Computational design of cyclic peptides for the customized oriented immobilization of globular proteins†
Abstract
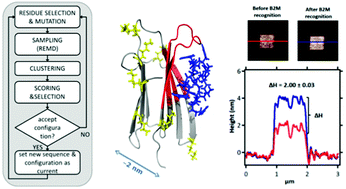
The oriented immobilization of proteins, key for the development of novel responsive biomaterials, relies on the availability of effective probes. These are generally provided by standard approaches based on in vivo maturation and in vitro selection of antibodies and/or aptamers. These techniques can suffer technical problems when a non-immunogenic epitope needs to be targeted. Here we propose a strategy to circumvent this issue by in silico design. In our method molecular binders, in the form of cyclic peptides, are computationally evolved by stochastically exploring their sequence and structure space to identify high-affinity peptides for a chosen epitope of a target globular protein: here a solvent-exposed site of β2-microglobulin (β2m). Designed sequences were screened by explicit solvent molecular dynamics simulations (MD) followed by experimental validation. Five candidates gave dose–response surface plasmon resonance signals with dissociation constants in the micromolar range. One of them was further analyzed by means of isothermal titration calorimetry, nuclear magnetic resonance, and 250 ns of MD. Atomic-force microscopy imaging showed that this peptide is able to immobilize β2m on a gold surface. In short, we have shown by a variety of experimental techniques that it is possible to capture a protein through an epitope of choice by computational design.


 Please wait while we load your content...
Please wait while we load your content...