Time-dependent bacterial transcriptional response to CuO nanoparticles differs from that of Cu2+ and provides insights into CuO nanoparticle toxicity mechanisms†
Abstract
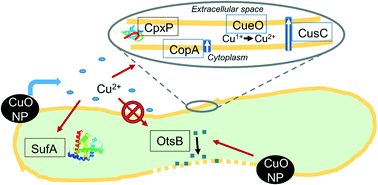
Better understanding of the time-resolved gene expression response of bacteria to CuO engineered nanomaterials (ENM) could lead to an improved mechanistic understanding of their antimicrobial effects. In this study, reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) has been used to characterize a time series of Escherichia coli gene expression after exposure to CuO nanoparticles (NP) (100 mg L−1 as Cu). Two dissolved Cu exposures in concentrations matching observed dissolution from the CuO NPs in the growth medium (1 ppm total Cu after 180 min) were included as a comparison against NP exposure. In the pulse dissolved Cu exposure, 1 ppm Cu was added at the beginning of the experiment. In the gradual dissolved Cu exposure, a total of 1 ppm dissolved Cu was divided into four doses (at 0, 10, 30, and 60 min) that matched the observed CuO NP dissolution. NP exposure led to induction of membrane damage gene expression, which aligns with hyperspectral imaging (HSI) results that identified a high NP affinity for cellular membranes shortly after exposure. Reactive oxygen species (ROS)-responsive genes were not induced for the NP-exposed E. coli within the 60 min time scale where Cu-(copA, cueO, and cusC) and protein damage gene (cpxP) expression were most induced – Cu2+ treatments led to minimal induction. Both Cu2+ treatments (pulse and gradual addition) led to higher levels of Cu- and protein damage-responsive gene induction than NP exposure, despite the lower total Cu exposure. For genes induced by all Cu exposures, pulse ion treatment led to temporally distinct transcriptional responses (peak inductions generally at 10 or 30 min) compared to both the gradual ion and NP treatments (peak inductions generally at 60 min). This time-resolved depiction of E. coli's transcriptional response to an antimicrobial soluble ENM identifies a response unique to ENM exposure and highlights the importance of sampling time when considering soluble ENM biological impacts. It also underscores the need, in nanotoxicity studies, to consider ionic controls that reflect the slow release of ion from soluble ENMs.
- This article is part of the themed collection: Sustainable Nanotechnology Organization


 Please wait while we load your content...
Please wait while we load your content...