DOI:
10.1039/C7AY01603D
(Technical Note)
Anal. Methods, 2017,
9, 5697-5702
Amino acid profiling from fingerprints, a novel methodology using UPLC-MS†
Received
3rd July 2017
, Accepted 7th August 2017
First published on 8th August 2017
Abstract
Fingermark evidence is extensively used in criminal investigations. Hence, there have been many investigations into the chemical compounds present in fingerprint deposits. In this technical note we describe the analysis of non-derivatised amino acid profiles obtained from fingerprints. We used UPLC with an amide stationary phase and subsequent detection using a triple quadrupole MS/MS and TOF-MS detector. The linearity (R2) was satisfactory for both MS detectors (>0.98 for all amino acids in the case of the triple quadrupole MS/MS and >0.96 in the case of the TOF-MS). Although the triple quadrupole had a higher sensitivity for most amino acids, both mass spectrometers were able to retrieve the amino acid profiles of fingerprints from 19 donors. Between these profiles, only minor differences were observed between the separate analyses on the different mass analyzers, mainly in L-proline, L-lysine and L-phenylalanine abundances. Surprisingly, the mean RSD in amino acid profiles from duplicate fingerprints turned out to be lower for the TOF-MS (18.6% ± 6.6% vs. 13.2% ± 3.8%), as did the mean RSD of the intraday reproducibility (8.22% ± 1.94% vs. 9.54% ± 3.07%).
Introduction
Fingermarks make up an important part of criminal investigations. The deposition is regarded as a proof of contact, with the odd exception, and the individualization has proven to contain a strong evidential value.1 And as such the fingermark found at a crime scene is regarded as an incriminating piece of evidence for the donor of the mark. The chemical composition of fingerprints has been investigated extensively, as comprehensively described by Girod et al. and Cadd et al.2,3 The analysis of the chemical compounds, in particular amino acids, has been investigated several times in the near past. Croxton et al. published the use of gas chromatography coupled with mass spectrometry (GCMS) for amino and fatty acids.4,5 Previously Girod and Weyermann have described the use of fatty acid analysis for donor classification,6 and recently Delgado-Povedano et al. described a method for enhancing metabolomics from sweat.7 By extracting the fatty components from fingerprints using CHCl3, Girod et al. succeeded in obtaining full profiles of a substantial amount of fingerprints. Even though it appeared later that the extraction methods used may not have been the optimum conditions,7,8 Girod et al. have shown the great potential of chemical profiling of fingerprints on the basis of exogenous and endogenous materials. And even though the fatty acids, squalene, cholesterol and triglycerides found in fingerprints may not be directly related to the finger or palm deposition, more information on what has been touched previous to the crime related deposition can be invaluable. One example was described by Weyermann et al., where the disappearance, or chemical breakdown, of certain compounds in a fingerprint is used as an indication for the age of a fingerprint.9 Liquid chromatography coupled with mass spectrometry (LCMS) has been applied previously for the analysis of amino acids10 and non-polar materials in fingerprints.11 Although the duration of the separation of amino acids in these first efforts is not optimal, parameters such as the resolution, repeatability and linearity are within an acceptable range. Only one attempt on the non-derivatised separation of amino acids from fingerprints, using electrophoresis, has been described.12 Although the separation of the analytes was satisfactory, the sensitivity of the separation method in combination with mass spectrometric detection was unsatisfactory. In this technical note, we describe the development of a method for the extraction and analysis of amino acids from fingerprints using ultra-high performance liquid chromatography (UHPLC or UPLC) with an amide stationary phase. Secondly, we will describe the differences between the use of a tandem quadrupole (QqQ) MS/MS and a time-of-flight (TOF) MS; both detectors are frequently used in metabolomic profiling studies.13 The TOF-MS offers high resolution and mass accuracy, whereas the triple quadrupole MS/MS provides structural information. The compared methods have been used for the analysis of the amino acid profiles from the fingerprints of 19 donors.
Materials and methods
Solvents and solutions
Amino acids L-alanine (98%), L-lysine (97%), L-leucine (98%), L-glutamic acid (99%), L-glutamine (99%), L-tyrosine hydrochloride monohydrate (98%), L-histidine hydrochloride monohydrate (98%), L-ornithine hydrochloride monohydrate (99%), hydroxy-L-proline (99%), L-asparagine (98%), L-threonine (98%), L-isoleucine (98%), L-serine (99%), L-methionine (98%), L-tryptophan (98%), L-valine (98%), L-phenylalanine (Phe, 98%), L-arginine hydrochloride (98%), L-cystine (98%), L-aspartic acid (98%) and L-proline (99%) were purchased from Sigma-Aldrich (Zwijndrecht, the Netherlands). Isotope labelled amino acids L-glutamic acid D5 (98.5 atom% D), L-glutamine D5 (98.8 atom% D), L-histidine hydrochloride monohydrate D3 (98.4 atom% D), L-threonine D2 (98.8 atom% D), L-serine D7 (98.6 atom% D), DL-cystine D6 (99.5 atom% D), hydroxy-L-proline D3 (99.2 atom% D), DL-valine D8 (99.1 atom% D) and L-ornithine D6 hydrochloride (99.1 atom% D) were purchased from CDN isotopes (Pointe-Claire, Canada). L-Aspartic acid D3 (98 atom% D), L-lysine D4 (98 atom% D), L-leucine D10 (98 atom% D), L-phenylalanine D5 (98 atom% D) and L-asparagine D8 (97 atom% D) were purchased from Sigma-Aldrich (Zwijndrecht, the Netherlands). L-Tyrosine D2 (98 atom% D), L-tryptophan D5 (97 atom% D) and L-methionine D3 (98 atom% D) were purchased from Isotec (Zwijndrecht, the Netherlands). L-Isoleucine 13C6 (99%) 15N (99%), L-alanine 13C3 (97–99%) D4 (97–99%) 15N (97–99%), L-arginine hydrochloride D7 (98%) 15N4 (98%), and L-proline 13C5 (99%) 15N (99%) were purchased from Cambridge Isotope Laboratories (Tewksbury, MA, USA). UPLC-grade acetonitrile, methanol and formic acid were purchased from Biosolve (Valkenswaard, the Netherlands).
A mixture of 21 amino acids was used to identify and quantify the amino acids with UPLC-MS. Every amino acid was individually dissolved in MeOH (containing 5 v/v% formic acid), making up a stock solution with a concentration of 100, 250 or 500 mg L−1. An amino acid working solution of 2.0 mg L−1 of the 21 amino acids was prepared by adding the required volumes of stock solutions of each individual amino acid, followed by dilution with MeOH (containing 5% v/v formic acid) to a final volume of 25 mL. Similarly, an internal standard working solution of 2.0 mg L−1 of the isotope labelled internal standards was prepared. A calibration range from 0.10 to 1.60 mg L−1 was produced, which corresponds to 5–80 ng of a single amino acid in a sample volume (50 μL) per fingerprint. These solutions were prepared by adding 2.5, 5, 10, 15, 30, and 40 μL of 2.0 mg L−1 working solution into a glass screw neck injection vial. Internal standard solution (20 μL) was added to each vial, and MeOH containing 5 v/v% formic acid was added to the total volume of 100 μL. Fingerprints were collected from the right and left hand index fingers from 19 donors, 10 females and 9 males, ranging from 20 to 66 years old (Table S1†). Donors were asked to wear nitrile gloves for 10 minutes and rub their hands together prior to the print deposition, to create homogeneity in the composition on both hands of the donor. Fingerprints were collected on 2.5 × 5 cm pieces of aluminum foil. Aluminum foil was chosen as a substrate as it is non-porous and flexible and hence it is convenient in the sample preparation process. The aluminum foil sheets were transferred into a 15 mL polypropylene conical tube (Fisherbrand) and internal standard solution (20 μL) was added. MeOH (2 mL, containing 5% v/v formic acid) was added and the solution was mixed using a vortex. After mixing, the aluminum foil was removed from the tube and the sample solution was evaporated under nitrogen flux. The material was dissolved in MeOH (100 μL, containing 5% v/v formic acid) and then transferred into an injection vial. A blank sample was prepared by the addition of 25 μL MeOH solvent to a clean aluminum foil sheet and a positive control was prepared by adding 25 μL of 2.0 mg L−1 amino acid working solution to a clean aluminum foil sheet. After drying in air, these samples were processed as described above.
UPLC-TOF MS
The separation was carried out using a 150 mm UPLC ethylene bridged hybrid (BEH) amide column (Waters, Milford, MA, USA). An Aqcuity I-class UPLC autosampler and binary solvent pump (Waters, Milford, MA, USA) were used to inject and elute the sample solution. The used flow rate was 0.500 mL min−1. The eluent of the column was sprayed into the ion source of the time-of-flight MS by electrospray ionization. The solvents used for LC were (A) 0.4% formic acid in acetonitrile (ACN) and (B) 0.4% formic acid in MilliQ. The gradient applied was 95% A for 3 minutes followed by a linear gradient from 5 to 50% B in 19 minutes and then 1 minute of 50% B. Finally, the column was reconditioned for 2 minutes with 95% A (the total run time for each sample was 25 minutes). For each analysis 2.5 μL of sample was injected. All samples were analyzed on an accurate mass TOF with a dual electrospray source (ESI) (Agilent 6220, Santa Clara, CA, USA). The system was operated in the positive ion mode and MS spectra from m/z 40–1200 were acquired at a resolution of 7500 at m/z 400. The spectra were acquired at a rate of 1 spectrum per second. The capillary voltage was set at 3.5 kV, the source gas temperature was set at 325 °C and a drying gas flow of 5 L min−1 was used. The nebulizer pressure used was 30 psig. The fragmentor, skimmer and octapole 1 RF voltages were set at 160, 65 and 250 V, respectively. MS full scan data were acquired with Agilent Mass Hunter Data Acquisition software (version B.04.00) and data were processed using Agilent Mass Hunter Qualitative Analysis software (version B.05.00) and Quantitative Analysis software (version B.05.00).
UPLC-QqQ MS/MS
The UPLC conditions were identical to those of the UPLC-TOF MS method described above. All samples were analyzed on a tandem quadrupole mass spectrometer (MicroMass Quattro Premier XE, Waters, Milford, MA, USA) which was operated in positive electrospray ionization (ESI) mode. The ESI-parameters were: capillary voltage 0.5 kV; source temperature 130 °C; desolvation gas temperature 400 °C at a flow rate of 1000 L h−1 (N2); and cone gas flow rate 100 L h−1 (N2). Argon was used as the collision gas at a flow rate of 0.27 mL min−1. Multiple reaction monitoring (MRM) transitions, cone voltages and collision energies for all analytes and internal standards are summarized in Table S2.† The data analysis software was MassLynx (version 4.1) and QuanLynx.
Fingerprint volunteers
For the experiments the fingerprints were donated voluntarily by colleagues, who all gave informed consent. No ethical approval was obtained as the material is gathered in a non-invasive manner and did not infringe on any privacy of the donors; no images were taken of the fingerprints. All experiments were carried out following institutional guidelines and are in compliance with relevant laws.
Results
The extracted ion chromatograms, as shown in Fig. 1, present an overview of the separation of the non-derivatised amino acid mixture in 17 minutes. For both the triple quadrupole MS/MS and the TOF-MS the limit of detection (LOD), limit of quantification (LOQ), linearity (R2) and intraday reproducibility were determined for all amino acids (Table 1). In the case of the triple quadrupole, the linearity for all amino acids exceeded 0.99 except for L-methionine and L-phenylalanine (>0.98). For the TOF-MS, L-iso-/leucine, L-lysine and L-methionine were the only amino acids with a linearity not exceeding 0.99 (0.988, 0.987 and 0.964, respectively). The respective LODs and LOQs achieved with the triple quadrupole as the detector were found to be lower for all amino acids, except L-alanine, L-phenylalanine, L-tryptophan, L-tyrosine, L-iso-/leucine and hydroxy-L-proline, than achieved with the TOF-MS. The intraday reproducibility determined for the 21 amino acids was found to be comparable in both cases. Overall, the mean intraday RSD of the peak areas was 9.54% ± 3.07% for the triple quadrupole and 8.22% ± 1.94% for the TOF-MS. The extraction efficiency of the amino acids was calculated and in general was between 96 and 117 percent (Table S3†). The fact that efficiencies higher than 100% are found might be due to methanol evaporation, slightly concentrating the samples. The extraction efficiency of L-methionine is somewhat lower (89.1%), which may be caused by the oxidation of L-methionine to L-methionine sulfoxide, although this was not investigated any further. Next, the amino acid profiles (relative amino acid abundances to serine, the most abundant amino acid in fingerprints)2 from fingerprints from 19 donors were analyzed on both mass spectrometers (Fig. 2). Four amino acids (L-cystine, L-methionine, L-tyrosine, and hydroxy-L-proline) were excluded from further analysis, as they were unsuccessfully quantified in either method. In general, the profiles obtained from the 19 donors appear to be very similar. However, some striking differences were found in the abundances of L-proline, L-lysine and L-asparagine, which appear to be slightly higher when analyzed with the triple quadrupole. The abundance of L-phenylalanine however, appears to be slightly higher in some donors when analyzed with the TOF-MS. To our surprise the mean RSD of the amino acid profiles based on duplicate fingerprints is generally higher in the case of the triple quadrupole analyses, when compared to the TOF-MS analyses (18.6% ± 6.6% vs. 13.2% ± 3.8%). The total amount of amino acid per fingerprint was calculated for each donated fingerprint (Fig. 3). Although the amino acid profiles of the 19 donors did not show large variability at first sight, the differences in the total amino acid deposited per fingerprint are significant, ranging from about 60 to 1200 ng per fingerprint deposition. Although a further analysis is outside of the scope of this technical note, we think that this may be due to the fact that the fingerprint deposition was not precisely controlled and factors such as deposition pressure, time and surface area may have varied between depositions. Despite this lack of controlled fingerprint deposition, prints from the same donors often yielded comparable amounts of total amino acid. While the absolute abundances in each fingerprint are largely similar, the calculated total amounts of amino acid were overall higher for the TOF-MS. This may be explained by the fact that the samples were analyzed on the TOF-MS after analysis on the triple quadrupole MS/MS and some methanol might have evaporated, yielding slightly higher concentrations in the fingerprint samples.
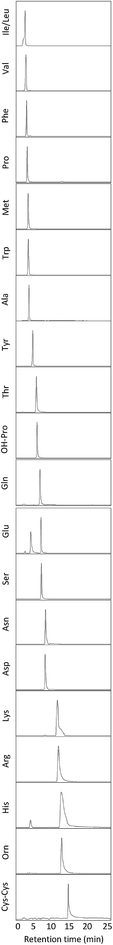 |
| | Fig. 1 Extracted ion chromatograms (XICs) of a typical separation of 21 amino acids on the time of flight MS. | |
Table 1 Linearity (R2), limit of detection (LOD), limit of quantification (LOQ) and intraday reproducibility of the 21 amino acids on both the triple quadrupole and the time of flight MS. Limits of detection and quantification were estimated based on S/N ratios (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10
1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Intraday reproducibility is based on the relative standard deviation of peak areas from isotopically labelled amino acids from 5 different standards
1). Intraday reproducibility is based on the relative standard deviation of peak areas from isotopically labelled amino acids from 5 different standards
| Compound |
Triple quadrupole MS/MS |
TOF-MS |
|
R
2
|
LOD (ng per fingerprint) |
LOQ (ng per fingerprint) |
Intraday reproducibility (RSD) |
R
2
|
LOD (ng per fingerprint) |
LOQ (ng per fingerprint) |
Intraday reproducibility (RSD) |
|
L-Alanine |
0.992 |
1.04 |
3.46 |
6.63 |
0.997 |
0.89 |
2.97 |
9.53 |
|
L-Arginine |
0.996 |
0.17 |
0.58 |
13.4 |
0.993 |
0.33 |
1.11 |
7.30 |
|
L-Asparagine |
0.998 |
0.06 |
0.19 |
13.9 |
0.994 |
0.35 |
1.15 |
7.26 |
|
L-Aspartic acid |
0.995 |
0.38 |
1.28 |
13.2 |
0.999 |
0.75 |
2.49 |
6.13 |
|
L-Cystine |
0.993 |
0.71 |
2.35 |
12.8 |
0.998 |
1.96 |
6.54 |
13.6 |
|
L-Glutamic acid |
0.993 |
0.16 |
0.55 |
7.98 |
0.994 |
0.21 |
0.70 |
7.86 |
|
L-Glutamine |
0.998 |
0.08 |
0.27 |
6.78 |
0.999 |
0.57 |
1.90 |
8.79 |
|
L-Histidine |
0.999 |
0.58 |
1.95 |
9.04 |
0.994 |
0.67 |
2.24 |
9.33 |
| Hydroxy-L-proline |
0.999 |
0.11 |
0.37 |
9.20 |
0.991 |
0.10 |
0.34 |
8.19 |
|
L-Iso-/leucine |
0.997 |
0.05 |
0.17 |
4.64 |
0.988 |
0.04 |
0.15 |
10.7 |
|
L-Lysine |
0.995 |
0.58 |
1.92 |
13.2 |
0.987 |
1.45 |
4.84 |
9.47 |
|
L-Methionine |
0.982 |
0.23 |
0.77 |
11.2 |
0.964 |
0.63 |
2.09 |
5.77 |
|
L-Ornithine |
0.997 |
0.19 |
0.64 |
10.9 |
0.997 |
0.74 |
2.48 |
6.97 |
|
L-Phenylalanine |
0.981 |
0.32 |
1.08 |
7.77 |
0.992 |
0.05 |
0.18 |
6.98 |
|
L-Proline |
0.992 |
0.03 |
0.11 |
13.6 |
0.992 |
0.04 |
0.14 |
6.57 |
|
L-Serine |
0.992 |
0.11 |
0.38 |
6.12 |
0.992 |
0.34 |
1.14 |
7.40 |
|
L-Threonine |
0.992 |
0.07 |
0.24 |
9.15 |
0.998 |
0.17 |
0.58 |
5.79 |
|
L-Tryptophan |
0.995 |
0.11 |
0.38 |
5.94 |
0.995 |
0.04 |
0.14 |
8.48 |
|
L-Tyrosine |
0.999 |
0.78 |
2.58 |
5.53 |
0.991 |
0.09 |
0.29 |
7.54 |
|
L-Valine |
0.991 |
0.03 |
0.12 |
9.93 |
0.992 |
0.11 |
0.35 |
10.8 |
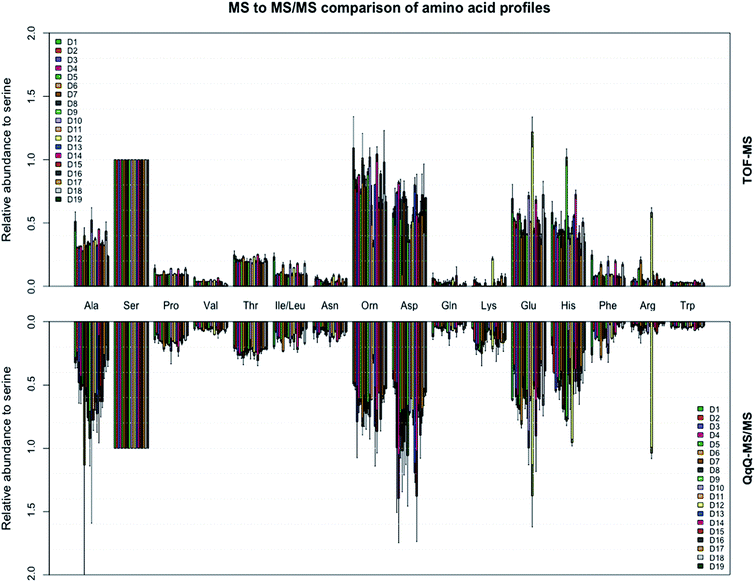 |
| | Fig. 2 Amino acid profiles of the 16 most abundant amino acids found in the fingerprints of 19 donors analyzed on both the triple quadrupole and time of flight MS. All amino acid abundances are relative to serine. Amino acid abundances for each donor were calculated as the mean of the two donated prints with standard deviation as shown. | |
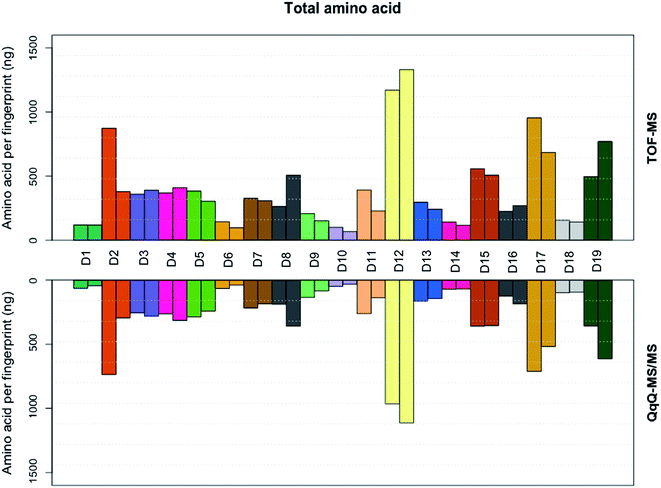 |
| | Fig. 3 Total amino acid (ng) per fingerprint, calculated by summing all amino acid abundances found in each fingerprint sample of the 19 donors, for both the triple quadrupole and time of flight MS. | |
Conclusion
In this technical note, we have described a method for the separation and quantification of amino acids from fingerprints using UPLC and both a triple quadrupole MS/MS and a TOF-MS. The use of the amide stationary phase circumvents the need to derivatise amino acids prior to LC-MS analysis, thereby shortening and simplifying the sample preparation process. This facilitates the analyses of the large numbers of samples needed to study the intra- and inter-variability of amino acids profiles found in fingerprints. Although the triple quadrupole had a higher sensitivity for most amino acids, the TOF-MS showed sufficient sensitivity to yield the same amino acid profiles obtained from 19 donors. Interestingly, the results obtained with the TOF-MS had lower mean RSDs in amino acid profiles from duplicate fingerprints.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
W. v. H. acknowledges a NWO research grant (no. 2014-01-124PRO) (RAAK-PRO).
References
-
C. Champod, C. J. Lennard, P. Margot and M. Stoilovic, Fingerprints and other ridge skin impressions, CRC press, 2016 Search PubMed.
- A. Girod, R. Ramotowski and C. Weyermann, Forensic Sci. Int., 2012, 223, 10–24 CrossRef CAS PubMed.
- S. Cadd, M. Islam, P. Manson and S. Bleay, Sci. Justice, 2015, 55, 219–238 CrossRef PubMed.
- R. S. Croxton, M. G. Baron, D. Butler, T. Kent and V. G. Sears, J. Forensic Sci., 2006, 51, 1329–1333 CrossRef CAS PubMed.
- R. S. Croxton, M. G. Baron, D. Butler, T. Kent and V. G. Sears, Forensic Sci. Int., 2010, 199, 93–102 CrossRef CAS PubMed.
- A. Girod and C. Weyermann, Forensic Sci. Int., 2014, 238, 68–82 CrossRef CAS PubMed.
- M. M. Delgado-Povedano, M. Calderon-Santiago, F. Priego-Capote and M. D. L. de Castro, Anal. Chim. Acta, 2016, 905, 115–125 CrossRef CAS PubMed.
- S. J. Cadd, L. Mota, D. Werkman, M. Islam, M. Zuidberg and M. de Puit, Anal. Methods, 2015, 7, 1123–1132 RSC.
- C. Weyermann, C. Roux and C. Champod, J. Forensic Sci., 2011, 56, 102–108 CrossRef CAS PubMed.
- M. de Puit, M. Ismail and X. M. Xu, J. Forensic Sci., 2014, 59, 364–370 CrossRef CAS PubMed.
- K. A. Mountfort, H. Bronstein, N. Archer and S. M. Jickells, Anal. Chem., 2007, 79, 2650–2657 CrossRef CAS PubMed.
- T. Atherton, R. Croxton, M. Baron, J. Gonzalez-Rodriguez, L. Gamiz-Gracia and A. M. Garcia-Campana, J. Sep. Sci., 2012, 35, 2994–2999 CrossRef CAS PubMed.
- K. Dettmer, P. A. Aronov and B. D. Hammock, Mass Spectrom. Rev., 2007, 26, 51–78 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ay01603d |
|
| This journal is © The Royal Society of Chemistry 2017 |
 Open Access Article
Open Access Article *cd
*cd

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10
1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Intraday reproducibility is based on the relative standard deviation of peak areas from isotopically labelled amino acids from 5 different standards
1). Intraday reproducibility is based on the relative standard deviation of peak areas from isotopically labelled amino acids from 5 different standards


