Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Solvent water interactions within the active site of the membrane type I matrix metalloproteinase†
Elena
Decaneto‡
ab,
Tatiana
Vasilevskaya‡
c,
Yuri
Kutin‡
 a,
Hideaki
Ogata‡
a,
Moran
Grossman
b,
Irit
Sagi
d,
Martina
Havenith
b,
Wolfgang
Lubitz
a,
Hideaki
Ogata‡
a,
Moran
Grossman
b,
Irit
Sagi
d,
Martina
Havenith
b,
Wolfgang
Lubitz
 a,
Walter
Thiel
a,
Walter
Thiel
 c and
Nicholas
Cox
c and
Nicholas
Cox
 *ae
*ae
aMax Planck Institute for Chemical Energy Conversion, Stiftstraße. 34-36, D-45470, Mülheim an der Ruhr, Germany. E-mail: nicholas.cox@cec.mpg.de
bDepartment of Physical Chemistry II, Ruhr-Universität Bochum, 44780 Bochum, Germany
cMax-Planck-Institut für Kohlenforschung, Kaiser-Wilhelm-Platz 1, D-45470, Mülheim an der Ruhr, Germany
dDepartment of Biological Regulation, The Weizmann Institute of Science, Rehovot, Israel
eResearch School of Chemistry, Australian National University, Canberra, ACT 2601, Australia. E-mail: nick.cox@anu.edu.au
First published on 14th September 2017
Abstract
Matrix metalloproteinases (MMP) are an important family of proteases which catalyze the degradation of extracellular matrix components. While the mechanism of peptide cleavage is well established, the process of enzyme regeneration, which represents the rate limiting step of the catalytic cycle, remains unresolved. This step involves the loss of the newly formed N-terminus (amine) and C-terminus (carboxylate) protein fragments from the site of catalysis coupled with the inclusion of one or more solvent waters. Here we report a novel crystal structure of membrane type I MMP (MT1-MMP or MMP-14), which includes a small peptide bound at the catalytic Zn site via its C-terminus. This structure models the initial product state formed immediately after peptide cleavage but before the final proton transfer to the bound amine; the amine is not present in our system and as such proton transfer cannot occur. Modeling of the protein, including earlier structural data of Bertini and coworkers [I. Bertini, et al., Angew. Chem., Int. Ed., 2006, 45, 7952–7955], suggests that the C-terminus of the peptide is positioned to form an H-bond network to the amine site, which is mediated by a single oxygen of the functionally important Glu240 residue, facilitating efficient proton transfer. Additional quantum chemical calculations complemented with magneto-optical and magnetic resonance spectroscopies clarify the role of two additional, non-catalytic first coordination sphere waters identified in the crystal structure. One of these auxiliary waters acts to stabilize key intermediates of the reaction, while the second is proposed to facilitate C-fragment release, triggered by protonation of the amine. Together these results complete the enzymatic cycle of MMPs and provide new design criteria for inhibitors with improved efficacy.
1. Introduction
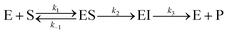
The membrane type I matrix metalloproteinase (MT1-MMP or MMP-14) is a zinc-dependent transmembrane endopeptidase that plays a major role in regulation of cellular migration and in the degradation and remodeling of extracellular matrix components, such as type I, II, III collagens, fibronectin, gelatin and proteoglycans.1,2 In recent years, interest in MT1-MMP has increased following observations that high expression levels of MT1-MMP are also associated with tumor progression and metastasis.3,4 An inhibitor candidate that meets pharmaceutically required specificity and affinity has yet to be identified.5,6The MMP family of proteins has been extensively studied. The catalytic Zn2+ ion is located near the protein surface, with the cleft above the metal site allowing binding of substrate peptides. In its resting state the Zn ion is coordinated by three His residues (His239, His243, His249 for MT1-MMP) and at least one water molecule which serves as the nucleophile during substrate hydrolysis.7,8 Mechanistic studies at close-to-neutral pH revealed that this water molecule is strongly polarized forming a bridge between the conserved glutamate base (Glu240) and the Lewis acidic Zn.9–11
A four-step catalytic mechanism of peptide hydrolysis has been proposed for MMPs.7,12–14 Upon peptide docking (Fig. 1a, ES, 1) the reaction starts with the nucleophilic attack of the carbonyl carbon of the scissile bond by the catalytic water molecule. The side chain of Glu240 acts as a proton shuttle, transferring a proton of the glutamate-polarized water to the nitrogen atom of the scissile amide forming a metal penta-coordinated intermediate complex (ES‡, 2). The deprotonated water nucleophile then attacks the C–O bond of the peptide leading to cleavage of the C–N bond (3). This step is coupled to or followed by the transfer of the second water-derived proton to the amine group via the glutamate base. Theoretical modeling suggests that a second auxiliary solvent water molecule acts as an electrophile, stabilizing the newly formed carboxylate product.7 The last step corresponds to the release of the carboxylate product and the re-uptake of a water molecule. (Fig. 1a, 4).
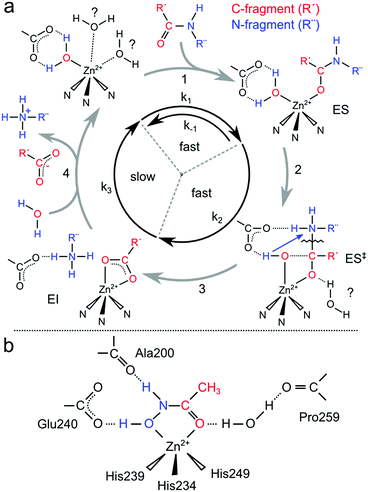 | ||
| Fig. 1 (a) Proposed catalytic mechanism of peptide hydrolysis of MMPs; (b) schematic illustration of the binding features of an acetohydroxamic acid (AHA) molecule to the catalytic zinc of MMPs. | ||
Crystallographic characterization of the intermediates of the reaction cycle has thus far been elusive. Bertini et al.15 have reported structures of two different MMPs where substrate was present, capturing the cleaved peptide fragments in the catalytic pocket. These are the earliest intermediates that can be characterized suggesting that the catalytic steps (Fig. 1a, 2, 3) are fast and the rate-limiting step instead represents product release (Fig. 1a, 4). This is in agreement with steady-state and pre-steady-state kinetics data from the tumor necrosis factor-a converting enzyme (TACE),16 another member of the MMP family. In addition, computational studies suggest that the loss of the bidentate C-terminus fragment from the Zn2+ during (Fig. 1a, 4) is the rate limiting step with an estimated barrier of 20 kcal mol−1;13 however this intermediate has yet to be characterized. The large barrier is derived from the negatively charged C-fragment coordinating the positive Zn2+ ion in a bidentate fashion. It is also not clear in what order the peptide fragments are released. The study by Bertini et al.15 characterized an intermediate with both C- and N-fragment in the catalytic pocket and a second intermediate with only the N-fragment suggesting, surprisingly, that C-fragment dissociation occurs before N-fragment dissociation. This has led to a concerted mechanism being proposed involving partial carboxylate detachment to initiate N-fragment release, coupled with solvent water insertion.
Pinpointing the protein structural elements that foster this progression has been challenging. Point mutations are found to strongly affect the rate of catalysis,17 particularly when replacing the glutamate base;18 however, as the relevant mutated residues are implicated in both the hydrolysis and product release steps, their precise role in enzyme reactivation is unclear. The role of auxiliary/solvent water in product release is also not resolved, nor is there good experimental evidence for auxiliary water molecule(s) invoked in theoretical treatments of the catalytic cycle. This is mainly due to the difficulty of crystallizing the protein in its native form owing to autocatalysis. As a consequence, MMPs are typically crystallized in the presence of various inhibitors including: regulator proteins (TIMPs),19,20 peptides and small synthetic molecules, such as hydroxamic acids, e.g. acetohydroxamic acid21 (AHA, Fig. 1b) with the latter chelating the catalytic zinc ion displacing the water nucleophile.22 To some extent the AHA-bound form of the enzyme models the ES‡ intermediate, at least in terms of the coordination of the Zn ion. However, as these inhibitors are small, they can be removed after crystallization by extensive washing to at least allow the solvation of the Zn ion in its resting state to be examined as achieved in the study of Bertini.15 Using this approach, both MMP-8 and MMP-12 have been characterized leading to two different descriptions of the resting state solvation sphere. In MMP-8 only one water ligand is resolved with full occupancy, while two further water molecules have only 20% occupancy. If the latter are neglected, the resulting geometry is a flattened tetrahedron. In contrast, the active site of MMP-12 contains three coordinating water molecules leading to an almost regular octahedral geometry of the Zn site.15 For MT1-MMP, the system used in this study, only an X-ray structure with a TIMP has been reported.19,20 The uninhibited coordination number has instead been inferred from stopped-flow X-ray absorption (XAS), which suggested that the metal is tetracoordinated, i.e. it only has one water ligand, the nucleophilic water.23
Regardless of the precise number of coordinating water molecules in the resting state, it is the presence of solvent in the ‘activated’ states of catalysis which is crucial to address function. Recent THz absorbance spectroscopy suggests that solvent water dynamics plays a crucial role for the enzymatic activity and protein recognition.23 Here we report the crystal structure of the catalytic domain of human MT1-MMP with a small peptide bound at its catalytic site, modeling the first product state after peptide cleavage. This novel structure identifies two auxiliary water molecules whose role in the enzymatic reaction was analyzed using optical, magnetic resonance and computational techniques. Together these data provide new information on the catalytic mechanism of MMPs and the importance of the immediate solvation sphere in directing catalysis.
2. Materials and methods
2.1 Preparation of Zn2+ and Co2+-MT1-MMP
The procedure for the expression, purification and preparation of Zn2+- and Co2+-MT1-MMP is described in the ESI,† S1, S2 and S3. Metal content of protein samples was measured by atomic absorption spectroscopy (Mikrolab Kolbe GmbH, Mülheim/Ruhr, Germany). Unless otherwise stated, all spectroscopic measurements were performed at a protein concentration of 100–300 μM in 50 mM Hepes pH 7.4, 100 mM NaCl and 5 mM CaCl2. Inhibited samples contained additionally 0.5–1 M of acetohydroxamic acid (AHA), (ESI,† S4). Inhibited samples were incubated for at least 2 hours in AHA prior to measurement. Low temperature measurements included glycerol in the buffer: 30% v/v for EPR, 80% v/v for MCD.2.2 Enzymatic assays
Enzymatic assays were performed at 25 °C by monitoring the increase in fluorescence intensity of methoxycoumarin-4-acetyl (Mca) due to the decrease of FRET quenching by 2,4-dinitrophenyl (Dnp) upon hydrolysis of the fluorogenic peptide Mca-KPLGKK(Dnp)AR-NH224,25 (λex: 340 nm, λem: 400 nm) with a 96-well plate reader connected to a fluorescence spectrophotometer (Varian Cary Eclipse). The Michaelis Menten constant for the hydrolysis of the collagen-like substrate was estimated by fitting the initial velocity (for the first ≈100 s) with the Michaelis Menten equation vi = vmax[S]/(KM + [S]), where vi is the initial rate (fluorescence unit per second), [S] is the substrate concentration (M), vmax is the maximum reaction rate attained at the saturating substrate concentration and KM is the Michaelis–Menten constant.2.3 Protein crystallography
Crystals were obtained from 10 mg ml−1 of MT1-MMP using the sitting drop/vapor-diffusion method upon mixing with an equal volume of 0.2 M ammonium nitrate, 20% (w/v) PEG 3350 (pH 6.2).26 For X-ray diffraction experiments, the crystals were soaked in buffer containing 0.3 M ammonium nitrate, 25% (w/v) PEG 3350 (pH 6.2) and 20% glycerol as a cryoprotectant. Diffraction data sets were collected on beamline BL14.2 at BESSYII (Hemholtz-Zentrum Berlin, Germany). A complete native data set was collected to 2.24 Å resolution at an X-ray wavelength of 0.91841 Å (PDB entry 5H0U). Diffraction images were indexed, integrated and scaled using the program XDS.27 The molecular replacement method was applied using the program MOLREP28 and the coordinates of the MT1-MMP-TIMP2 complex (PDB entry 1BQQ)19 were used as an initial search model. Refinement was carried out by the program PHENIX.REFINE.29 The stereochemical properties were checked by RAMPAGE.30 Data collection and refinement statistics are listed in the ESI,† S5.2.4 Room temperature absorption and MCD measurements
Electronic absorption spectra were recorded with a Cary Model 50 Spectrophotometer. MCD spectra were recorded with 1 nm bandwidth on a Jasco J-715 spectropolarimeter. The 1 cm pathlength quartz cuvette was inserted in a permanent magnet of 1.4 T field strength (Olis, Inc., Atlanta, GA). MCD spectra were collected to estimate the molar extinction coefficient of wild type MT1-MMP using the protein's tryptophan residues.31,32 A calibration curve was determined by measuring MCD spectra of different concentrations (25, 50, 75, 100, 150 and 200 μM) of L-tryptophan in 50 mM Tris (pH 7.4), 100 mM NaCl and 5 mM CaCl2. The MCD spectrum of 18 μM of MT1-MMP in the same buffer was acquired and compared with the calibration curve to calculate the molar extinction coefficient (ESI,† Fig. S2).2.5 Low temperature MCD measurements
MCD experiments at low temperature were carried out with an Olis DSM17 CD spectropolarimeter while the sample was placed in the Oxford cryostat Spectromag SM4000. The temperature range was from 1.7 K to 20 K and the energy range from 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 330 cm−1 (300 nm) to 12
330 cm−1 (300 nm) to 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 cm−1 (800 nm). The solutions were transferred to pre-cooled MCD holders with pre-cooled syringes, and then frozen in liquid nitrogen.
500 cm−1 (800 nm). The solutions were transferred to pre-cooled MCD holders with pre-cooled syringes, and then frozen in liquid nitrogen.
2.6 EPR measurements
X-band CW EPR measurements were performed in the temperature range of 5 to 20 K using a Bruker E500 spectrometer equipped with a Bruker ER 4116DM resonator, Oxford Instruments ESR 935 cryostat and ITC503 temperature controller. Microwave power levels used were in the range of 630 μW to 20 mW. Magnetic field modulation amplitude was 7.5 G. Q-band pulse EPR, ESEEM/HYSCORE and Davies ENDOR measurements were performed between 2 and 6.5 K using a Bruker ELEXSYS E580 Q-band pulse EPR/ENDOR spectrometer equipped with a home-built TE011 microwave resonator33 and cryogen free variable temperature cryostat from Cryogenic Ltd.34 The RF amplifier used for ENDOR measurements was AR 2500L (Amplifier Research). Field-swept EPR spectra were obtained via integration of the electron spin echo (ESE) signal. The length of the π pulse was 80 ns and the delay between the two pulses was 348 ns.2.7 Setup of the computational model
To create an all-atom three-dimensional model of MT1-MMP, the initial coordinates of the heavy atoms were taken from the crystal structure. Other conceivable structures of metal coordination spheres at the structural and catalytic zinc sites were built using the Discovery Studio Visualizer. To model the inhibited enzyme, an acetohydroxamate molecule was manually inserted into the active site using the Discovery Studio Visualizer software, using the crystal structure of MMP-9 complexed with a similar inhibitor (PDB ID: 2OW1)35 as a guide. The model systems were solvated with 9678 water molecules using the VMD software36 and equilibrated by means of 500 ps molecular dynamics (MD) simulations with periodic boundary conditions.2.8 Molecular dynamics simulations
The CHARMM37 force field was used for the protein atoms and the TIP3P model38 for water. Electrostatic interactions were computed using the Particle-Mesh-Ewald method with a grid spacing of 1.0 Å. Electrostatic and van der Waals interactions were smoothly switched to zero between 10 and 12 Å. A temperature of 300 K was controlled with a Langevin thermostat. The bond lengths of water molecules were constrained using the SHAKE algorithm.39 All MD simulations were carried out using the NAMD program.40 During these classical MD simulations, the coordinates of amino acid side chains that form the coordination environment of both zinc ions were frozen, i.e. Asp71, His69, Thr73, His84, Tyr86, His97, His122, His126, and His132. This helped to prevent drastic reorganizations in the structural site (keeping the zinc coordination number of 4). At the catalytic site the zinc coordination number became 6 (3 histidine residues and 3 water molecules), which is favored due to the force field parameterization of the zinc ion. Thereafter the solvation water shell was reduced by keeping only water molecules within 12 Å from the protein. The subsequent MM minimization employed 1000 steepest-descent steps, 5000 conjugate-gradient steps, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps using the adopted basis Newton–Raphson method. During these minimizations, the atoms from the coordination environment of the two zinc ions were fixed. The resulting model systems were taken as starting structures for QM/MM calculations. In the studies on the Co-substituted enzyme, the metal was changed from zinc to cobalt.
000 steps using the adopted basis Newton–Raphson method. During these minimizations, the atoms from the coordination environment of the two zinc ions were fixed. The resulting model systems were taken as starting structures for QM/MM calculations. In the studies on the Co-substituted enzyme, the metal was changed from zinc to cobalt.
2.9 QM/MM geometry optimizations
The QM region included the coordination sphere of the metal ions: the side chains of Asp71, His69, His84, His97, and the zinc/cobalt ion for the structural site; the side chains of His122, His126, His132, the acetate fragment of Glu123, the zinc/cobalt ion, 1–3 water/hydroxide ligands and/or an inhibitor for the catalytic site. The energy and gradients in the QM part were computed at the DFT level with the hybrid functional B3LYP41,42 in combination with the 6-31G** basis set.43 The rest of the protein and the other water molecules constituted the MM region, which was described by the CHARMM force field. All reported calculations on the Co-substituted enzyme were done on the high-spin (S = 3/2) state of Co2+. The QM/MM calculations were performed with the ChemShell software44,45 using the TURBOMOLE 6.3 program46 for the DFT calculations and the DL-POLY program47 for the MM calculations. Geometries were optimized using hybrid delocalized internal coordinates.48 The optimized region comprised all amino acid residues and water molecules containing atoms within a distance of 6 Å from any atom of the QM part; the rest of the protein and other solvent molecules were kept fixed. In all QM/MM calculations QM-MM electrostatic interactions were treated with electronic embedding and without any cutoff for QM-MM electrostatics. The chosen QM/MM setup and protocol have been found to be adequate in a previous QM/MM study of the MMP-2 catalytic cycle.132.10 Property calculations
All single-point property calculations were carried out with the ORCA program49 at QM(B3LYP/6-31G**)/CHARMM optimized geometries. The hyperfine A tensor was obtained from single-point calculations with the meta-GGA TPSSh functional50 and the triply polarized CP(PPP) basis set51 on cobalt; the other atoms were described by the uncontracted def2-TZVP basis set.52,53 We used an enhanced integration grid and integration accuracy of 11.0 for cobalt and of 9.0 for the cobalt-coordinating nitrogen atoms.51 Scalar relativistic effects were accounted for by using the zeroth-order regular relativistic approximation (ZORA) methodology.54 Point charges on the MM atoms were taken from the CHARMM force field. We calibrated the chosen QM(TPSSh/CP(PPP)) level of theory for the calculation of cobalt(II) isotropic hyperfine coupling constants against experimental data (ESI,† S12).The state-averaged complete active space self-consistent field (SA-CASSCF) method was used for calculation of on-site zero-field splitting (ZFS) parameters and of d–d transitions in Co(II) coordination complexes. The active space in the SA-CASSCF calculations included seven electrons and five 3d-based molecular orbitals. 10 (18) roots were considered for the quartet (doublet) state. Dynamic correlation effects were included by using second-order N-electron valence perturbation theory (NEVPT2).55,56 The ZORA Hamiltonian was employed in all these calculations, and the ZORA-recontracted version of the def2-TZVP basis set was used for all atoms. The spin–orbit coupling was represented by the spin–orbit mean field operator.57
3. Results and discussion
3.1 X-ray structure of catalytic domain of MT1-MMP
We report a high-resolution crystal structure of the catalytic domain of wild type MT1-MMP at a resolution of 2.2 Å26 (Fig. 2 and ESI,† S5). Unlike previous crystallographic studies, no exogenous inhibitor (e.g. TIMPs, AHA) was included in the crystallization procedure. Globally, our MT1-MMP structure is similar to structures previously reported for MT1-MMP complexed with a TIMP-1 mutant20 and TIMP-2.19 No significant differences are seen in the surrounding of both the metal active site and the specificity pockets controlling substrate recognition, representing surface interactions in the immediate vicinity of the catalytic Zn site. The only exception was the conformation of the MT-Loop (Fig. 2). The MT-Loop is a region with the sequence 163PYAPIREG170 characteristic of MT-MMPs. While its function is not entirely clear, its deletion does not affect cell surface expression and proteolytic activity but does result in inefficient localization and pro-MMP-2 activation on the cell surface.58 Recently, this region has been identified as an essential region to promote cellular invasion and a potential exosite target region to develop selective MT1-MMP inhibitors.59 The different conformation of the loop detected by X-ray crystallography may reflect the flexibility of this area, with its precise orientation in the crystal structure due to crystal packing affects. A similar observation was made for the membrane protein MT3-MMP,60 further supporting the notion that the conformation of the MT-loop is important for the divergent function of this family of enzymes.20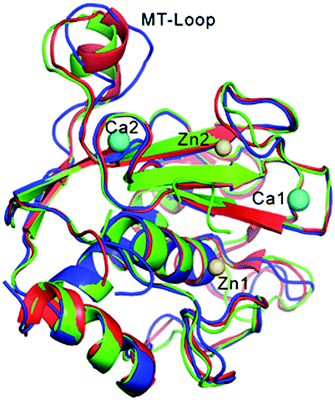 | ||
| Fig. 2 Superposition of the structures of the obtained MT1-MMP (blue) and the enzyme inhibited by TIMP-1 (red, PDB: 3MA2)20 and by TIMP-2 (green, PDB: 1BUV).19 The protein inhibitor TIMPs are not shown for clarity. Zn ions (catalytic Zn1 and structural Zn2) are colored in gold; Ca ions (Ca1 and Ca2) in cyan. | ||
The approximately spherical supercomplex contains four metal sites. Two sites contain zinc, whereas the other two sites contain calcium. The catalytic zinc is bound (Fig. 3) in the central part of the active-site cleft coordinated by the imidazole Nε2 nitrogens of His239, His243 and His249.7,12 As no inhibitor was included in the crystallization, the remaining ligands of the catalytic site should be solvent derived. This however was not the case. Additional electron density was found close to the active site pocket which could be fitted with a sequence of at least three histidines. This additional density is suspected to be associated with the His-tag engineered protein to allow purification (ESI,† S6). This His-tag was inserted into the flexible “hinge” region which links the catalytic domain to a C-terminal hemopexin domain.61In vitro maturation of the protein construct leads to autoproteolysis of the hinge region, which is not necessary for catalytic function.26 We suspect that the inadvertent presence of the residual His-tag in crystal preparations is required to stabilize the enzyme during the crystallization process as no His-tag free population was observed. We note that there is no evidence for the His-tag associating with the complex in solution and that the Zn/His-tag complex is not an intermediate of the hydrolysis reaction. Nevertheless, the structure obtained for the catalytic Zn site is novel. While it shares features with earlier inhibited structures using small synthetic molecules and peptides, it is the first to resolve multiple water interactions with the putative C-terminus product fragment, which computational modeling has predicted for the first product intermediate of the reaction cycle. A complete list of all reported MMP structures with a carboxylate bound at the catalytic Zn site is given in the ESI,† S6, see Table S2 and Fig. S13.
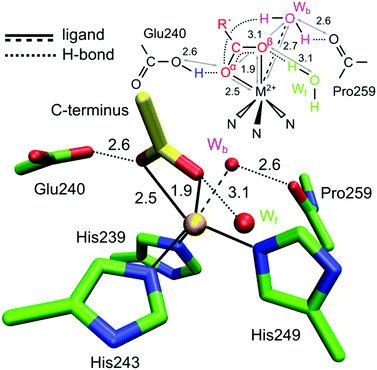 | ||
| Fig. 3 Crystal structure of the catalytic zinc ion (Zn1) of MMP-MT1 with a small peptide (His-tag, yellow stick) bound as a bi-dentate ligand via its C-terminus (PDB entry 5H0U). The Zn has three protein derived ligands: His239, His243 and His249 and distant water ligands: a water in front of the page (Wf) and a water behind the page (Wb). | ||
The interaction of the His-tag with the protein cleft is shown in the ESI,† S6. The MMPs active site is characterized by three unprimed pockets (S1 to S3) and three primed pockets (S1′ to S3′) which drive the binding of the substrates. The primed pockets interact with the N-terminus fragment, while the C-terminus fragment instead interacts with the unprimed pockets. The interaction of the C-terminus fragment of the His-tag was found in the unprimed pockets (S1 and S2).
The two additional Zn sites are described in the ESI:† a structural zinc coordinated by the imidazole Nε2 nitrogens of His186 and His201, the Nδ1 nitrogen of His214, and one carboxylate oxygen of Asp188; and a third adventitious Zn binding site that was found on the surface of the protein exposed to the solvent and corresponds to a crystallization artifact. It is bound via the carboxylate oxygen of Asp248 and stabilized by one of the histidine residues from the His-tag (ESI,† S5 and S6).
3.2 A novel structure modelling the first product state
Theoretical modeling of the reaction cycle (see Fig. 1) suggests that protein hydrolysis occurs via a nucleophilic attack of the α carbon of the peptide bond by a water molecule. The transition state for this reaction is a “gem-diol” type structure; crystal structures that contain a small synthetic molecule bound to the Zn to some extent model this intermediate, with a two-oxygen bidentate ligand motif which corresponds to the peptide backbone oxygen and the water oxygen nucleophile. The structure reported in this study is different. It instead contains a bidentate carboxylate ligand, which is a product of the reaction. Indeed the first product intermediate proposed is that of a Zn2+ complexed with an asymmetric bidentate carboxylate ligand. Theoretical models predict that such a motif is stabilized by:7,12,13(1) a hydrogen bond between the α oxygen of the carboxylate and the transiently protonated Glu240 residue;
(2) a hydrogen bond between the β oxygen of the carboxylate and a near-by auxiliary water molecule, which acts as an electrophile.
Both of these features have been seen in crystal structures of MMPs;62–64 in our structure the carboxylate C-terminus of the His-tag is in H-bonding distance to one oxygen of the Glu240 (2.6 Å) and an auxiliary water molecule, termed throughout this study as water in front (Wf) as it is located on the front face of the Zn ion in the projection where the three His residues are located below the Zn and the Glu240 sits behind the Zn on the left hand side (Fig. 3 and 4a). A survey of all structures reveals that the presence of Wf is correlated with the asymmetric bidentate carboxylate ligand motif (Zn–O carboxylate distances of ≈2.0 and ≈2.5 Å). In the complete set of MMP structures with an exogenous carboxylate ligand, the carboxylate coordination mode is plastic, binding as a symmetric bidentate ligand all the way to a monodentate ligand, see ESI,† S6, Table S2.
The more novel feature of our crystal structure is an additional auxiliary water molecule, termed throughout this study as water back (Wb) as it is located on the back face of the Zn ion. It interacts with both the Zn2+ ion and the 2nd coordination sphere proline residue. Such an auxiliary water molecule has only been observed once before, in a structure of MMP3 with a bound peptide.65 This earlier structure, however, did not contain Wf, nor did it stabilize a bidentate motif for the carboxylate. It instead more closely resembled the 1st product state that could be trapped in the study of Bertini, in which the carboxylate has already partially detached from the Zn2+ ion (mono-dentate). Wb in our structure is located such that it provides H-bonds to the α-oxygen of the C-terminus, the same as Wf. Owing to its location near Glu240, it represents the best candidate to displace/replace the bound C-fragment. We hypothesize that this process is triggered upon proton transfer from the Glu240 to the amine, breaking the H-bond between the Glu240 and C-terminus. A small rearrangement of Wb can then act to recover the H-bond network between Glu240 and the C-fragment, stabilizing an intermediate containing a monodentate carboxylate ligand (see Section 4). Within this interpretation, the MMP3 structure65 described above would then be representative of a later timepoint in this progression.
Lastly, we note that the position of the C-terminus/Glu240 couple in our structure also provides insight into how the C-fragment and N-fragment of the hydrolysis reaction likely interact immediately after peptide cleavage. The existing crystal structure of Bertini et al. locates the position of N-fragment (IAG) after the hydrolysis reaction. In our structure additional electron density is seen in approximately the same region, which we assigned to two cryoprotectant (glycerol) molecules. If instead this density is fitted to the IAG molecule, which is approximately the same size, the position of the amine head group in the 1st product state can be proposed. The nitrogen of the amine is within H-bonding distance of both the C-terminus (Oα) and the Glu240 (both 2.7 Å). We also note that in this model the Glu240 interacts with both the N-terminus and C-terminus fragments via a single oxygen (Fig. 4b). This is the same orientation for the Glu240 as seen in the earlier Bertini structures and in other MMPs with bound carboxylates (ESI,† S5 and S6). This is an important deviation from theoretical models of the first product state, which have instead assumed that the C- and N-fragments interact via a bidentate Glu240 (Fig. 4c) as is proposed for the gem-diol intermediate state. This result then implies that for the last proton transfer event, involving proton shuttling between the transiently protonated Glu240 and the amine, only requires one oxygen (the protonated one) of the Glu240, as recently suggested by Vasilevskaya et al.,13 making good chemical sense in so far that the proton transfer distance is significantly decreased using this motif.
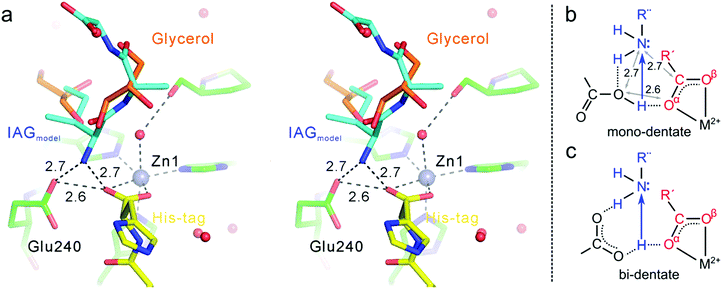 | ||
| Fig. 4 (a) Modeled position of a putative N-fragment associated with the catalytic site (steroview). The position of the N-fragment (IAG) was reoptimized from the structure(s) of Bertini et al.15 in the location of cryoprotectant molecules seen in our structure. (b and c) Possible proton transfer intermediates, with model b consistent with our crystallographic data. | ||
3.3 Detecting interactions between the auxiliary/solvent water and cofactor throughout the catalytic cycle – replacement of the spectroscopically silent Zn2+ ion with Co2+
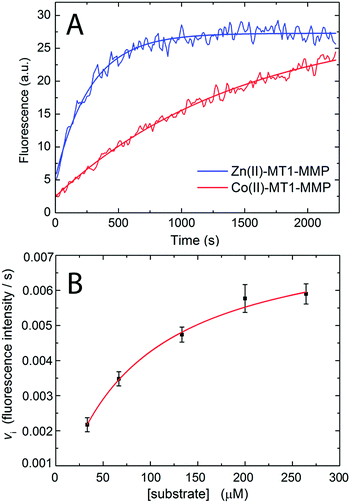
Magnetic spectroscopic techniques, such as electron paramagnetic resonance (EPR) and magnetic circular dichroism (MCD), are valuable biophysical tools for the study of metalloproteins allowing the local structure (coordinating ligands), global structure (protein conformation) and catalytic mechanism to be examined in detail, under conditions similar to the physiological environment.66–68 Although Zn2+ is a d10 closed shell ion and thus magnetically silent, magnetic spectroscopic studies can be made possible by replacing it with Co2+, which is instead a paramagnetic d7 ion. Co2+ is used in preference to other transition metals owing to its size and coordination chemistry being similar to Zn2+ with earlier literature studies demonstrating Zn/Co exchange does not significantly affect protein activity.66–68Importantly, for MMP homologues (e.g. MMP-1 and MMP-12) Zn/Co replacement does not change the coordination number/geometry of the active site.69,70 The protocol for replacing the “spectroscopically silent” Zn2+ ions in MT1-MMP with Co2+ is described in the ESI,† S3. The modified enzyme is fully active as demonstrated with a standard assay developed for MMPs based on a FRET mechanism.24,25 In this assay, the consumption of a fluorogenic collagen-like peptide leads to an increase of fluorescence at 400 nm allowing the reaction kinetics to be monitored. Interestingly the cobalt enzyme has a lower substrate turnover frequency of approximately 20% and a 2 fold increase of the Michaelis Menten constant KM, (36 ± 15 vs. 85 ± 8 μM, Fig. 5A and B). The increase in KM can be interpreted in a number of ways. As the rate limiting step of the reaction is the loss of the two peptide fragments from the protein cavity, the simplest enzymatic model requires four states (see Fig. 1): (i) free enzyme (E) + substrate (S); (ii) the enzyme–substrate complex (ES); (iii) the enzyme–product complex (EI); and (iv) free enzyme + product(s). Such a scenario can still be rationalized within the Michaelis Menten model, but KM includes a pre-factor (KM′) which takes into account the product dissociation step.
 | (1) |
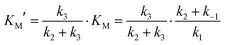 | (2) |
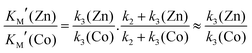 | (3) |
3.4 Dynamic interactions of auxiliary waters: electronic absorption and low temperature MCD of Co2+-MT1-MMP
Electronic absorption spectra were collected for the MMP protein at room temperature, in the presence and absence of the inhibitor AHA, resolving broad, weak transitions in the 400–600 nm (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–24
000–24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cm−1) region with extinction coefficients of the order of ≈100 M−1 cm−1, which were relatively insensitive to AHA addition. The inferred weak extinction coefficient (50 and 150 M−1 cm−1) suggests that both Co2+ sites of the protein are either 5 or 6-coordinate74–85 as tetrahedral complexes display significantly larger values ε ≥ 150 M−1 cm−1.74,85–93 This includes literature data from Co substituted MMPs: MMP-1,69 which has a tetrahedral catalytic site,94 exhibits an intense band with ε580 ≈ 1000 M−1 cm−1, whereas MMP-12, which has a octahedral catalytic site,15 shows a weaker band with ε545 ≈ 80 M−1 cm−1. Note that owing to the large scattering background seen in all samples, presumably resulting from partial enzyme aggregation, it is difficult to estimate with confidence the extinction coefficient or the effect of the inhibitor (Fig. 6a).
000 cm−1) region with extinction coefficients of the order of ≈100 M−1 cm−1, which were relatively insensitive to AHA addition. The inferred weak extinction coefficient (50 and 150 M−1 cm−1) suggests that both Co2+ sites of the protein are either 5 or 6-coordinate74–85 as tetrahedral complexes display significantly larger values ε ≥ 150 M−1 cm−1.74,85–93 This includes literature data from Co substituted MMPs: MMP-1,69 which has a tetrahedral catalytic site,94 exhibits an intense band with ε580 ≈ 1000 M−1 cm−1, whereas MMP-12, which has a octahedral catalytic site,15 shows a weaker band with ε545 ≈ 80 M−1 cm−1. Note that owing to the large scattering background seen in all samples, presumably resulting from partial enzyme aggregation, it is difficult to estimate with confidence the extinction coefficient or the effect of the inhibitor (Fig. 6a).
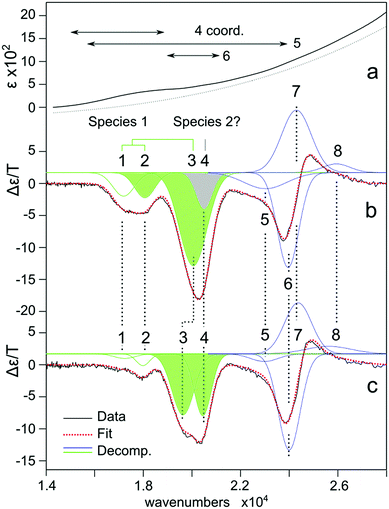 | ||
| Fig. 6 (a) Electronic absorption spectra of Co2+ substituted MT1-MMP. The dashed line represents an estimate of the scattering background due to protein aggregation. (b) Corresponding low temperature MCD spectrum (1.8 K, 7 T). (c) With AHA inhibitor. The red dashed lines represent a fitting of the spectra assuming eight bands modeled in terms of a Gaussian profile. Individual bands are colored green (catalytic), blue (structural) and grey (uncertain). See also ESI,† S7. | ||
To refine this picture, low temperature MCD of Co2+-substituted MT1-MMP was performed. These measurements identify multiple spin-allowed d–d transitions consistent with two or more protein-bound Co2+ ions (Fig. 6b). The profile could be modeled in terms of eight transitions. The addition of the inhibitor AHA was used to assign these transitions to either the catalytic site or the structural site; with the latter not being influenced by the binding of AHA (Fig. 6c). Bands 1–4 are all modified by the inhibitor and are thus assigned to the catalytic site: band 1 (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) band 2 (18
200) band 2 (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) are significantly diminished or completely lost upon inhibitor binding whereas band 3 (20
000) are significantly diminished or completely lost upon inhibitor binding whereas band 3 (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) decreases in intensity by 50% and is shifted to the red (500 cm−1), and band 4 doubles in intensity (its position is not well defined in the non-inhibited form). Bands 5–8 are unchanged and thus assigned to the structural site (see ESI,† S7). It is noted that the structural site should also have some contribution in the 17
000) decreases in intensity by 50% and is shifted to the red (500 cm−1), and band 4 doubles in intensity (its position is not well defined in the non-inhibited form). Bands 5–8 are unchanged and thus assigned to the structural site (see ESI,† S7). It is noted that the structural site should also have some contribution in the 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–21
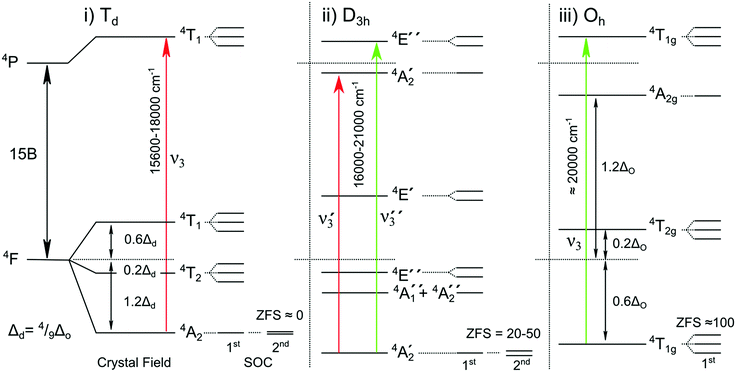
000–21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cm−1 region and thus the fitted bands should only be assigned dominantly to one species or the other. The electronic transitions observed represent the highest spin-allowed d–d transition of Co2+: i.e. transitions between electronic states derived from the ground state 4F and excited state 4P terms.95 For the free ion these levels are separated by 16
000 cm−1 region and thus the fitted bands should only be assigned dominantly to one species or the other. The electronic transitions observed represent the highest spin-allowed d–d transition of Co2+: i.e. transitions between electronic states derived from the ground state 4F and excited state 4P terms.95 For the free ion these levels are separated by 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 cm−1 (15B), with the crystal/ligand field (10 Dq < 5000 cm−1) tuning their exact positions (Fig. 7).95 The bands assigned to the catalytic site in its uninhibited form best resemble those seen for 5 coordinate Co2+ complexes. Such complexes typically resolve two MCD features of negative ellipticity, derived from the splitting of the 4P term, here split by ≈2500 cm−1, supporting the assignment of bands 1–2 to 4A2′(F) → 4A2′(P) and band 3 to 4A2′(F)→ 4E′′(P).
800 cm−1 (15B), with the crystal/ligand field (10 Dq < 5000 cm−1) tuning their exact positions (Fig. 7).95 The bands assigned to the catalytic site in its uninhibited form best resemble those seen for 5 coordinate Co2+ complexes. Such complexes typically resolve two MCD features of negative ellipticity, derived from the splitting of the 4P term, here split by ≈2500 cm−1, supporting the assignment of bands 1–2 to 4A2′(F) → 4A2′(P) and band 3 to 4A2′(F)→ 4E′′(P).
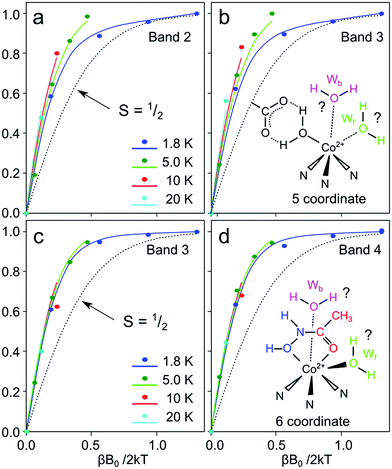
The induction of MCD signals as a function of the temperature and applied field provides further support for this assignment.66,96,97 The MCD C-term activity of Co2+ complexes is dependent on the ordering and separation of the two Kramers doublets that describe its ground magnetic spin state. In the instance where the separation is large, only one Kramers state is populated at cryogenic temperatures (0–20 K) and thus the induction of the MCD signal – described by its magnetization curve – is temperature invariant (the magnetization curves overlay). In contrast, if the separation is small, the relative populations of the two Kramers states will significantly vary as a function of temperature (<100 cm−1) and as such its magnetization curves will vary with temperature; a phenomenon called ‘nesting’. The separation between the two Kramers states is defined by the zero-field splitting of the complex, which is defined by the parameter D. For Co2+ complexes the zero-field splitting comes about due to spin–orbit coupling (SOC) and its magnitude broadly correlates with the coordination number.66,96,97 For tetrahedral complexes, the singlet ground electronic state (4A2) is not strongly affected by SOC, whereas for octahedral complexes, the orbitally degenerate ground electronic state will split (to 1st order) due to the SOC mechanism; 5 coordinate complexes fall in between, with the SOC mechanism operating to 2nd order.97 This sets up a hierarchy of D values which Co2+-substituted enzymes typically follow: D is small (or negative) for tetrahedral complexes, moderate for 5 coordinate complexes and large for octahedral complexes, although there is significant overlap within these three ligand fields.66,96,97
Magnetization curves for these two bands (2 and 3) assigned to the catalytic state show nesting behavior consistent with a fine structure splitting value of approximately D = +25 cm−1, and thus consistent with a 5 and 4 coordinate Co2+ complex. We note that band 1 has a non-physical magnetization curve presumably due to the overlap with another unresolved feature (i.e. structural site). It is less clear what band 4 represents. This band does not display the same field and temperature dependence lacking the nesting behavior of bands 2 and 3, suggesting a large, positive fine structure value (D > +100 cm−1). This simple, weak feature may represent a second population of the catalytic site in which it adopts an octahedral coordination with a 4T1g(F) → 4T1g(P) transition. The presence of both these signals would then suggest that the resting state solvation of the Co2+ substituted enzyme is best described in terms of a dynamic 1st coordination sphere with two or three water molecules. Whether both the front and back waters can bind independently of each other in a 5 coordinate geometry, or if instead there is a preferred solvation complex in the 5-coordinate form cannot be deduced from this data.
Upon binding of the inhibitor, the spectrum collapses into a doublet centered at 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 624 cm−1, although a shoulder may be present at 18
624 cm−1, although a shoulder may be present at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cm−1, which could be an underlying structural site signal. This feature shows no nesting behavior (D > 130 cm−1), suggesting that the catalytic site with the inhibitor bound is best described in terms of an octahedral coordination. Both bands are assigned to a single complex, with the splitting due to a lowering of perfect octahedral symmetry. This requires the inhibited active site to have one water ligand. It is not clear whether the additional water binds on the front or back face of the complex (Fig. 8).
000 cm−1, which could be an underlying structural site signal. This feature shows no nesting behavior (D > 130 cm−1), suggesting that the catalytic site with the inhibitor bound is best described in terms of an octahedral coordination. Both bands are assigned to a single complex, with the splitting due to a lowering of perfect octahedral symmetry. This requires the inhibited active site to have one water ligand. It is not clear whether the additional water binds on the front or back face of the complex (Fig. 8).
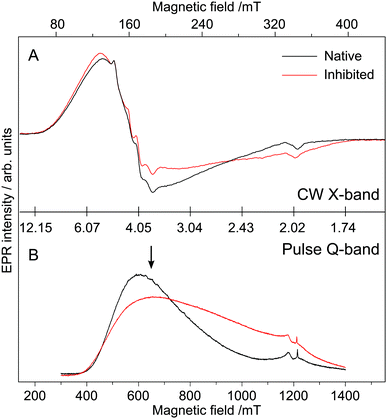
3.5 Dynamic interactions of auxiliary waters: EPR of Co2+-MT1-MMP
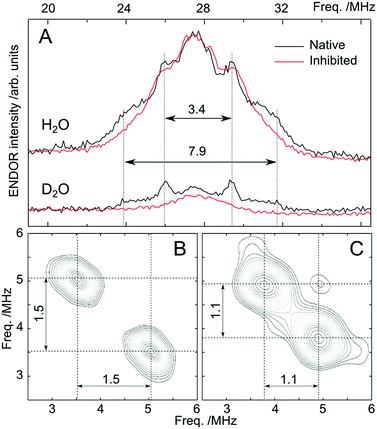
The above magnetic-optical analysis was refined using EPR. Fig. 9 shows CW X-band and pulse Q-band EPR spectra of the Co-substituted enzyme with and without AHA. The EPR signal at g ≈ 4.3 is indicative of Co2+ complex(es) with large positive zero-field splitting (D > +1 cm−1), i.e. the magnetic ground state is MS = |±½〉.68,98 Temperature-dependent data shown in the ESI,† S7 confirm this assignment. The low-field signal consists of a broad feature with no resolved hyperfine structure and a second, relatively narrow feature, with an additional splitting. These two components show different temperature and power dependencies, with the broad signal saturating more readily (see ESI,† S7). The eight-line hyperfine pattern, which originates from the interaction with the 59Co nucleus (I = 7/2), is often obscured at X-band (and higher frequencies) due to the effects of g- and D-strain.68 The ground state assignment suggests that both Co sites of the protein are either pentacoordinated or octahedrally coordinated, consistent with MCD data described above (see also ESI,† S7).68 The small signal at g = 2 (B0 ≈ 340 mT at X-band) is part of the baseline (resonator) and another background signal (adventitious Fe3+) observed at g = 4.33 (B0 = 159 mT at X-band) varied from batch to batch. Inhibition of the enzyme with AHA changes the EPR line shape, indicating that the broad signal envelope represents (at least in part) the catalytic site, as the structural site is not expected to change upon inhibition. Interestingly, the hyperfine splitting observed is unaffected by inhibition, suggesting that this may be solely associated with the structural site. We note however that the hyperfine structure was not readily observed in all samples. The line shape of the main component indicates that the Co2+ ion has an approximately axial ZFS tensor (E/D ≈ 0.1). The rhombicity of the signal increases (E/D ≈ 0.13) upon AHA binding. Although a small E/D is usually thought to be reflective of octahedral coordination, in our case owing to high level symmetry (see Section 3.5 Fig. 11) we do not consider this parameter to give a definitive assessment of the coordination number.68Subsequently pulse double resonance EPR experiments71 were performed to directly assess the number of water ligands that bind at the catalytic Co2+ ion and to examine whether these are displaced by AHA. Davies 1H-ENDOR measurements show that there is at least one auxiliary water ligand in both the inhibited and non-inhibited state, with hyperfine coupling (Adip) of about 3 MHz typical for aqueous transition metal complexes.99,100 It has a ‘Pake-like’ pattern indicative of a dipolar-coupled species (sitting perpendicular to the principal axis of the D tensor) that is lost upon deuteration. Both properties are only consistent with the presence of one or more water molecules in the 1st coordination sphere. These results immediately imply that the AHA form of the cofactor is 6 coordinate, with three His ligands, two ligands from the inhibitor and 1 auxiliary water, in agreement with MCD data. A second, more strongly coupled water, is resolved by 2H-ESEEM/HYSCORE (see Fig. 10B). The observed hyperfine coupling of 1.5 MHz for 2H corresponds to Aobs ≈ 9.8 MHz for 1H due to the difference in the gyromagnetic ratios. We reason that the signal is too broad to be readily observed in the corresponding 1H-ENDOR experiment. Indeed we encountered a similar situation for the Mn/Fe cofactor of the R2lox protein.101 The hyperfine coupling of this 1st coordination sphere water is approximately the same as observed earlier for the Zn finger protein Transcription Factor IIIA98 and is assigned to the catalytic water. These results imply that the cofactor must be at least 5 coordinate in the non-inhibited state, with three His ligands, the polarized/catalytic water and at least 1 auxiliary water. We find that the 1.5 MHz signal is lost upon AHA binding (see Fig. 10C), replaced by a smaller coupling of 1.1 MHz, presumably the OH of the AHA. As the 1.5 MHz coupling is completely abolished, AHA is bound to all catalytic sites at this concentration. One further observation can be made from this dataset. A non-exchangeable proton signal with a splitting of 3.4 MHz is also resolved in the 1H-ENDOR spectrum of the non-inhibited sample; this signal is not lost upon deuteration. It is likely derived from the non-exchangeable proton(s) of the histidine ligands. This signal is however lost upon binding of AHA, suggesting that the ligand field of the catalytic Co2+ changes upon binding of the inhibitor. This final result supports the notion that the non-inhibited state is different from the inhibited state, i.e. 5-coordinate, in agreement with MCD data. However as with the MCD measurements, EPR results do not show how the auxiliary water(s) binds in either the non-inhibited or inhibited state.
3.6 QM/MM modeling of the structural and active site of MT1-MMP
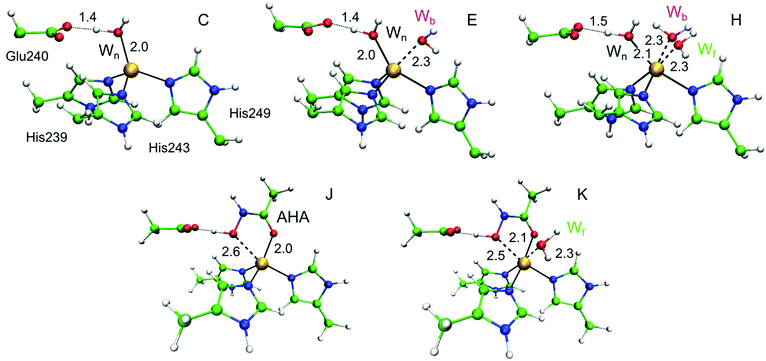
To further explore the role of the auxiliary water molecules during catalysis, QM/MM calculations were performed on a set of plausible structural models A–K (see ESI,† S8–S10 for detailed results and discussion). For selected models, the QM/MM optimized geometries of catalytic zinc coordination spheres with coordination numbers 4, 5 and 6 are shown in Fig. 11(C, E, and H). In addition, structural site models (A, B) and catalytic site models with extra water molecules in the second coordination shell (between Glu240 and the nucleophilic water) were examined (see ESI,† S8–S10). As expected, in tetrahedral models the single water ligand that binds to the metal ion also interacts with the nearby Glu240, and thus represents the nucleophilic water (Wn) of the reaction mechanism. All 5-coordinate models contain the same water ligand (Wn), and a second water that binds on the back face of the cofactor (Wb), presumably because this allows for a hydrogen bonding interaction with the Pro259 residue. By definition, in the 6-coordinate models all three water molecules act as ligands (Wn, Wb, and Wf on the front face of the cofactor).The validity of these models was tested by comparing the calculated D value for each model with experimental values determined from MCD measurements (Table 1). As expected, the tetrahedral models (C, D) yield D values of the wrong sign and magnitude and can thus be discarded. One 5-coordinate model (E), in which Wn directly interacts with Glu240 yields a positive intermediate D value (+33 cm−1) in agreement with the dominant species observed by MCD. The other 5-coordinate models in which an additional water is located between Wn and Glu240 (F and G) yield negative D values and are thus not supported. Finally, the 6-coordinate models (H, I) yield large positive D values, larger than 80 cm−1. These models could therefore explain the second species that may be present in the MCD data. We note that other calculated magnetic parameters (e.g.59Co and 14N hyperfine constants) were not discriminatory in terms of coordination/solvation number assignment.
| CASSCF | NEVPT2 | |||
|---|---|---|---|---|
| D | E/D | D | E/D | |
| C | +11.8 | 0.27 | +10.0 | 0.28 |
| D | −16.8 | 0.10 | −14.2 | 0.11 |
| E | +39.1 | 0.01 | +33.4 | 0.04 |
| F | −20.3 | 0.30 | ±16.2 | 0.33 |
| G | −27.3 | 0.12 | −23.3 | 0.10 |
| H | +119.2 | 0.07 | +110.5 | 0.10 |
| I | +96.1 | 0.19 | +84.0 | 0.21 |
| J | −22.2 | 0.29 | −18.9 | 0.28 |
| K | +89.2 | 0.29 | ±83.2 | 0.33 |
| Exp. (native) | D = +24.5 cm−1; E/D = 0.10 | |||
| Exp. (inhibited) | D > +100 cm−1; E/D = 0.13 | |||
We considered two further computational models (J, K) to investigate how auxiliary water interacts with the cofactor in the presence of bound AHA: model J does not include an additional first coordination sphere (auxiliary) water, whereas model K does. The hydroxamate anion (CONHO−) binds bidentately to the catalytic Zn2+ ion leading to a distorted trigonal-bipyramidal geometry around the metal, and it also forms a hydrogen bond with the carboxylate group of Glu240. The –NH– group forms another hydrogen bond with the carbonyl oxygen of Ala200, and van der Waals interactions and hydrophobic contacts play a critical role in stabilizing the inhibitor-enzyme complex.102 In the QM/MM optimized geometry for model K, water binds only to the front face (Wf). Comparison between computed and experimental MCD parameters shows that, as expected, model J is inconsistent with MCD data yielding a small negative D value. In contrast, structure K is consistent with the experimentally observed D value, i.e. >100 cm−1, derived from the MCD measurements and is in agreement with ENDOR results that showed the AHA complex has one first coordination sphere water. Structure K represents a genuine octahedral complex, with a water ligand stabilizing the bidentate ligation of the AHA. Thus all magneto-optical and magnetic resonance data presented above can be interpreted as follows:
(1) the solvation of the non-inhibited state is such that the Co/Zn ion is 5-coordinate (with only the Wb auxiliary water bound) which may be in equilibrium with a 6-coordinate population (with both auxiliary waters bound);
(2) the solvation of the inhibited state is such that the Co/Zn ion has one auxiliary water bound (Wf) rendering the complex 6-coordinate.
Finally it should be emphasized that QM/MM geometry optimizations with Co2+ instead of Zn2+ converged to qualitatively identical structures with a maximum absolute difference in bond length of 0.18 Å (ESI,† S9).
4. Conclusions – implications for the mechanism
Our study provides new criteria for the mechanism of MMPs, modelling the first product state of the reaction, which has thus far only been accessed theoretically. In conjunction with the ground breaking study of Bertini et al.,15 our study completes the reaction cycle and provides a chemical basis for C-terminus loss and the role that auxiliary/solvent water plays in this key reaction step. A complete reaction cycle of MMPs is described below and summarized in Fig. 12.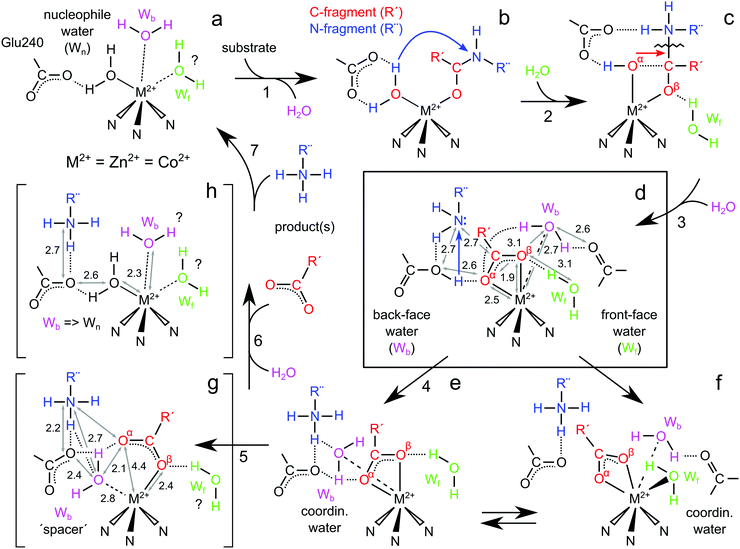 | ||
| Fig. 12 Complete reaction cycle for MMPs including the sequence of steps involved in product release and the role of auxiliary/solvent water in this process. Structures (a, resting), (g and h) represent intermediates trapped in the Bertini study.15 Structures (b) and (c) show the peptide binding and cleavage steps. Structure (d) represents the 1st product state of the reaction following peptide cleavage, developed in this study. Structures (e) and (f) show two possible resolvation pathways, with only (e) leading to C-fragment loss. | ||
We favour a resting state structure that has at least one additional, non-catalytic water molecule (Wb, Fig. 12a). A third water ligand (Wf) may also associate with the Zn2+ rendering the site octahedral. The dynamic nature of the solvation sphere – under physiological conditions – is not surprising and has literature precedent.8,23 Importantly, we find that Wb more readily associates with the metal site than Wf. In all 5-coordinate DFT models the second solvating water is Wb rather than Wf, presumably because Wb forms an H-bond to the second sphere Pro259 residue, whereas no such interaction with the protein is available for Wf. Both waters are displaced by the bulky substrate, which is found to coordinate in a bidentate fashion leading to the gem-diol intermediate (Fig. 12b and c). Following the cleavage of the peptide backbone the first predicted intermediate contains a newly formed C-terminus of one product fragment coordinating the Zn2+ as an asymmetric bidentate ligand (Fig. 12d). The bound carboxylate is stabilized by two interactions: (i) an H-bond between the α-oxygen of the carboxylate and transiently protonated Glu240; and (ii) an H-bond to the β-oxygen of the carboxylate and an auxiliary water. Both these structural elements are found in the crystal structure we report here, with the auxiliary water bound on the front face (Wf). Our structure deviates from the original computational proposal7 in two important ways:
(1) The position of the Glu240 prohibits it from forming a bidentate H-bonding network to the amine product, as is proposed in the gem-diol intermediate.
Instead, we can model a monodentate H-bonding bridge between the two protein fragments. We speculate that, for this step in the cycle, an H-bonding interaction involving only the transiently protonated oxygen of the Glu240 would assist the subsequent proton transfer to the amine fragment as the tunneling distance is significantly smaller.
(2) The structure includes a second water (Wb) which is also within H-bonding distance to the α-oxygen of the carboxylate, further participating in stabilizing the asymmetric carboxylate binding motif.
We suspect that this water represents the water used as the nucleophile (Wn) in the next reaction cycle as it is uniquely placed to displace the α-oxygen of the product carboxylate. We speculate that the trigger for this event is the proton movement to the amine. Upon proton transfer, Wb moves within the coordination sphere of the Zn2+ (approx. 2 Å) to replace the lost H-bonding network between the α-oxygen of the product and the Glu240. Simultaneously it may also gain an H-bond to the amine. At this point the water can displace the α-oxygen forming the first experimentally characterized intermediate of the reaction, a mono-dentate 5-coordinate complex reported by Bertini and coworkers (Fig. 12g). This structure also exhibits a favourable H-bonding network between both product fragments facilitated by the incoming/relocated Wb and the Glu240. The mono-dentate C-terminus fragment is then released from the Zn2+ site, perhaps in competition with solvent, i.e. water binding on the back face, as seen in our resting state models (Fig. 12h). We note though that such a water molecule was not observed in the Bertini structure.15 Nevertheless, carboxylate loss would strengthen the Zn2+ water (Wn) interaction, shortening the Wn–Zn2+ bond and disrupting its interaction with the amine. The amine, which at this point only weakly associates with the Glu240 (via H-bonding) is then also released.
The above sequence suggests that the C-fragment loss can be short-circuited slowing enzymatic turnover. Following proton transfer to the amine destabilizing the Zn-carboxylate complex, the solvation sphere can respond in more than one way. Instead of Wb moving to restore the H-bonding network involving the Glu240 as described above (Fig. 12e), Wf could instead directly ligate the Zn2+ ion rendering the complex 6-coordinate, thus preventing the release of the carboxylate (Fig. 12f). This is indeed what we observe for the Co enzyme inhibited with AHA – an additional water binds on the front face rendering the complex 6-coordinate. Presumably both re-solvation pathways are in equilibrium, however we suspect that Wf binding is more likely to occur for the Co2+ enzyme as opposed to the native Zn2+ form, owing to the higher coordination number preference of Co2+. This simple rationale would then explain the slower turnover of the Co2+ enzyme. Thus, the solvation and inhibition of the cofactor via small synthetic molecules are likely interrelated providing another crucial element needed for understanding the affinity of inhibitors of MMPs and their rational design. The strategy would be to ensure that the inhibitor mimics substrate/solvent interactions, either by replicating 1st solvation sphere interactions, or by the inclusion of functional groups which can replace the 1st solvation shell and its H-bonding to the protein surface. It is less clear how solvent/substrate interactions can be exploited toward selective inhibition of MMPs. This forms ongoing work of our laboratory.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Andreas Göbels and Dr Eckhard Bill are gratefully acknowledged for the acquisition of MCD spectra. We thank the staff of BESSYII (Helmholtz-Zentrum Berlin, Germany) for their help during the diffraction data collection. Financial support was provided by the Max Planck Gesellschaft and the Cluster of Excellence RESOLV (EXC 1069) funded by the Deutsche Forschungsgemeinschaft. I. S. is supported by the Binational Science Foundation (BSF) and Israel Science Foundation (ISF). I. S. and M. H. are also supported by the ERC Advanced Grant 695437 THz-Calorimetry. M. G. is an Awardee of the Weizmann Institute of Science National Postdoctoral Award Program for Advancing Women in Science and a recipient of an A. v. Humboldt Fellowship. N. C. acknowledges the support of the Australian Research Council: Future Fellowship (FT140100834). Open Access funding provided by the Max Planck Society.Notes and references
- M. Kamioka, T. Ishibashi, H. Ohkawara, R. Nagai, K. Sugimoto, H. Uekita, T. Matsui, S. I. Yamagishi, K. Ando, T. Sakamoto, N. Sakamoto, Y. Takuwa, I. Wada, M. Shiomi, Y. Maruyama and Y. Takeishi, J. Cell. Physiol., 2011, 226, 1554–1563 CrossRef CAS PubMed.
- P. A. Eisenach, P. C. de Sampaio, G. Murphy and C. Roghi, J. Biol. Chem., 2012, 287, 11533–11545 CrossRef CAS PubMed.
- S. Pahwa, M. J. Stawikowski and G. B. Fields, Cancers, 2014, 6, 416–435 CrossRef CAS PubMed.
- K. B. Hotary, E. D. Allen, P. C. Brooks, N. S. Datta, M. W. Long and S. J. Weiss, Cell, 2003, 114, 33–45 CrossRef CAS PubMed.
- S. Gupta and V. Patil, in Matrix Metalloproteinase Inhibitors, ed. S. P. Gupta, Springer Basel, 2012, vol. 103, ch. 2, pp. 35–56 Search PubMed.
- R. E. Vandenbroucke and C. Libert, Nat. Rev. Drug Discovery, 2014, 13, 904–927 CrossRef CAS PubMed.
- V. Pelmenschikov and P. E. M. Siegbahn, Inorg. Chem., 2002, 41, 5659–5666 CrossRef CAS PubMed.
- E. Decaneto, S. Suladze, C. Rosin, M. Havenith, W. Lubitz and R. Winter, Biophys. J., 2015, 109, 2371–2381 CrossRef CAS PubMed.
- L. L. Johnson, A. G. Pavlovsky, A. R. Johnson, J. A. Janowicz, C.-F. Man, D. F. Ortwine, C. F. Purchase II, A. D. White and D. J. Hupe, J. Biol. Chem., 2000, 275, 11026–11033 CrossRef CAS PubMed.
- R. L. Stein and M. Izquierdomartin, Arch. Biochem. Biophys., 1994, 308, 274–277 CrossRef CAS PubMed.
- B. Arza, M. De Maeyer, J. Felez, D. Collen and H. R. Lijnen, Eur. J. Biochem., 2001, 268, 826–831 CrossRef CAS PubMed.
- N. Díaz and D. Suárez, J. Phys. Chem. B, 2008, 112, 8412–8424 CrossRef PubMed.
- T. Vasilevskaya, M. G. Khrenova, A. V. Nemukhin and W. Thiel, J. Comput. Chem., 2015, 36, 1621–1630 CrossRef CAS PubMed.
- B. W. Matthews, Acc. Chem. Res., 1988, 21, 333–340 CrossRef CAS.
- I. Bertini, V. Calderone, M. Fragai, C. Luchinat, M. Maletta and K. J. Yeo, Angew. Chem., Int. Ed., 2006, 45, 7952–7955 CrossRef CAS PubMed.
- A. Solomon, B. Akabayov, A. Frenkel, M. E. Milla and I. Sagi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 4931–4936 CrossRef CAS PubMed.
- T. Crabbe, S. Zucker, M. I. Cockett, F. Willenbrock, S. Tickle, J. P. O'Connell, J. M. Scothern, G. Murphy and A. J. P. Docherty, Biochemistry, 1994, 33, 6684–6690 CrossRef CAS PubMed.
- J. Cha and D. S. Auld, Biochemistry, 1997, 36, 16019–16024 CrossRef CAS PubMed.
- C. Fernandez-Catalan, W. Bode, R. Huber, D. Turk, J. J. Calvete, A. Lichte, H. Tschesche and K. Maskos, EMBO J., 1998, 17, 5238–5248 CrossRef CAS PubMed.
- M. Grossman, D. Tworowski, O. Dym, M.-H. Lee, Y. Levy, G. Murphy and I. Sagi, Biochemistry, 2010, 49, 6184–6192 CrossRef CAS PubMed.
- H. Nar, K. Werle, M. M. T. Bauer, H. Dollinger and B. Jung, J. Mol. Biol., 2001, 312, 743–751 CrossRef CAS PubMed.
- S. P. Gupta, Chem. Rev., 2007, 107, 3042–3087 CrossRef CAS PubMed.
- M. Grossman, B. Born, M. Heyden, D. Tworowski, G. B. Fields, I. Sagi and M. Havenith, Nat. Struct. Mol. Biol., 2011, 18, 1102–1108 CAS.
- E. Decaneto, S. Abbruzzetti, I. Heise, W. Lubitz, C. Viappiani and M. Knipp, Photochem. Photobiol. Sci., 2015, 14, 300–307 CAS.
- U. Neumann, H. Kubota, K. Frei, V. Ganu and D. Leppert, Anal. Biochem., 2004, 328, 166–173 CrossRef CAS PubMed.
- H. Ogata, E. Decaneto, M. Grossman, M. Havenith, I. Sagi, W. Lubitz and M. Knipp, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2014, 70, 232–235 CrossRef CAS PubMed.
- W. Kabsch, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 125–132 CrossRef CAS PubMed.
- A. Vagin and A. Teplyakov, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 22–25 CrossRef CAS PubMed.
- P. D. Adams, P. V. Afonine, G. Bunkoczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L.-W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger and P. H. Zwart, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 213–221 CrossRef CAS PubMed.
- S. C. Lovell, I. W. Davis, W. B. Arendall, P. I. W. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson and D. C. Richardson, Proteins, 2003, 50, 437–450 CrossRef CAS PubMed.
- B. Holmquist and B. L. Vallee, Biochemistry, 1973, 12, 4409–4417 CrossRef CAS PubMed.
- A. I. Nalepa, J. J. Taing, A. Savitsky and M. Knipp, Protein Expression Purif., 2013, 88, 33–40 CrossRef CAS PubMed.
- E. Reijerse, F. Lendzian, R. Isaacson and W. Lubitz, J. Magn. Reson., 2012, 214, 237–243 CrossRef CAS PubMed.
- V. Krewald, M. Retegan, N. Cox, J. Messinger, W. Lubitz, S. DeBeer, F. Neese and D. A. Pantazis, Chem. Sci., 2015, 6, 1676–1695 RSC.
- A. Tochowicz, K. Maskos, R. Huber, R. Oltenfreiter, V. Dive, A. Yiotakis, M. Zanda, W. Bode and P. Goettig, J. Mol. Biol., 2007, 371, 989–1006 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- A. D. MacKerell, D. Bashford, M. Bellott, R. L. Dunbrack, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiórkiewicz-Kuczera, D. Yin and M. Karplus, J. Phys. Chem. B, 1998, 102, 3586–3616 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- J.-P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, J. Comput. Phys., 1977, 23, 327–341 CrossRef CAS.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- P. J. Stephens, F. J. Devlin, C. F. Chabalowski and M. J. Frisch, J. Phys. Chem., 1994, 98, 11623–11627 CrossRef CAS.
- V. A. Rassolov, J. A. Pople, M. A. Ratner and T. L. Windus, J. Chem. Phys., 1998, 109, 1223–1229 CrossRef CAS.
- P. Sherwood, A. H. de Vries, M. F. Guest, G. Schreckenbach, C. R. A. Catlow, S. A. French, A. A. Sokol, S. T. Bromley, W. Thiel, A. J. Turner, S. Billeter, F. Terstegen, S. Thiel, J. Kendrick, S. C. Rogers, J. Casci, M. Watson, F. King, E. Karlsen, M. Sjøvoll, A. Fahmi, A. Schäfer and C. Lennartz, THEOCHEM, 2003, 632, 1–28 CrossRef CAS.
- S. Metz, J. Kästner, A. A. Sokol, T. W. Keal and P. Sherwood, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2014, 4, 101–110 CrossRef CAS.
- R. Ahlrichs, M. Bär, M. Häser, H. Horn and C. Kölmel, Chem. Phys. Lett., 1989, 162, 165–169 CrossRef CAS.
- W. Smith and T. R. Forester, J. Mol. Graphics, 1996, 14, 136–141 CrossRef CAS PubMed.
- S. R. Billeter, A. J. Turner and W. Thiel, Phys. Chem. Chem. Phys., 2000, 2, 2177–2186 RSC.
- F. Neese, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2011, 2, 73–78 CrossRef.
- V. N. Staroverov, G. E. Scuseria, J. Tao and J. P. Perdew, J. Chem. Phys., 2003, 119, 12129 CrossRef CAS.
- F. Neese, Inorg. Chim. Acta, 2002, 337, 181–192 CrossRef.
- D. A. Pantazis, X.-Y. Chen, C. R. Landis and F. Neese, J. Chem. Theory Comput., 2008, 4, 908–919 CrossRef CAS PubMed.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
- C. van Wüllen, J. Chem. Phys., 1998, 109, 392 CrossRef.
- C. Angeli, R. Cimiraglia and J.-P. Malrieu, J. Chem. Phys., 2002, 117, 9138–9153 CrossRef CAS.
- C. Angeli, R. Cimiraglia and J.-P. Malrieu, Theor. Chem. Acc., 2006, 116, 434–439 CrossRef CAS.
- B. A. Hess, C. M. Marian, U. Wahlgren and O. Gropen, Chem. Phys. Lett., 1996, 251, 365–371 CrossRef CAS.
- W. R. English, B. Holtz, G. Vogt, V. Knäuper and G. Murphy, J. Biol. Chem., 2001, 276, 42018–42026 CrossRef CAS PubMed.
- A. M. Woskowicz, S. A. Weaver, Y. Shitomi, N. Ito and Y. Itoh, J. Biol. Chem., 2013, 288, 35126–35137 CrossRef CAS PubMed.
- R. Lang, M. Braun, N. E. Sounni, A. Noel, F. Frankenne, J. M. Foidart, W. Bode and K. Maskos, J. Mol. Biol., 2004, 336, 213–225 CrossRef CAS PubMed.
- H. Nagase and G. B. Fields, Biopolymers, 1996, 40, 399–416 CrossRef CAS PubMed.
- I. Bertini, V. Calderone, M. Fragai, C. Luchinat, S. Mangani and B. Terni, Angew. Chem., Int. Ed. Engl., 2003, 42, 2673–2676 CrossRef CAS PubMed.
- L. Devel, F. Beau, M. Amoura, L. Vera, E. Cassar-Lajeunesse, S. Garcia, B. Czarny, E. A. Stura and V. Dive, J. Biol. Chem., 2012, 287, 26647–26656 CrossRef CAS PubMed.
- E. Nuti, D. Cuffaro, F. D'Andrea, L. Rosalia, L. Tepshi, M. Fabbi, G. Carbotti, S. Ferrini, S. Santamaria, C. Camodeca, L. Ciccone, E. Orlandini, S. Nencetti, E. A. Stura, V. Dive and A. Rossello, ChemMedChem, 2016, 11, 1626–1637 CrossRef CAS PubMed.
- B. D. Belviso, R. Caliandro, D. Siliqi, V. Calderone, F. Arnesano and G. Natile, Chem. Commun., 2013, 49, 5492–5494 RSC.
- J. A. Larrabee, C. M. Alessi, E. T. Asiedu, J. O. Cook, K. R. Hoerning, L. J. Klingler, G. S. Okin, S. G. Santee and T. L. Volkert, J. Am. Chem. Soc., 1997, 119, 4182–4196 CrossRef CAS.
- W. Maret and B. L. Vallee, Methods Enzymol., 1993, 226, 52–71 CAS.
- B. Bennett, in Metals in Biology: Applications of High-Resolution EPR to Metalloenzymes, ed. G. Hanson and L. Berliner, Springer New York, New York, NY, 2010, pp. 345–370 DOI:10.1007/978-1-4419-1139-1_10.
- S. P. Salowe, A. I. Marcy, G. C. Cuca, C. K. Smith, I. E. Kopka, W. K. Hagmann and J. D. Hermes, Biochemistry, 1992, 31, 4535–4540 CrossRef CAS PubMed.
- I. Bertini, M. Fragai, Y. M. Lee, C. Luchinat and B. Terni, Angew. Chem., Int. Ed., 2004, 43, 2254–2256 CrossRef CAS PubMed.
- N. Cox, A. Nalepa, M.-E. Pandelia, W. Lubitz and A. Savitsky, in Method Enzymol., ed. Z. Q. Peter and W. Kurt, Academic Press, 2015, vol. 563, pp. 211–249 Search PubMed.
- I. Bertini, V. Calderone, M. Cosenza, M. Fragai, Y. M. Lee, C. Luchinat, S. Mangani, B. Terni and P. Turano, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 5334–5339 CrossRef CAS PubMed.
- P. J. Hajduk, G. Sheppard, D. G. Nettesheim, E. T. Olejniczak, S. B. Shuker, R. P. Meadows, D. H. Steinman, G. M. Carrera, P. A. Marcotte, J. Severin, K. Walter, H. Smith, E. Gubbins, R. Simmer, T. F. Holzman, D. W. Morgan, S. K. Davidsen, J. B. Summers and S. W. Fesik, J. Am. Chem. Soc., 1997, 119, 5818–5827 CrossRef CAS.
- B. L. Vallee and R. J. P. Williams, Proc. Natl. Acad. Sci. U. S. A., 1968, 59, 498–505 CrossRef CAS.
- F. Tonello, G. Schiavo and C. Montecucco, Biochem. J., 1997, 322, 507–510 CrossRef CAS PubMed.
- C. E. Sabel, R. Carbone, J. R. Dabous, S. Y. Lo and S. Siemann, Biochem. Biophys. Res. Commun., 2011, 416, 106–110 CrossRef PubMed.
- J. A. Larrabee, C. H. Leung, R. L. Moore, T. Thamrong-nawasawat and B. S. H. Wessler, J. Am. Chem. Soc., 2004, 126, 12316–12324 CrossRef CAS PubMed.
- R. Bicknell, G. R. Hanson, B. Holmquist and C. Little, Biochemistry, 1986, 25, 4219–4223 CrossRef CAS PubMed.
- D. C. Brown and K. D. Collins, J. Biol. Chem., 1991, 266, 1597–1604 CAS.
- J. B. Bjarnason and J. W. Fox, Biochemistry, 1983, 22, 3770–3778 CrossRef CAS PubMed.
- S. L. Rose, L. C. Dickinson and E. W. Westhead, J. Biol. Chem., 1984, 259, 4405–4413 CAS.
- G. S. Baldwin, A. Galdes, H. A. O. Hill, S. G. Waley and E. P. Abraham, J. Inorg. Biochem., 1980, 13, 189–204 CrossRef CAS PubMed.
- B. Holmquist, T. A. Kaden and B. L. Vallee, Biochemistry, 1975, 14, 1454–1461 CrossRef CAS PubMed.
- W. Stocker, R. L. Wolz, R. Zwilling, D. J. Strydom and D. S. Auld, Biochemistry, 1988, 27, 5026–5032 CrossRef.
- B. Holmquist and B. L. Vallee, J. Biol. Chem., 1974, 249, 4601–4607 CAS.
- B. L. Vallee, R. T. Simpson, R. D. Kobes, R. W. Erbe and W. J. Rutter, Biochemistry, 1971, 10, 2466–2470 CrossRef CAS.
- W. Maret, I. Andersson, H. Dietrich, H. Schneiderbernlohr, R. Einarsson and M. Zeppezauer, Eur. J. Biochem., 1979, 98, 501–512 CrossRef CAS PubMed.
- A. J. Sytkowski and B. L. Vallee, Biochemistry, 1978, 17, 2850–2857 CrossRef CAS PubMed.
- S. A. Latt and B. L. Vallee, Biochemistry, 1971, 10, 4263–4270 CrossRef CAS PubMed.
- F. A. Cotton and R. H. Soderberg, J. Am. Chem. Soc., 1962, 84, 872–873 CrossRef CAS.
- F. F. Morpeth and V. Massey, Biochemistry, 1982, 21, 1318–1323 CrossRef CAS PubMed.
- M. Vasak and J. H. R. Kagi, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 6709–6713 CrossRef CAS.
- W. Maret, Biochemistry, 1989, 28, 9944–9949 CrossRef CAS PubMed.
- I. Bertini, V. Calderone, L. Cerofolini, M. Fragai, C. F. G. C. Geraldes, P. Hermann, C. Luchinat, G. Parigi and J. M. C. Teixeira, FEBS Lett., 2012, 586, 557–567 CrossRef CAS PubMed.
- L. Banci, A. Bencini, C. Benelli, D. Gatteschi and C. Zanchini, Structures versus Special Properties, Springer Berlin Heidelberg, Berlin, Heidelberg, 1982, pp. 37–86 DOI:10.1007/BFb0111296.
- L. C. Kuo and M. W. Makinen, J. Am. Chem. Soc., 1985, 107, 5255–5261 CrossRef CAS.
- M. W. Makinen, L. C. Kuo, M. B. Yim, G. B. Wells, J. M. Fukuyama and J. E. Kim, J. Am. Chem. Soc., 1985, 107, 5245–5255 CrossRef CAS.
- C. J. Walsby, D. Krepkiy, D. H. Petering and B. M. Hoffman, J. Am. Chem. Soc., 2003, 125, 7502–7503 CrossRef CAS PubMed.
- N. Cox, W. Lubitz and A. Savitsky, Mol. Phys., 2013, 111, 2788–2808 CrossRef CAS.
- X. Tan, M. Bernardo, H. Thomann and C. P. Scholes, J. Chem. Phys., 1993, 98, 5147–5157 CrossRef CAS.
- H. S. Shafaat, J. J. Griese, D. A. Pantazis, K. Roos, C. S. Andersson, A. Popović-Bijelić, A. Gräslund, P. E. M. Siegbahn, F. Neese, W. Lubitz, M. Högbom and N. Cox, J. Am. Chem. Soc., 2014, 136, 13399–13409 CrossRef CAS PubMed.
- R. P. Verma, in Matrix Metalloproteinase Inhibitors: Specificity of Binding and Structure-Activity Relationships, ed. P. S. Gupta, Springer Basel, Basel, 2012, pp. 137–176 DOI:10.1007/978-3-0348-0364-9_5.
Footnotes |
| † Electronic supplementary information (ESI) available: (S1) Expression and purification of the active catalytic domain of MT1-MMP; (S2) molar extinction coefficient determination for wild type MT1-MMP; (S3) preparation of apo-MT1-MMP (metal-free material)/preparation of Co2+-MT1-MMP; (S4) inhibition of Co2+-MT1-MMP by acetohydroxamic acid (AHA); (S5) X-ray data-collection statistics for MT1-MMP; (S6) additional crystallographic figures. Comparison of the X-ray structure of MT1-MMP to literature MMP structures; (S7) analysis of Co2+ EPR and MCD spectra from wild type and inhibited Co2+-MT1-MMP; (S8) QM/MM geometry optimizations of structural models A-K; (S9) comparison of computed key distances in Co2+ and Zn2+ QM/MM models; (S10) optimized QM/MM geometries (QM part); (S11) optimized model geometries used for EPR parameter calculations; (S12) calibration of computed cobalt isotropic hyperfine coupling constants; (S13) references. See DOI: 10.1039/c7cp05572b |
| ‡ ED (biochemistry), TV (computations), YK (EPR) and HO (crystallography) made complementary and equal contributions to the manuscript. |
| This journal is © the Owner Societies 2017 |