Oligomerization of FVFLM peptides and their ability to inhibit beta amyloid peptides aggregation: consideration as a possible model†
Abstract
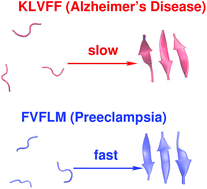
Preeclampsia, a pregnancy-specific disorder, shares typical pathophysiological features with protein misfolding disorders including Alzheimer's disease. Characteristic for preeclampsia is the involvement of multiple proteins of which fragments of SERPINA1 and β-amyloid co-aggregate in urine and placenta of preeclamptic women. To explore the biophysical basis of this interaction, we investigated the multidimensional efficacy of the FVFLM sequence in SERPINA1, as a model inhibitory agent of β-amyloid aggregation. After studying the oligomerization of FVFLM peptides using all-atom molecular dynamics simulations with the GROMOS43a1 force field and explicit water, we report that FVFLM can aggregate and its aggregation is spontaneous with a remarkably faster rate than that recorded for KLVFF (aggregation “hot-spot” from β-amyloid). The fast kinetics of FVFLM aggregation was found to be driven primarily by core-like aromatic interactions originating from the anti-parallel orientation of complementarily uncharged strands. The conspicuously stable aggregation mechanism observed for FVFLM peptides is found not to conform to the popular 'dock-lock' scheme. We also found high propensity of FVFLM for KLVFF binding. When present, FVFLM disrupts the β-amyloid aggregation pathway and we propose that FVFLM-like peptides might be used to prevent the assembly of full-length Aβ or other pro-amyloidogenic peptides into amyloid fibrils.


 Please wait while we load your content...
Please wait while we load your content...