The design of a mechanical wave-like DNA nanomachine for the fabrication of a programmable and multifunctional molecular device†
Abstract
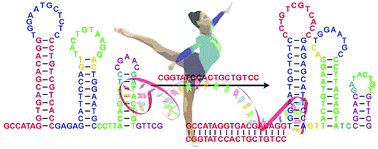
We report a novel mechanical wave-like DNA nanomachine. A successive stem-loop structure is involved, which will rearrange successively from one side to the opposite side upon binding with an input activator just like the motion of a mechanical wave, and thereby achieving diverse functions.


 Please wait while we load your content...
Please wait while we load your content...