); the total ammonium nitrogen (TAN; NH4+/NH3) concentration was determined by ion chromatography (ICS-3000, IonPacCS16 column) with electrical conductivity detection (Dionex, Switzerland); phosphate, sulfate and chlorine concentrations were measured by ion chromatography (ICS-3000, IonPac AS11-HC column); magnesium, calcium, potassium and sodium by inductively coupled plasma optical emission spectrometry (ICP-OES, Ciros, Spectro Analytical Instruments, Kleve, Germany); soluble chemical oxygen demand (SCOD) with cuvette tests (Hach-Lange, Berlin, Germany) and total inorganic carbon (TIC) by means of a TOC-TN Analyser (IL 550, Hach-Lange, Berlin, Germany). For TAN measurements, samples were diluted in 0.01 M HCl and for TIC measurements in 0.01 M NaOH to avoid loss of NH3 and CO2, respectively. For all other ions, samples were diluted in MilliQ water. Additionally, total solids (TS) were determined in sludge and manure according to standard methods.21 Given the low TS (<7%), one Liter of sludge or manure was considered as one kg. This characterization was generally performed in quadruplicate for stored urine and in triplicate for sludge and manure (see below for details). For male stored urine (2012) (U1–U11, see Table 1), the ion content was determined in the undiluted sample (U1) only, and was extrapolated for the rest of the samples composed of this urine (U2–U11). Only pH and TAN were determined under each experimental condition (U1–U11; see Table S1†).
000 × g for 10 minutes. For sludge and manure characterization, 5–10 ml of MilliQ water were added, and the sample was shaken for 10–15 minutes and centrifuged at 4000 × g for 15 minutes. Stored urine, sludge and manure physical and chemical characteristics (summarized in Tables 1 and S1†) were determined as follows: pH was measured at experimental temperature (780 pH Meter with primatrode with NTC no. 6.0228.010, Metrohm, Herisau, Switzerland); the total ammonium nitrogen (TAN; NH4+/NH3) concentration was determined by ion chromatography (ICS-3000, IonPacCS16 column) with electrical conductivity detection (Dionex, Switzerland); phosphate, sulfate and chlorine concentrations were measured by ion chromatography (ICS-3000, IonPac AS11-HC column); magnesium, calcium, potassium and sodium by inductively coupled plasma optical emission spectrometry (ICP-OES, Ciros, Spectro Analytical Instruments, Kleve, Germany); soluble chemical oxygen demand (SCOD) with cuvette tests (Hach-Lange, Berlin, Germany) and total inorganic carbon (TIC) by means of a TOC-TN Analyser (IL 550, Hach-Lange, Berlin, Germany). For TAN measurements, samples were diluted in 0.01 M HCl and for TIC measurements in 0.01 M NaOH to avoid loss of NH3 and CO2, respectively. For all other ions, samples were diluted in MilliQ water. Additionally, total solids (TS) were determined in sludge and manure according to standard methods.21 Given the low TS (<7%), one Liter of sludge or manure was considered as one kg. This characterization was generally performed in quadruplicate for stored urine and in triplicate for sludge and manure (see below for details). For male stored urine (2012) (U1–U11, see Table 1), the ion content was determined in the undiluted sample (U1) only, and was extrapolated for the rest of the samples composed of this urine (U2–U11). Only pH and TAN were determined under each experimental condition (U1–U11; see Table S1†).
Ion activities in each matrix were determined as a function of experimental temperature, pH and solution composition using PHREEQC (version 2.18.00)22 and a database using the Pitzer approach for calculating ion activities.23 The concentrations expressed in mol kg−1 in PHREEQC were considered equivalent to mol L−1. SCOD was transformed to acetate equivalents, because acetate was shown to represent 47% of the SCOD in stored urine.24 The transformation was determined according to the following stoichiometric relation:
| C2H3O2 + 1.75O2 → 2CO2 + 1.5H2O |
Manure was additionally tested for the presence of somatic and F-RNA coliphages. Only somatic coliphages were detected in pig manure at >103 PFU g−1.
2.4 Experimental setup
ΦX174 and T4 were tested in one batch of stored urine, one batch of sludge and one batch of manure; HAdV in two batches of stored urine, one batch in sludge and one batch of manure; and MS2 in all the batches described in Table 1. An overview over the experimental setup is provided in the supplementary material (Scheme S1†). Prior to each inactivation experiment, the matrices were stored at the targeted experimental temperature during one (urine) or five to ten (manure, sludge) days to establish a stable temperature and microbial activity as well as to avoid drastic changes in solution composition during the inactivation phase.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
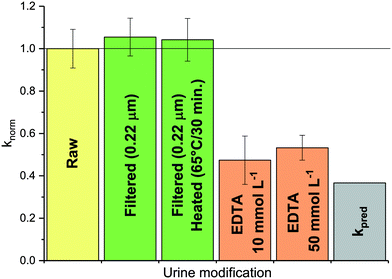
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 diluted stored urine (U10) with MS2. To test the role of microbial activity, MS2 was exposed to samples filtered at 0.22 μm to remove live microorganisms. To inhibit enzyme activity, samples were first filtered at 0.22 μm to remove microorganisms and the remaining enzymes were heat-inactivated at 65 °C for 30 minutes. To minimize the influence of cations on inactivation, the complexing agent ethylenediaminetetraacetic acid (EDTA; Acros) was added to stored urine to obtain a final concentration of 10 and 50 mmol L−1. The role of microbial and enzyme activity in manure (M4) was tested by exposing MS2 to heat-sterilized manure (65 °C for 1 h).
9 diluted stored urine (U10) with MS2. To test the role of microbial activity, MS2 was exposed to samples filtered at 0.22 μm to remove live microorganisms. To inhibit enzyme activity, samples were first filtered at 0.22 μm to remove microorganisms and the remaining enzymes were heat-inactivated at 65 °C for 30 minutes. To minimize the influence of cations on inactivation, the complexing agent ethylenediaminetetraacetic acid (EDTA; Acros) was added to stored urine to obtain a final concentration of 10 and 50 mmol L−1. The role of microbial and enzyme activity in manure (M4) was tested by exposing MS2 to heat-sterilized manure (65 °C for 1 h).
2.5 Data analysis
Inactivation kinetics were determined by least-square fit of the data to a first-order model:| C(t) = C0e−kobst | (1) |
| C(t) = C0,faste−kobs,fastt + C0,slowe−kobs,slowt | (2) |
2.6 Model to predict MS2 inactivation
MS2 inactivation rate constants were determined in all stored urine, sludge, and manure samples, over a wide range of solution conditions and experimental temperatures. This data set was used to challenge the predictive model of MS2 inactivation discussed in Decrey et al.,15 which was established for well-controlled solutions. The model demonstrated that MS2 infectivity loss can be related to the degradation (transesterification) of the viral genome by different bases. Specifically, the inactivation rate constants of MS2 in laboratory solutions could be predicted as follows: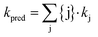 | (3) |
log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kj = β·pKa + D kj = β·pKa + D | (4) |
In contrast to β, pKa, kj and D are dependent on temperature. For any given temperature, D(T) was estimated based on available experimental data15 for j = NH3 as follows:
D(T) = log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kNH3(T) − 0.41·pKaNH3(T) kNH3(T) − 0.41·pKaNH3(T) | (5) |
All pKa(T) values were calculated according to eqn (S1) (see ESI,† determination of pKa), and kNH3(T) was determined by the Arrhenius relationship reported in Decrey et al.:15
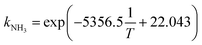 | (6) |
Finally, it is apparent that only those bases contribute to inactivation of MS2 that are present at a significant activity (eqn (3)), and that have a conjugated acid with a relatively high pKa (eqn (4)). Given the composition of the matrices used herein (Tables 1 and S1†), the only bases (j) considered were therefore OH−, NH3, CO32−, HCO3−, PO43− and HPO42−. Note that a user-friendly interface for the predictive model is available for free online (https://lodecrey.shinyapps.io/MS2inactivation/).28
3 Results and discussion
3.1 Inactivation kinetics
Example virus inactivation curves obtained in one batch of human urine, sludge and manure are shown in Fig. 1 (see ESI,† Fig. S2 and S3 for the complete set of inactivation curves). Viruses in all matrices studied exhibited a loss of infectivity with time, and inactivation followed first-order kinetics, except in the case of ΦX174 and T4 in manure (Fig. 1 and S3†). First-order inactivation rate constants and coefficients of determination are listed in Table S2 (ESI†). First-order inactivation kinetics were also reported by Höglund et al.29 and Vinnerås et al.30 for Salmonella phage 28B and rotavirus in stored urine. In contrast to the findings herein, however, Vinnerås et al.30 observed a two phase inactivation behavior for MS2 and ΦX174, with an initial fast inactivation followed by a slow first-order reduction.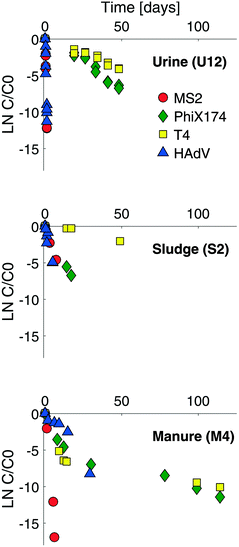 | ||
| Fig. 1 Inactivation curves for MS2 (red circles), ΦX174 (green diamonds), T4 (yellow squares) and HAdV (blue triangles) in stored urine (U12), sludge (S2) and manure (M4). The complete set of inactivation curves obtained in this study are shown in the ESI† (Fig. S2 and S3). | ||
The first-order inactivation kinetics observed herein indicate that solutions conditions were stable over the course of the inactivation experiments. Correspondingly, a characterization of the composition of stored urine over time showed that both pH and ion concentrations were stable when stored at 20 °C or 35 °C. For sludge and manure, however, changes in solution conditions were observed (ESI,† Table S3). For example, in the case of sludge (S2), the concentration of TAN doubled over the course of 14 days, and the concentrations of Ca2+, Mg2+, SO42− decreased more than two-fold whereas the pH and other ions were stable; in the case of manure (M1), the pH increased from 7.58 to 8.0 and TIC doubled whereas other ions were stable. The effect of the changing solution conditions on inactivation, however, remained small: predicted inactivation rate constants of MS2 (see section 2.6) differed by less than a factor of 1.4 for most matrices over the duration of an experiment (see ESI,† Table S3). Consequently, for any downstream analysis, the average of the initial and final composition was used for S2, S3, M1, M2 and M3 (Tables 1 and S1†); for S1 and M4, only the composition at initial time was assessed.
3.2 Validation of MS2 inactivation prediction in stored urine, sludge and manure
The comparison between predicted and observed inactivation rate constants is shown in Fig. 2. Over half of the predictions fell within 80–120% of kobs, and three-fourths of the predictions were within 60–140% of kobs. This ability of the model to accurately predict MS2 inactivation supports the notion that the dominant underlying mechanism of inactivation is base-catalyzed transesterification of the ssRNA genome.15,16 This appears to be the case in both synthetic solutions and in real matrices such as stored urine, and sludge and manure. Consistent with our results, Gao et al.31 recently reported that during mesophilic anaerobic treatment of Coxsackievirus, the viral genome was the main inactivating target.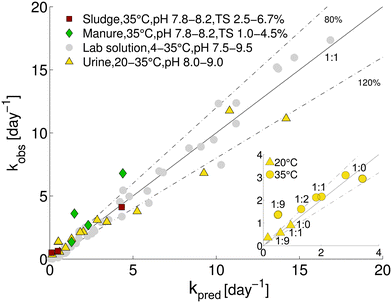 | ||
Fig. 2 Comparison of measured and predicted MS2 inactivation rate constants for stored urine, sludge and manure. For comparison, data from lab solutions (Decrey et al.;15 grey circles) are also shown. Values of kpred were determined from eqn (3) and (4). The solid line represents a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 relation between measurement and prediction (kpred/kobs = 1). Dashed lines indicate 80% and 120% of kpred/kobs (i.e., kpred/kobs = 0.8 and 1.2 respectively). The inset shows kpredversus kobs for different dilutions of urine. The urine 1 relation between measurement and prediction (kpred/kobs = 1). Dashed lines indicate 80% and 120% of kpred/kobs (i.e., kpred/kobs = 0.8 and 1.2 respectively). The inset shows kpredversus kobs for different dilutions of urine. The urine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water ratio for each data point is indicated. water ratio for each data point is indicated. | ||
A model sensitivity analysis was conducted to assess the influence of pH and temperature, and the inclusion of measured ion concentrations in the model, on the accuracy of the prediction (Fig. S4†). Specifically, we re-assessed the model prediction for all 22 samples at either pH values of 0.1 units surrounding the measured value or at temperatures of 1 °C surrounding the measured temperature. In addition, predictions were carried out that included only TAN, or TIC and TAN, but none of the other ions in solution. This analysis revealed similar sensitivity to shifts resulting from changes in pH by 0.1 units and from changes in temperature by 1 °C (Fig. S4 and Table S2†). A relatively minor error in the measurement may thus lead to an inaccurate kpred. Interestingly, no relevant differences were observed if all ions were taken into account in the prediction, as compared to only the TIC and TAN (Fig. S4 and Table S2†). Thus, measurements of temperature, pH, TIC and TAN are sufficient to obtain a reasonably accurate prediction of MS2 inactivation rate constants. Removal of TIC from the prediction led to a lower accuracy (Fig. S4 and Table S2†). This highlights the importance of carbonate in MS2 inactivation in HEAM. Carbonate and bicarbonate contributed between 15 and 40% to the total kobs in stored urine, sludge and manure (Fig. 3). The contribution of carbonate species was even higher (>50%) in sludge S2, which had a pH < 8.0 and equivalent amounts of TIC and TAN.
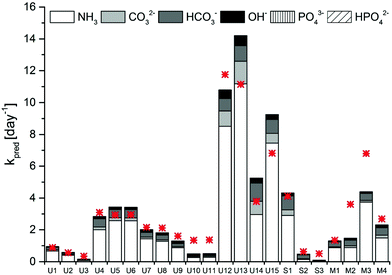 | ||
| Fig. 3 Contribution of the main bases present in stored urine, sludge and manure to the kpred of MS2. The measured kobs is depicted by the red asterisk. | ||
3.3 Influence of HEAM-associated parameters on MS2 inactivation
Despite the quite high accuracy of the predicted inactivation rate constants for MS2 in all matrices considered, some small deviations from the measured kobs were observed: for some stored urine samples, the model yielded a slight overestimate of the rate constant compared to the observation, while the model tended to underestimate inactivation in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 diluted urine, sludge and manure (Fig. 2). These deviations may be in part linked to imprecise measurements of the ion concentrations and the PHREEQC estimation of the ion activities. Alternatively, underestimations could indicate that additional inactivating processes occur that are not accounted for in the model. In this context, we assessed the role of two HEAM-associated parameters in inactivation, namely biological (microbial or enzymatic) activity and metal ions, and we discussed the influence of additional bases not considered by our model.
9 diluted urine, sludge and manure (Fig. 2). These deviations may be in part linked to imprecise measurements of the ion concentrations and the PHREEQC estimation of the ion activities. Alternatively, underestimations could indicate that additional inactivating processes occur that are not accounted for in the model. In this context, we assessed the role of two HEAM-associated parameters in inactivation, namely biological (microbial or enzymatic) activity and metal ions, and we discussed the influence of additional bases not considered by our model.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 diluted urine. Heat-sterilization of manure (M4) even led to an increase in inactivation, resulting in a kobs of 4.6 (±0.7) day−1 (data not shown) compared to 2.7 (±0.8) day−1 in untreated manure. Thus, MS2 seemed insensitive to biological activity. Similar results were observed in soil saturated with secondary effluent by Nasser et al.,33 and in nitrifying urine by Bischel et al.34 In contrast, Mondal et al.35 reported that MS2 could be inactivated by both commercial and sludge-derived protease solutions. At sufficiently high protease concentrations, biological activity may thus compete with chemically-mediated MS2 inactivation in HEAM, though this situation was not encountered herein.
9 diluted urine. Heat-sterilization of manure (M4) even led to an increase in inactivation, resulting in a kobs of 4.6 (±0.7) day−1 (data not shown) compared to 2.7 (±0.8) day−1 in untreated manure. Thus, MS2 seemed insensitive to biological activity. Similar results were observed in soil saturated with secondary effluent by Nasser et al.,33 and in nitrifying urine by Bischel et al.34 In contrast, Mondal et al.35 reported that MS2 could be inactivated by both commercial and sludge-derived protease solutions. At sufficiently high protease concentrations, biological activity may thus compete with chemically-mediated MS2 inactivation in HEAM, though this situation was not encountered herein.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 diluted urine was amended with EDTA, a metal complexing agent. This was found to decrease kobs to a value close to kpred (see Fig. 4, orange bars). Thus, the presence of EDTA suppressed the action of the urine constituents responsible for the higher kobs. Interestingly, if ammonia (as NH4Cl) was added to (EDTA-free) 1
9 diluted urine was amended with EDTA, a metal complexing agent. This was found to decrease kobs to a value close to kpred (see Fig. 4, orange bars). Thus, the presence of EDTA suppressed the action of the urine constituents responsible for the higher kobs. Interestingly, if ammonia (as NH4Cl) was added to (EDTA-free) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 diluted urine, kobs increased as expected based on the increase in {NH3} (see ESI,† Fig. S5). The effect of ammonia was thus additive to that of the metal ions, resulting in an overall higher inactivation than in other urine matrices with the same NH3 activity. We currently cannot explain, however, why urine dilution, followed by ammonia addition results in higher inactivation than an equivalent ammonia activity in undiluted urine.
9 diluted urine, kobs increased as expected based on the increase in {NH3} (see ESI,† Fig. S5). The effect of ammonia was thus additive to that of the metal ions, resulting in an overall higher inactivation than in other urine matrices with the same NH3 activity. We currently cannot explain, however, why urine dilution, followed by ammonia addition results in higher inactivation than an equivalent ammonia activity in undiluted urine.
The role of metal ions was not explicitly studied in sludge and manure. Nevertheless, literature reports indicate that particularly manure contains metal cations up to the mmol L−1 concentration range.38,39 It is thus reasonable to conclude that metal ions in these matrices also contribute to MS2 inactivation.
3.4 Influence of genome type and microbial activity on virus inactivation kinetics in urine, sludge and manure
As discussed above, considerable heterogeneity exists among the inactivation kinetics of viruses with different genome types. Specifically, in well-controlled solutions, DNA viruses were shown to be more persistent than ssRNA viruses. To validate this trend in real matrices, we compared the inactivation of ssRNA MS2 phage to that of three DNA viruses, specifically phage ΦX174 (ssDNA), phage T4 (dsDNA) and HAdV (dsDNA). Inactivation kinetics were assessed in a subset of stored urine, sludge and manure samples at 35 °C, and were compared to those obtained in laboratory solution with and without NH3.15,16As expected, the ssRNA virus MS2 was inactivated more readily than the DNA viruses in most matrices, (Fig. 1 and ESI† Table S2). This confirmed the higher sensitivity of ssRNA viruses to NH3 and mildly alkaline pH. In undiluted stored urine at 35 °C (U12), a four log10 (99.99%) inactivation was achieved within one day for MS2, whereas it took more than 100 days to reach the same level of inactivation for T4. In sludge and manure the differences among ssRNA and DNA viruses were smaller, with a four log10 loss being achieved within 15 and 207 days in sludge (S2) and 3.5 and 40 days in manure (M4) at 35 °C for MS2 and T4 respectively. A possible reason for the narrower range of inactivation kinetics among ssRNA and DNA viruses in sludge and manure is the contribution of microbial or enzymatic activity to inactivation. While microbiological inactivation is not relevant for MS2 compared to its rapid chemical inactivation (see section 3.3.1), it may accelerate the inactivation of DNA viruses beyond the slow chemical inactivation kinetics in HEAM, in particular in matrices with high microbiological activity.
Consistent with our previous findings, HAdV was the most readily inactivated among the DNA viruses, and it was more resistant than MS2 in stored urine (U4) at pH 8.2 and approximately 20 mmol L−1 NH3 and in manure (M4) (Fig. 1 and ESI† Table S2). However, in stored urine with a higher pH and NH3 content (U12) and in sludge (S2), the inactivation of HAdV increased, resulting in similar to greater inactivation rate constants compared to MS2 (Fig. 1 and ESI† Table S2). A comparison with controlled solutions of similar physical–chemical properties revealed that pH could account for the fast inactivation observed in U12 (Fig. 5). In contrast, inactivation in S2 could not be explained by the known physical–chemical parameters, indicating a potential contribution of microbiological processes.
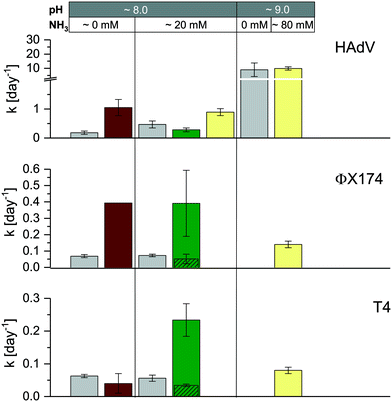 | ||
| Fig. 5 Comparison of virus inactivation in laboratory solution (grey bars), stored urine (U4 [pH 8.0, 20 mmol L−1 NH3], U12 [pH 9.0, 80 mmol L−1 NH3]), sludge (S2) and manure (M4) at 35 °C. Kinetics in laboratory solution (from left ot right: phosphate carbonate buffer [pH 8.0, 50 mmol L−1 carbonate, 60 mmol L−1 phosphate] and ammonium carbonate [pH 8.0, 50 mmol L−1 carbonate, 20 mmol L−1 NH3]) were derived from Decrey et al.,16 except for HAdV at pH 9.0 (phosphate buffer [pH 9.0, 50 mmol L−1 carbonate, 40 mmol L−1 phosphate]). The exact rate constants for U4, U12, S2 and M4 are listed in Table S2.† For ΦX174 and T4 in M4, columns with diagonal patterns depict kobs_slow. Error bars depict 95% confidence interval associated with kobs. | ||
Faster than expected inactivation was also observed for ΦX174 and T4 in manure. Specifically, these two phages exhibited biphasic inactivation behavior in manure with an initial fast inactivation followed by a secondary, slower phase (eqn (2); ESI,† Fig. S6). For both phages, kobs_fast was 4–5 times larger than kobs_slow (Fig. 5, green bars) and the kobs_slow generally corresponded to the kobs in a controlled solution with the same physical–chemical properties (Fig. 5, grey bars). The first, rapid decrease in infective virus thus is caused by parameters associated with manure that are not present in laboratory solutions, whereas the second, slower phase may be attributed to the effect of solution conditions only. We propose that the biphasic inactivation kinetics is associated with virus adsorption onto the manure. Specifically, while irreversible adsorption can be ruled out as a removal mechanism (given the high recovery of infective virus in our experimental protocol; see Materials and methods), adsorption to solids may lead to the protection of viruses from microbial and enzymatic activity. The fast, initial inactivation could thus be dominated by microbiological inactivation of suspended viruses, whereas the slower phase results from physical–chemical inactivation of viruses adsorbed to solids. This hypothesis is consistent with the findings by others that demonstrate faster inactivation in the liquid- than in the solid-associated virus fraction during anaerobic digestion42 and in wastewater.43
Microbiological contributions to virus inactivation have previously been reported for HAdV, Hepatitis A virus, norovirus and enteroviruses in a range of matrix types.33,44–51 In most cases, inactivation could only be partly attributed to biological processes, whereas physical–chemical matrix components also played a role. Unfortunately, the matrix composition was usually poorly characterized, and it is therefore difficult to parameterize its influence in inactivation. It appears, however, that the most important matrix components and their resulting virucidal activities vary widely depending on both the matrix and virus type. For example, it was shown that septic tank effluent digestion was more efficient at inactivating viruses when mixed with dairy cattle or swine manure slurry.50,51 The same authors observed that among 31 bacterial strains isolated from animal manure, only 10 proved to be efficient at inactivating virus. Here, we propose inactivating microbial or enzymatic activity in sludge but not in manure for HAdV, in manure but not in sludge for T4, and in both for ΦX174 (Fig. 5). The distinct matrix and virus parameters that result in inactivation, however, remain to be determined.
The complex nature of virus inactivation in real matrices is further reflected in the literature, where a large variation in virus inactivation kinetics in excreta is reported. Consequently, results contradicting our data can be found: for example, others have shown ΦX174 to be inactivated as fast as MS2 in stored urine12 and stored fecal sludge,14 and HAdV to be inactivated faster than MS2 in fecal sludge.52 Similarly, ssRNA phage f2 and coxsackievirus exhibited comparable inactivation to rotavirus (dsRNA) during mesophilic anaerobic digestion of sludge with, albeit at low pH (~7.3).53 On the other hand, other studies provided observations consistent with our data. For example, F-RNA specific coliphages were shown to be more sensitive than somatic (DNA) coliphage and dsDNA Salmonella phage 28B during mesophilic digestion of raw sewage sludge54 and the organic fraction of municipal solid waste.55 Furthermore, somatic coliphages were found to exhibit low sensitivity to the addition of urea, calcium carbonate and sodium percarbonate used to sanitize composted sewage sludge, which is consistent with our finding that ΦX174 and T4 are not affected by the main chemical components of stored urine and sludge.56 Overall, the influence of solids content on the inactivation of different virus types remains poorly understood and needs to be further elucidated. In particular, more research is needed on matrices with lower liquid fractions, such as fecal sludge (solids content between 20–95% (ref. 14)).
4 Conclusions
Among enteric viruses, the vast majority have an ssRNA genome. A good understanding of the factors that promote the inactivation of ssRNA viruses during waste treatment is therefore particularly important. The data presented herein demonstrate that for ssRNA phage MS2, inactivation kinetics and mechanisms established in laboratory solutions are transferable to real matrices. We furthermore established that carbonate, ammonia, pH and temperature are important drivers of inactivation that need to be maximized to enhance treatment efficiency. Future studies should be extended to additional ssRNA viruses to more conclusively establish the optimal treatment conditions.While viruses with other genome types are not as common, some enteric viruses with public health relevance have DNA and dsRNA genomes (e.g., adenovirus, polyomavirus, rotavirus). For DNA and dsRNA viruses, kinetic insights developed in controlled solutions may not be as readily transferable to real matrices, due to the potential contribution of microbiologically driven inactivation processes in HEAM. The kinetics based on physical–chemical parameters established in laboratory solutions can therefore only be considered as worst case scenarios for the inactivation of DNA and dsRNA viruses in HEAM.
Discrepancies exist between our results and literature reports, as well as among different literature reports. To reconcile those, a better understanding of the mechanisms involved in virus inactivation in HEAM is needed. While we believe that we have a good handle on main mechanism involved in the inactivation of ssRNA viruses, those responsible for DNA and dsRNA virus inactivation remain to be determined. In this context, we again emphasize the potential contribution of microorganisms on virus inactivation. Determining the virus properties that render it susceptible to microbiological inactivation will be an important next step in understanding inactivation in HEAM.
Finally, this study helps identify appropriate proxies to monitor virus inactivation in HEAM. Specifically, our data confirms that somatic coliphage such T4 or ΦX174 are conservative indicators of resistant ssDNA, dsDNA and dsRNA viruses. However, they are too stable to serve as indicators for the inactivation of the more labile HAdV. The utility of these indicators as suitable proxies for further DNA or dsRNA viruses should thus be confirmed.
Acknowledgements
This work was supported by the Swiss National Science Foundation [grant no. 200021_146829/1] and by EPFL discretionary funding. This study was conducted within the framework of the VUNA (Valorization of Urine Nutrients in Africa) project.References
- A. Albihn, K. Nyberg, J. Ottoson and B. Vinneras, Sustain. Agric. Ecosyst. Heal. Sustain. Agric. B. 1, 2012 Search PubMed.
- WHO, Guidelines for the safe use of wastewater, excreta and greywater - v. 4. Excreta and greywater use in agriculture, World Health Organisation, 2006 Search PubMed.
- E. Emmoth, Virus inactivation – evaluation of processes used in biowaste management, PhD thesis, Diss. Swedish Univ. Agric. Sci., Uppsala, Sweden, 2010, ISBN 978-91-576-9001-2 Search PubMed.
- A. N. Pell, J. Dairy Sci., 1997, 80, 2673–2681 CrossRef CAS PubMed.
- L. Strande, M. Ronteltap and D. Brdjanovic, Faecal Sludge Management — Systems Approach Implementation and Operation, IWA Publishing London, 2014 Search PubMed.
- S. S. Lim, et al. , Lancet, 2012, 380, 2224–2260 CrossRef.
- D. Mara, J. Lane, B. Scott and D. Trouba, PLoS Med., 2010, 7(11), e1000363 Search PubMed.
- F. M. Ngure, B. M. Reid, J. H. Humphrey, M. N. Mbuya, G. Pelto and R. J. Stoltzfus, Ann. N. Y. Acad. Sci., 2014, 1308, 118–128 CrossRef PubMed.
- J. Bartram and S. Cairncross, PLoS Med., 2010, 7(11), e1000367 Search PubMed.
- P. Hawkins, I. Blackett and C. Heymans, The missing link in sanitation service delivery: a review of fecal sludge management in 12 cities, Water and Sanitation Program, 2014 Search PubMed.
- E. Tilley, L. Ulrich, C. Lüthi, P. Reymond and C. Zurbrügg, Compendium of Sanitation Systems and Technologies (2nd Revised Edition), Swiss Fed. Inst. Aquat. Sci. Technol. (Eawag), Dübendorf, Switzerland, 2014, ISBN: 978-3-906484-57-0 Search PubMed.
- B. Vinneras, A. Nordin, C. Niwagaba and K. Nyberg, Water Res., 2008, 42, 4067–4074 CrossRef PubMed.
- J. Fidjeland, M. E. Magri, H. Jönsson, A. Albihn and B. Vinnerås, Water Res., 2013, 47, 6014–6023 CrossRef CAS PubMed.
- M. E. Magri, L. S. Philippi and B. Vinnerås, Appl. Environ. Microbiol., 2013, 79, 2156–2163 CrossRef CAS PubMed.
- L. Decrey, S. Kazama, K. M. Udert and T. Kohn, Environ. Sci. Technol., 2015, 49, 1060–1067 CrossRef CAS PubMed.
- L. Decrey, S. Kazama and T. Kohn, Appl. Environ. Microbiol., 2016, 82, 4909–4920 CrossRef CAS PubMed.
- F. Bosshard, F. Armand, R. Hamelin and T. Kohn, Appl. Environ. Microbiol., 2013, 79, 1325–1332 CrossRef CAS PubMed.
- B. M. Pecson, L. V. Martin and T. Kohn, Appl. Environ. Microbiol., 2009, 75, 5544–5554 CrossRef CAS PubMed.
- T. Kohn and K. L. Nelson, Environ. Sci. Technol., 2007, 41, 192–197 CrossRef CAS PubMed.
- K. Gallandat, Evaluation of pathogens inactivation during thermophilic anaerobic digestion of faecal sludge, Msc thesis, EPF Lausanne, 2014 Search PubMed.
- APHA, AWWA and WEF, Standard Methods for the Examination of Water and Wastewater, 1999 Search PubMed.
- D. L. Parkhurst and C. A. J. Appelo, Model. Tech. B. 6, 2013, p. 497 Search PubMed.
- S. D. Hafner and J. J. Bisogni, Water Res., 2009, 43, 4105–4114 CrossRef CAS PubMed.
- T. A. Larsen, K. M. Udert and J. Lienert, Source Separation and Decentralization for Wastewater Management, IWA Publishing, 2013 Search PubMed.
- C. Guzmán, J. Jofre, A. R. Blanch and F. Lucena, J. Virol. Methods, 2007, 144, 41–48 CrossRef PubMed.
- E. C. Baughan and R. P. Bell, Proc. R. Soc. London, Ser. A, 1937, 158, 464–478 CrossRef CAS.
- S. Liotta and V. K. La Mer, J. Am. Chem. Soc., 1938, 60, 1967–1974 CrossRef CAS.
- L. Decrey, Available on: https://lodecrey.shinyapps.io/MS2inactivation/, 2015.
- C. Höglund, N. Ashbolt, T. A. Stenstrom and L. Svensson, Adv. Environ. Res., 2002, 6, 265–275 CrossRef.
- B. Vinnerås, A. Nordin, C. Niwagaba and K. Nyberg, Water Res., 2008, 42, 4067–4074 CrossRef PubMed.
- T. Gao, Y. Tong, M. Cao, X. Li and X. Pang, Water Res., 2013, 47, 4259–4264 CrossRef CAS PubMed.
- A. Chandran, S. K. Pradhan and H. Heinonen-Tanski, J. Appl. Microbiol., 2009, 107, 1651–1657 CrossRef CAS PubMed.
- A. M. Nasser, R. Glozman and Y. Nitzan, Water Res., 2002, 36, 2589–2595 CrossRef CAS PubMed.
- H. N. Bischel, A. Schertenleib, A. Fumasoli, K. M. Udert and T. Kohn, Environ. Sci.: Water Res. Technol., 2015, 1, 65–76 CAS.
- T. Mondal, D. A. Rouch, N. Thurbon, S. R. Smith and M. A. Deighton, J. Water Health, 2015, 13, 459–472 CrossRef PubMed.
- M. Forconi and D. Herschlag, Methods Enzymol., 2009, 468, 91–106 CAS.
- D. O. Corona-Martínez, P. Gomez-Tagle and A. K. Yatsimirsky, J. Org. Chem., 2012, 77, 9110–9119 CrossRef PubMed.
- F. A. Nicholson, B. J. Chambers, J. R. Williams and R. J. Unwin, Bioresour. Technol., 1999, 70, 23–31 CrossRef CAS.
- M. B. McBride and G. Spiers, Commun. Soil Sci. Plant Anal., 2001, 32, 139–156 CrossRef CAS.
- B. Marti, K. M. Udert and E. Morgenroth, Bioelectrochemical Removal of Sulfur from Source-Separated Urine, Msc Thesis, ETH Zurich, 2010, p. 69 Search PubMed.
- J. Lin, J. Aoll, Y. Niclass, M. I. Velazco, L. Wünsche, J. Pika and C. Starkenmann, Environ. Sci. Technol., 2013, 47, 7876–7882 CrossRef CAS PubMed.
- D. A. Sanders, J. F. Malina, B. E. Moore, B. P. Sagik and C. A. Sorber, J. - Water Pollut. Control Fed., 1979, 51, 333–343 CAS.
- Y. Ye, R. M. Ellenberg, K. E. Graham and K. R. Wigginton, Environ. Sci. Technol., 2016, 50, 5077–5085 CrossRef CAS PubMed.
- R. L. Ward, Appl. Environ. Microbiol., 1982, 43, 1221–1224 CAS.
- D. O. Cliver and J. E. Herrmann, Water Res., 1972, 6, 797–805 CrossRef CAS.
- D. R. Knowlton and R. L. Ward, Appl. Environ. Microbiol., 1987, 53, 621–626 CAS.
- A. Carratalà, M. Rusiñol, J. Rodriguez-Manzano, L. Guerrero-Latorre, R. Sommer and R. Girones, Food Environ. Virol., 2013, 5, 203–214 CrossRef PubMed.
- R. M. Chaudhry, K. L. Nelson and J. E. Drewes, Environ. Sci. Technol., 2015, 49, 2815–2822 CrossRef CAS PubMed.
- M. Y. Deng and D. O. Cliver, Microb. Ecol., 1995, 30, 43–54 CrossRef CAS PubMed.
- M. Y. Deng and D. O. Cliver, Appl. Environ. Microbiol., 1995, 61, 87–91 CAS.
- M. Y. Deng and D. O. Cliver, Appl. Environ. Microbiol., 1992, 58, 2016–2021 CAS.
- M. E. Magri, J. Fidjeland, H. Jönsson, A. Albihn and B. Vinnerås, Sci. Total Environ., 2015, 520, 213–221 CrossRef CAS PubMed.
- S. K. Spillmann, F. Traub, M. Schwyzer and R. Wyler, Appl. Environ. Microbiol., 1987, 53, 2077–2081 CAS.
- S. Astals, C. Venegas, M. Peces, J. Jofre, F. Lucena and J. Mata-Alvarez, Water Res., 2012, 46, 6218–6227 CrossRef CAS PubMed.
- J. R. Ottoson, A. Schnurer and B. Vinneras, Lett. Appl. Microbiol., 2008, 46, 325–330 CrossRef CAS PubMed.
- F. Häfner, Optimised Ammonia Sanitisation of Sewage Sludge, Msc thesis, SLU Uppsala, 2014 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ew00311g |
| This journal is © The Royal Society of Chemistry 2017 |