Open Access Article
Open Access ArticleDirect screening for ribozyme activity in mammalian cells†
Yoko
Nomura
 ,
Hsiao-Chiao
Chien
,
Hsiao-Chiao
Chien
 and
Yohei
Yokobayashi
and
Yohei
Yokobayashi
 *
*
Nucleic Acid Chemistry and Engineering Unit, Okinawa Institute of Science and Technology Graduate University, Onna, Okinawa 904 0495, Japan. E-mail: yohei.yokobayashi@oist.jp
First published on 1st November 2017
Abstract
Engineered ribozymes are powerful tools for manipulating gene expression in living cells. However, identification of active ribozymes in mammalian cells has relied on empirical assays of a limited number of arbitrarily chosen ribozymes. Here, we synthesized 376 natural and 2625 synthetic variants of pistol ribozymes, and screened for active variants directly in mammalian cells, greatly expanding the ribozyme toolbox for biological applications.
Although many of their biological roles remain elusive, self-cleaving ribozymes have been found in viruses and in all kingdoms of life.1 The well-defined and compact structures of self-cleaving ribozymes have inspired chemists to engineer ribozymes with new functions such as allosteric response.2 Some of these engineered ribozymes have been shown to function in living cells to control gene expression.3 However, these efforts also revealed that many ribozymes do not always function as expected when the context changes, for example, from in vitro to cells, or from bacteria to mammalian cells. This may be due to the sensitivity of ribozymes to chemical environment (pH, metal ions, etc.) or the neighbouring sequences. On the other hand, there are thousands of self-cleaving ribozymes that have evolved to function in diverse organisms living in different environments1 which should serve as a rich source of ribozymes for biological applications.
However, exploiting this natural diversity is complicated by the difficulty associated with screening a large number of different ribozyme sequences for their catalytic activity in living cells of interest. This is especially the case with mammalian cells in which “one cell-one ribozyme” library generation and screening is technically demanding. Chen et al. reported direct selection of ribozyme activity in mammalian cells.4 However, this method requires complex RNA/DNA manipulations including ligation of an adapter DNA to minute quantities of self-cleaved ribozymes in the presence of complex cellular RNAs, as well as multiple rounds of selection. They also discovered that the low stringency of intracellular selection resulted in inefficient selection.
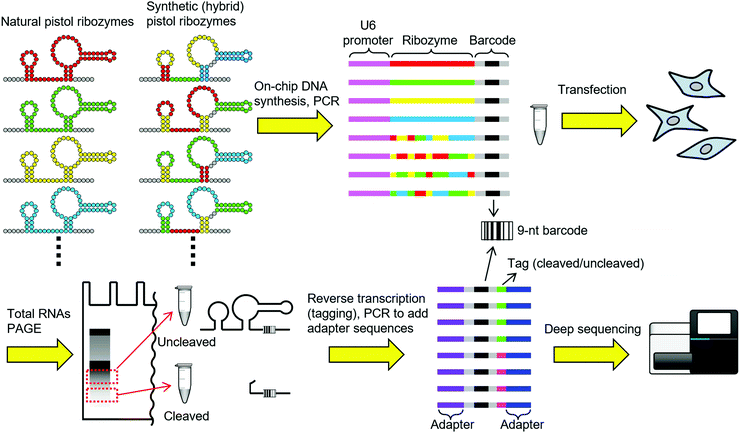
Here, we report a general methodology to screen thousands of arbitrary ribozyme sequences for activity in mammalian cells. Our strategy (Fig. 1) is to first generate a library of ribozymes of natural origin or designed sequences by on-chip parallel DNA synthesis. Each DNA sequence encodes a ribozyme and a unique 9-nt barcode sequence associated with the ribozyme. To minimize any potential interference between the ribozyme and the barcode sequences, each ribozyme is represented by 4 distinct barcodes. Next, the mammalian U6 promoter sequence is attached upstream of the synthetic DNA pool by PCR to obtain a linear dsDNA library. The dsDNA library is then transfected into mammalian cells, and the cells are allowed to transcribe the ribozyme ensemble. After 6 h of incubation, total RNAs are extracted from the cells and separated by polyacrylamide gel electrophoresis (PAGE). The parts of the gel that correspond to the sizes of uncleaved and cleaved (the 3′ fragment) transcripts are excised and RNAs are recovered. The RNAs are then separately reverse transcribed with primers containing different tag sequences, combined, and PCR amplified to add adapter sequences for deep sequencing. By counting the number of barcodes from the uncleaved and the cleaved populations, the ribozymes can be sorted according to the apparent intracellular activity.
To demonstrate and optimize this strategy, we chose to screen pistol ribozymes.5 Pistol ribozymes were discovered by bioinformatic search of genome sequences by the Breaker group.5a We recently engineered allosteric ribozymes (aptazymes) by fusing a guanine aptamer in tandem with a pistol ribozyme in vitro.6 However, we discovered that this particular pistol ribozyme did not function well in the human embryonic kidney cell line HEK293. To our knowledge, pistol ribozymes have not been shown to function in living cells outside their native context. Therefore, discovery of pistol ribozymes that function in a heterologous host may enable further biological applications of pistol ribozymes.
Weinberg et al. identified 676 pistol sequences from the genomes of various organisms and environmental samples.5a After removing duplicated sequences of the core ribozyme, we obtained 376 unique pistol ribozyme sequences. While the genomes of natural organisms are useful sources of ribozyme sequences, DNA synthesis is not limited by what are available in nature. Therefore, we designed synthetic variants of pistol ribozymes (sp-NNNNN) by shuffling the structural elements of the natural pistol ribozymes. Specifically, we selected 3–7 unique sequences in each of the stems P1, P2, P3, the linker J1, and the L1–L2 pseudoknot based on the frequencies in natural ribozymes and sequence diversity (Table S1, ESI†). These structural elements were then combinatorially fused to design synthetic pistol variants (Fig. 2). In total, we designed 376 natural and 2625 synthetic pistol ribozyme sequences each associated with 4 distinct barcodes, and synthesized them as a pool of DNAs.
After addition of the U6 promoter sequence, transfection, RNA extraction, and sequencing library preparation as described above, the sample was analyzed by MiSeq (Illumina). By counting the number of barcode reads from the cleaved (nclv) and uncleaved (nunclv) populations, apparent fraction cleaved (FC) values were calculated for all ribozyme variants as: FC = nclv/(nclv + nunclv). The FC values ranged from 0.312 to 0.653, which was somewhat narrower than we expected. It should be noted that there are many factors that may alter the FC values from the true intracellular cleavage efficiency. For example, differences in the RNA stability or recovery efficiency of the cleaved and the uncleaved fragments can affect their relative abundance. Other steps during the library preparation such as reverse transcription and PCR could also introduce additional bias. Furthermore, errors in oligonucleotide synthesis can lower the fraction of active ribozymes. Nevertheless, we proceeded to evaluate some individual variants to examine the correlation between FC and conventional reporter gene assay.
We chose the top 10 and the worst 5 pistol variants according to the deep sequencing based FC values. In addition, we chose 8 variants with intermediate FC values (Table S3, ESI†). Each ribozyme sequence was synthesized and inserted into the 3′ UTR of a fluorescent protein (EGFP) reporter gene in a plasmid. When transfected in mammalian cells, high ribozyme activity results in the scission of mRNA and repression of EGFP expression.
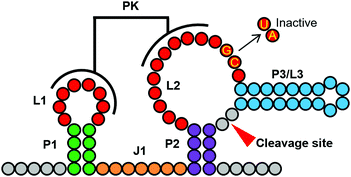
As shown in Fig. 3A, the top 10 ribozymes consistently downregulate EGFP expression by ∼90%, and the worst 5 ribozymes all show EGFP expression comparable to that of the empty (no ribozyme) plasmid. There is some variability in the intermediate FC range. However, it appears that ribozymes with FC above a threshold (e.g. ∼0.60 in this case) are highly likely to be active in mammalian cells. To ensure that the observed EGFP repression is due to the ribozyme activity and not due to the inserted sequence, we mutated the two conserved GC bases in L2 to UA (Fig. 2) in sp-11251, sp-11141, and sp-56441. Crystallographic studies have shown that these nucleotides are important for catalysis with the guanine functioning as a general base to abstract the proton from the 2′-OH group of the nucleotide where the scission occurs.5c,d Consequently, these mutations abolish the ribozyme activity.5a As expected, the mutations significantly increased EGFP levels (Fig. 3B).
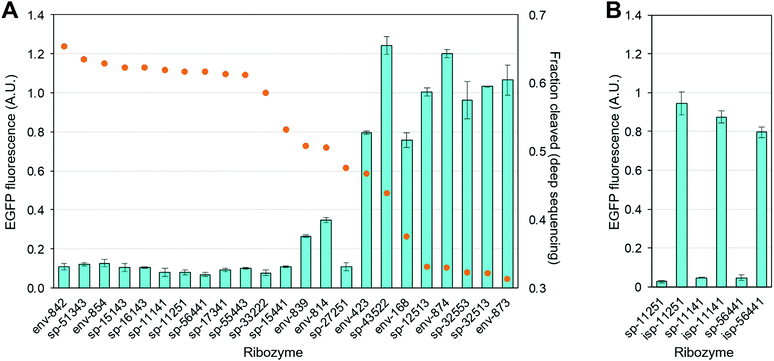 | ||
| Fig. 3 Activities of pistol ribozymes in mammalian cells. See ESI,† for experimental details. (A) Ribozyme activity based on EGFP assay in HEK293 cells (light blue bars) and FC values derived from deep sequencing (orange circles). EGFP fluorescence of HEK293 cells transfected with an EGFP–ribozyme plasmid was normalized to the fluorescence of the cells transfected with an empty (no ribozyme) plasmid. (B) Inactivation of the ribozyme by two-base mutation (isp-NNNNN) restores EGFP expression in 3 selected ribozymes. | ||
Some structural elements are more conserved among the most active (top 10) variants identified by sequencing. For example, 7/10 and 8/10 share the same P1 and P2 sequences, respectively (Table S3, ESI†). Moreover, all 5 of the least active variants contain 5- or 6-base linker (J1) whereas the most active variants all contain 7 or 8 bases (Table S3, ESI†).
Another interesting observation is that many of the synthetic pistol ribozymes are indeed active. Eight out of the top 10 sequences are of synthetic origin. Consequently, this “structural element shuffling” may be an effective strategy to expand the repertoire of ribozymes beyond the naturally occurring sequences for various applications.
Our previous deep sequencing assays of ribozymes in vitro were limited to localized randomization or statistical doping of a reference sequence.6–8 The cleavage site also affected where the mutations could occur and be analyzed. The use of on-chip parallel DNA synthesis and barcoding of each ribozyme sequence allowed us to study arbitrary sequences regardless of the cleavage pattern. Therefore, this approach significantly improves the generality of the deep sequencing assay of ribozyme activity.
In summary, we developed an efficient method to rapidly and directly screen for ribozyme activity in mammalian cells. We also described a new strategy to design novel ribozyme variants by shuffling structural elements of natural ribozymes to create a large set of synthetic variants. Together, these results should advance our ability to engineer ribozymes with new functions in mammalian and other cells.
The research was supported by Okinawa Institute of Science and Technology (OIST) Graduate University. We also thank OIST DNA Sequencing Section for technical support on deep sequencing.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) A. R. Ferré-D'Amaré and W. G. Scott, Cold Spring Harbor Perspect. Biol., 2010, 2, a003574 CrossRef PubMed; (b) R. M. Jimenez, J. A. Polanco and A. Lupták, Trends Biochem. Sci., 2015, 40, 648–661 CrossRef CAS PubMed.
- (a) R. R. Breaker, Curr. Opin. Biotechnol., 2002, 13, 31–39 CrossRef CAS PubMed; (b) R. Penchovsky, Biotechnol. Adv., 2014, 32, 1015–1027 CrossRef CAS PubMed.
- M. Felletti and J. S. Hartig, WIREs RNA, 2017, 8, e1395 CrossRef PubMed.
- X. Chen, L. Denison, M. Levy and A. D. Ellington, RNA, 2009, 15, 2035–2045 CrossRef CAS PubMed.
- (a) Z. Weinberg, P. B. Kim, T. H. Chen, S. Li, K. A. Harris, C. E. Lünse and R. R. Breaker, Nat. Chem. Biol., 2015, 11, 606–610 CrossRef CAS PubMed; (b) K. A. Harris, C. E. Lünse, S. Li, K. I. Brewer and R. R. Breaker, RNA, 2015, 21, 1852–1858 CrossRef CAS PubMed; (c) L. A. Nguyen, J. Wang and T. A. Steiz, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 1021–1026 CrossRef CAS PubMed; (d) A. Ren, N. Vušurović, J. Gebetsberger, P. Gao, M. Juen, C. Kreutz, R. Micura and D. J. Patel, Nat. Chem. Biol., 2016, 12, 702–708 CrossRef CAS PubMed.
- S. Kobori, K. Takahashi and Y. Yokobayashi, ACS Synth. Biol., 2017, 6, 1283–1288 CrossRef CAS PubMed.
- S. Kobori, Y. Nomura, A. Miu and Y. Yokobayahsi, Nucleic Acids Res., 2015, 43, e85 CrossRef PubMed.
- S. Kobori and Y. Yokobayashi, Angew. Chem., Int. Ed., 2016, 55, 10354–10357 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, sequence information, and deep sequencing results. See DOI: 10.1039/c7cc07815c |
| This journal is © The Royal Society of Chemistry 2017 |