Open Access Article
Open Access ArticleQuantum dot probes for cellular analysis
Dahai
Ren
*,
Bin
Wang
,
Chen
Hu
and
Zheng
You
State Key Laboratory of Precision Measurement Technology and Instruments, Department of Precision Instrument, Tsinghua University, Beijing, 100084, China. E-mail: rendh@tsinghua.edu.cn
First published on 26th January 2017
Abstract
Highly fluorescent and robust semiconductor nanocrystals (known as quantum dots or QDs) play a pivotal role in biological applications. In particular, the excellent optical properties of QDs make them more suitable for immunolabeling, molecular imaging, and multiplexed biological detection for cellular analysis than conventional fluorescent dyes. Many studies have applied QD probes for in vitro and in vivo assays, showing great improvements with respect to gaining insight into bioanalytical chemistry, target specificity, and cytotoxicity. In this review, we discuss the optical properties, specificity, and cytotoxic effects of QDs as well as the progress achieved in multicolor cellular imaging, immunolabeling for signaling pathways, and molecular detection at the cellular level. In addition, carbon QDs as alternatives to the toxic cadmium-based QDs and their applications in biotechnology are discussed. Despite the rapid development and recent progress of QD probes, much more work is required to determine the toxicity of cadmium-containing QDs used in live cells and animals.
1. Introduction
When studying cell metabolic and genetic behaviors at the cellular level, researchers usually fluorescently label the specific targets such as proteins, nucleic acids (DNA/RNA), and drugs to be screened.1–3 Under the particular excitation light wavelength employed, target molecules marked with fluorescent dyes can be observed using a fluorescence microscope. Thereby, the concentration and location of the target can be analyzed and quantified, providing substantial information of molecular and cellular physiology. Fluorescent dyes have gradually replaced radioisotopes as the target markers in biological applications because of their high sensitivities and simple procedures.However, the excitation wavelength ranges of conventional organic fluorophores are generally narrow. In addition, different fluorescent dyes have specific excitation wavelengths; for example, green fluorescent protein (GFP)4 emits fluorescence in the blue wavelength range, rhodamine 123 (ref. 5) emits at 507 nm, fluorescence isothiocyanate6 emits at 488 nm, and cyanine-labeled dye (Cy5)7 emits at 633 nm. This situation has made it difficult to realize the simultaneous detection of multicolor immunolabels for multiple targets, since the excitation filters must be constantly switched for the variety of fluorescent probes required. Moreover, organic dyes typically have asymmetric emission spectra broadened by a red tail,8 which usually leads to severe signal overlap between different dyes. Common organic dye molecules also suffer from background autofluorescence, weak resistance to photobleaching, and low brightness.9 For example, under the exposure rate of 105 photons per second, GFP can produce 104 to 105 photons with a fluorescence lifetime of 0.1 to 1 s, whereas organic fluorescent dyes can produce 105 to 106 photons during 1 to 10 s of the fluorescence lifetime, which is variable and depends on the chosen fluorophore. Consequently, it is difficult to achieve imaging and tracking of multiple molecular targets simultaneously or over the long-term. Quantum dots (QDs) provide new possibilities to overcome these problems owing to their unique optical properties.10,11
When binding to the cytomembrane and intracellular targets for cellular analysis, QDs' stability and toxicity are crucial for their performance. Coating and labeling are closely related to the stability of QDs in live cells, which are dependent on the synthesis and conjugation of QDs, respectively. Also, it is reasonable to take QDs' cytotoxicity into consideration as QDs may cause damage to cells. Therefore, we first discuss the optical properties and synthesis, conjugation and cytotoxic effects of QDs, laying the foundation for their cellular applications. Further, we discuss the recent progress achieved in quantum dot probes at the cellular level, including multicolor cellular imaging, immunolabeling for signaling pathways and molecular detection.
2. Probe synthesis, conjugation, and cytotoxicity
2.1 Optical properties and synthesis
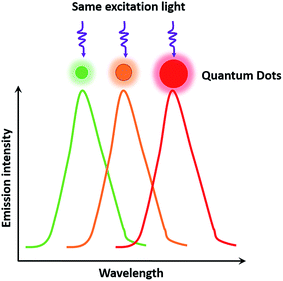
As semiconductor nanocrystals, QDs have wide excitation wavelength ranges and narrow emission ranges due to the quantum scale effect,12 which makes it possible to excite multicolor QDs under the same light source, thereby satisfying the needs of multiplexed detection and observation of different targets in biological samples. In addition, QDs possess many other unique properties that match well with cellular analysis, such as a high quantum yield ranging from 40% to 90%,12 high fluorescence intensity (20 times brighter than that of a single molecule of rhodamine 6G11), strong resistance to photobleaching (under the exposure of 105 photons per s, the continuous detection time of QDs is more than 1000 min, which is 100 times longer than that of rhodamine 6G11). Compared with organic dyes, an excellent property of QDs is the size-tunable symmetrical fluorescent emission as a function of core size (Fig. 1). The QD size is inversely proportional to the excited-state energy, so that larger-sized QDs result in lower energy, which consequently increases the emission wavelength.13 The ability to size-tune emission is typical for binary semiconductor materials such as CdSe. Alternatively, some groups developed ternary alloyed QDs in place of CdSe and succeeded in tuning the optical properties continuously without having to vary the particle size. The fluorescence wavelength of alloyed QDs such as CdSexS1−x and HgxCd1−xSe can be tuned through variations in the chemical composition and internal structure, which normalizes the brightness and size to solve the problem of inhomogeneity in multicolor comparisons.14,15 QDs show great potential and possibilities as remarkable probes in dynamic molecular imaging, target tracking, and highly sensitive detection for cellular analysis.16,17 Despite these advantages, QDs also have some drawbacks. In particular, QDs suffer from luminescence intermittency, referred to as blinking, which impedes their use in certain applications requiring single-molecule detection. The blinking phenomenon is related to the mobile charge on the surfaces of QDs, which can be suppressed through shell coating and decreasing the excitation intensity.18,19According to the synthetic conditions, synthesis methods of QDs can be classified into two categories: those conducted in an organic solvent and those conducted in an aqueous solution. Steigerwald et al.20 used the reaction of Cd(CH3)2 with (TMS)2S, (TMS)2Se, and (TMS)2Te in different solvents to obtain CdS, CdSe, and CdTe QDs, respectively. Bawendi et al.21 chose Cd(CH3)2 and TOPSe as the precursors, and synthesized CdSe QDs of different sizes by adjusting the temperature and reaction time to vary the CdSe growth rate with TOPO organic ligands. Another experiment was conducted by Zhang et al.22 to control the sizes of Ag2S during synthesizing. They used Ag(DDTC) and DT to grow Ag2S, and changed the reaction temperature and time to obtain Ag2S QDs in different sizes. However, the complicated preparation procedure of CdSe QDs mentioned before requires the use of harsh experimental conditions, and could even increase the risk of explosion. Peng et al.23 used CdO instead of the dangerous explosive Cd(CH3)2, and obtained CdSe QDs with good properties. Weller et al.24 synthesized CdSe/CdS/ZnS and CdSe/ZnSe/ZnS core–shell–shell nanocrystals, and the creative sandwich structure with the middle shell (CdS or ZnSe) improved the photoluminescence efficiency and photostability compared with ordinary CdSe/ZnS QDs. Deng et al.25 reported a new approach of synthesizing CdSe QDs in high-boiling-point solvents without poisonous or costly reagents such as octadecene and TOPO. With respect to synthesis methods in aqueous solution, CdTe QDs were synthesized in an aqueous solution from the reaction of Cd2+ and NaHTe precursors using thioglycolic acid and 1-mercapto glycerol as stabilizers.26,27 Zhang et al.28 used medroxyprogesterone acetate and thioglycolic acid as stabilizers, and obtained CdTe QDs with an enhanced quantum yield. Recently, Cao et al.29 developed a thiolate-mediated photoinduced synthesis method for Ag2S QDs in nonpolar solvents at low temperature for the first time.
The surface of QDs synthesized in aqueous solution is modified with a carboxyl group owing to the adoption of thioglycolic acid or other organic molecules with biological functional groups as stabilizers, which confers the QDs with the ability to easily conjugate with biomolecules and to exhibit excellent biocompatibility. However, the quantum yield of QDs arising from aqueous systems is relatively low. In comparison, QDs prepared in organic systems have a high quantum yield and excellent spectral characteristics, and are therefore more suitable as fluorescent materials. Nevertheless, the organic synthesis method is relatively complicated and demands harsh conditions. More importantly, QDs synthesized in organic systems are covered with hydrophobic molecules on their surface, which need to be modified by different coating and capping agents in order to mitigate their insolubility and improve their biocompatibility.
Furthermore, the high costs and toxicity of the precursors used in the conventional synthesis procedure are also regarded as shortcomings. Recently, novel QD synthesis methods referred to as green synthesis30–32 or biosynthesis,33–35 which do not require any hazardous reagents, have attracted extensive attention and are gradually emerging as satisfactory alternative methods of non-toxic QD synthesis.
2.2 Surface coating and cytotoxicity
Shell and surface coating play critical roles in determining the water-solubility, toxicity, and stability of QDs. As mentioned in Section 2.1, the QD synthesis procedure usually requires a step of surface modification using surfactants to make the QDs hydrophobic, which prevents their use in biomedical applications. To resolve this, researchers originally exchanged the surfactant coating with bifunctional ligand molecules carrying a hydrophobic anchoring group that can bind to the nanocrystal surface on one side and a hydrophilic group on another side.11,36–38 However, this method suffered from producing QDs that had relatively low stability and consequent weak hydrolytic resistance in biological samples. An alternative method is to coat the hydrophobic QDs with a cross-linked amphiphilic polymer, which greatly enhanced the stability of the nanocrystals.39,40 Based on the cross-linking coating chemistry, Díaz et al. developed a new surface architecture to generate stable and biocompatible photoswitchable QDs.41 They first prepared QDs coated with a photochromic polymer and then covalently modified the highly cross-linked polymer with photochromic diheteroarylethenes, which revealed thermally stable photoconversions in two manners with different spectral properties in the organic solvents. Finally, the obtained QDs maintained and even improved the original optical properties, and uniquely exhibited the tunable brightness of their emission when the light source was altered. The photoswitchable QDs showed remarkable fatigue resistance and photostability in many cycles of photoconversion.41 In the following studies, they combined photoswitchable QDs with a secondary dye (Alexa Fluor647 cadaverine) as the acceptor to form fluorescence resonance energy transfer (FRET), allowing for highly sensitive detection and dual-color imaging.42In general, introducing organic surface coatings will increase the hydrodynamic size of QDs, which is harmful for live cells and not suitable for endocytosis. There is also a strong demand for minimization of the hydrodynamic size for in vivo applications, and it was reported that QDs with a small hydrodynamic diameter (<5.5 nm) could be rapidly and efficiently eliminated from the body through urinary excretion.43 To minimize the hydrodynamic size, multidentate polymers have been typically employed as capping reagents.44,45 Gui et al. reported a method for the temperate aqueous synthesis of Ag2S QDs coated with multidentate polymers, which presented bright and sharp photoluminescence emission that can be tuned from the red to near-infrared region. The multidentate polymers can self-assemble and form a closed loops-and-trains structure on the QD surface, resulting in a thin polymer shell and consequent small hydrodynamic size. They finally obtained small hydrodynamic diameters of QDs (4.5–5.6 nm) close to the size of fluorescent proteins, indicating the feasibility and superiority of these QDs for reliable and effective imaging in live cells and animals.45
When used in live cells and animals, the toxicity of QDs containing heavy metal elements, including silver and cadmium, cannot be ignored. Although many researchers have investigated the distribution and toxicity of QDs in live cells or tissues,45–48 their toxic effects remain controversial and there still exists an urgent need for further investigations into the potential toxicity of heavy metal-containing QDs. It is widely believed that core-only QDs are more toxic than core–shell QDs, because heavy metal elements are trapped in the shell layer and the cytotoxicity of different surface coatings of the QDs vary substantially.49 Soenen et al.50 reported a positive correlation between the toxic effects and intracellular QD amount, and QDs localizing in endosomes could be more easily hydrolyzed due to the presence of local reactive oxygen species, demonstrating that polymer-coated QDs are not suitable for applications in proliferation-restricted cells. They further investigated the non-toxic concentration of QDs and the cell type-dependent changes in toxic end points for intracellular imaging over the long term.51,52 It was found that the asymmetric partitioning of QDs during recurrent cell division led to a rapid loss of intracellular nanocrystals, which impeded their use in long-term imaging and for molecular detection in live cells. However, Zhang et al.53 tested Ag2S QDs (15 & 30 mg kg−1) in vivo and demonstrated that the PEGylated-Ag2S QDs in the reticuloendothelial system can be gradually cleared, and did not show significant toxicity to mice for two months. Multiparametric evaluation of the cytotoxicity and accurate quantification of intracellular QDs are still challenging; therefore, more in-depth studies on the degradation of QDs in the cellular microenvironment are required.
2.3 Bioconjugation and specificity
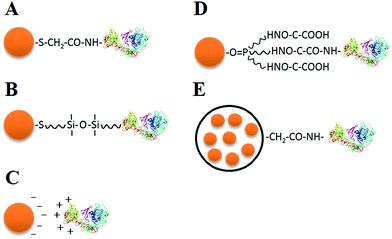
To employ QDs in cell analysis, bioconjugation is necessary to attach biological molecules to the QD coating surface while maintaining the biological function of these molecules. Current strategies for QD bioconjugation include electrostatic interaction, covalent linkage, and the use of functionalized microbeads as conjugation mediators (Fig. 2).Thioglycolic acid and other surfactants are usually used to modify the surface of QDs to improve their solubility and make them negatively charged with a pH of 6–8. Negatively charged QDs can be directly connected with positively charged proteins owing to electrostatic interactions (Fig. 2C),54,55 forming QD–antibody conjugates. The QD–protein conjugates could be very stable, but electrostatic interactions may suffer from nonspecificity when there are other positively charged molecules in the microenvironment.
There is a wide range of covalent linkage methods available, including connection between carboxyl and amino (–COOH and –NH2) functional groups of bifunctional molecules on the QD surface (Fig. 2A),11 functional group linkage of typical silane compounds (Fig. 2B),56 linkage between hydrophilic and hydrophobic groups (Fig. 2D),57 and use of a biotin-(strept)avidin system.58 Covalent linkage exhibits robust stability, whereas the intracellular enzymatic reaction may affect the covalent bond between QDs and biomolecules when probing live cells.
Both water-soluble and fat-soluble QDs can be encapsulated into polymeric microbeads, and then typical functional groups on the surfaces of the microbeads can be used for high-affinity bioconjugation (Fig. 2E). Several QDs can be embedded into a single microbead, which greatly enhances the fluorescence intensity and retards the hydrolysis of QDs and Cd2+ release, reducing the QD cytotoxicity. Nie et al. reported a novel multicolor optical coding method for parallel biological applications, which was based on multiplexed coding, combining the wavelength and intensity by encapsulating multicolor QDs into polymer microbeads at typical precise ratios.14,59–61 Despite the relatively large size (i.e., at the micrometer scale) of the polymer bead, this multiplexed coding method unprecedentedly enlarges the coding range of QDs and shows great potential for parallel multiplexed analysis in biomedical assays.
Target specificity strictly depends on the conjugation method and electrical properties of QDs. Nonspecific binding leads to a low signal-to-noise ratio and reduces the detection sensitivity, which is likely to result in false-positive staining for biomarkers in cells, thereby reducing the detection accuracy and reliability. In general, QDs with large electric charges exhibit a greater tendency to nonspecifically bind to proteins, cellular membranes, and other biomarkers. A common method is to replace the surface coating that brings the free electric charges with polyethylene glycol39,62 or small hydroxyl-containing molecules63 to make the QD surface electrically neutral. As the hydrodynamic size is often enlarged when replacing the surface coating, there is a trade-off between specificity and QD dimension according to different biological applications.
2.4 New generation of QDs
Recently, carbon-based QDs (C-dots) have attracted intensive research attention owing to their excellent biocompatibility and low toxicity. Compared with conventional heavy metal-containing QDs, C-dots maintain almost all of the favorable attributes of QDs and possess unique advantages in production, bioconjugation, and cytotoxicity.64 Graphene QDs (GQDs) are a new category of C-dots and additionally show excellent performance of graphene, such as eminent chemical inertness and fine surface grafting using the π–π conjugated network, making them highly effective in bioimaging, detection of sensitive molecules, and bioelectronics.65 C-dots are attracting more attention due to their particular advantages over traditional QDs in cellular analysis. However, the detailed mechanism for the photoluminescence of GQDs and efficient synthesis of small-scale GQDs with uniform morphology require further study.3. Cell imaging and tracking
Owing to their numerous advantages described above, QDs have been commonly employed in a diverse group of scientific fields over the last 20 years, and their biological applications have been extensively reviewed.8,9,16,17,60,66–74 Here, we will focus specifically on QD applications for cellular analysis, which plays a vital role in genetic and phenotypic studies to comprehend tumor cell heterogeneity and other challenging issues at the cellular level.3.1 Cell imaging
The size-related tunable fluorescence emission of QDs provides promising methods for multicolor cellular imaging, which has become one of the most widely explored applications of QDs, since QDs with different colors can be excited simultaneously. Moreover, the excellent photostability of QDs enables continuous observation for a long period. Cell imaging with QDs has attracted great interest, and considerable improvements have been achieved in developing QDs for multicolor cellular imaging, long-term imaging, or tracing and labeling of intercellular substances.Kang et al.75 used QD-conjugated aptamers to observe the expression of different cancer markers simultaneously in the same cancer cell. They conjugated three different QDs (with three distinct emission wavelengths: 605, 655, and 705 nm) using three aptamers (AS1411, TTA1, MUC-1, targeting specific cancers), respectively, and then observed the multiplex imaging picture. The fluorescence images for several cells were obtained and analyzed, and the triple cellular imaging of three different QDs was successfully performed. QD-aptamers with high fluorescence intensity showed comparatively good fluorescence signals on the cellular surfaces.
To obtain qualified profiles, high-quality QDs are necessary for cellular imaging. Non-blinking QDs such as individual lanthanide-doped upconverting nanoparticles76 and zinc-blended CdSe/CdS core/shell QDs77 were reported, which solved the long-standing on/off emission problem and provided strong upconverted luminescence when applied in cellular imaging. Furthermore, CdSe–CdS core–shell QDs reported by Chen et al.78 meet the universal requirement of high particle uniformity, high photoluminescence quantum yields, narrow emission spectral line shapes, and minimal single-dot emission intermittence. Taking advantage of the high-quality dots reported, significant improvement has been made in the application of QDs for cellular imaging and other optical applications.
The new generation of QDs has been attracting more and more attention. Since the accidental discovery of C-dots in 2004 by Xu et al.79 while purifying single-wall carbon nanotubes by electrophoresis methods, they have been widely applied in bioimaging applications such as in multiphoton or high-resolution imaging. Subsequently, GQDs were rapidly developed as a new kind of low-toxic C-dot, igniting tremendous research interest in the past few years, and playing an important role in bioimaging.
Cao et al.80 passivated C-dots by poly(propionylethyleneimine-co-ethyleneimine); ethyleneimine fraction, 20%. The treated C-dots became readily soluble in water, and were also found to be strongly emissive in the visible wavelength range when excited by using a 458 nm argon ion laser and an 800 nm femtosecond pulsed laser. Experiments showed that the C-dots could effectively label both the cell membrane and cytoplasm of MCF-7 cells, but did not reach the nucleus. C-dots exhibited two-photon luminescence images at 800 nm excitation, showing fairly promising potential for cellular imaging.
Zrazhevskiy et al.81 developed an improved method named multicolor multicycle molecular profiling (M3P). The M3P technology can create elaborate quantitative molecular profiles in parallel for living cells. They designed a universal platform based on QD–protein A (named QD–SpA), and used the platform to prepare a library of functional QD–antibody (called QD–SpA–Ab) probes quickly and flexibly. To realize the multicycle, they developed new specimen regeneration and target re-staining techniques. They also performed a multitarget and multicycle analysis to demonstrate that the M3P method has an extensive molecular profiling capacity (Fig. 3). As a fast-preparation method with straightforward evaluation, this promising technology is widely expected to create new applications in molecular analysis and systems biology.
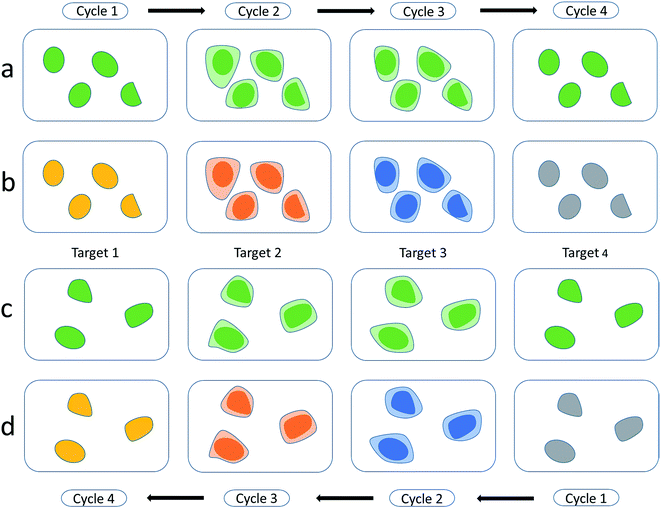 | ||
| Fig. 3 Multitarget and multicycle sequential staining procedure based on protein A and QDs. Single-color dyeing of several molecular targets on the same cell subpopulation is carried out based on QD545–SpA–Ab probes in a sequential manner (a and b) and in a reverse (c and d) manner. Reproduced from ref. 81 with permission. Copyright © 2013, Rights Managed by Nature Publishing Group. | ||
Compared to current organic dyes, fluorescent proteins, and other molecular fluorophores, the photostability of QDs is one of its most specific characteristics, which provides a great benefit for long-term imaging. Many groups have focused on this property and have made significant progress as a result. Fluorescent silicon QDs82 show steady photostability, pH stability, excellent aqueous dispersibility, strong fluorescence, and good biocompatibility. Furthermore, silicon nanoparticles83 were reported to show robust photostability, strong fluorescence, and favorable biocompatibility. Because of their good pH stability and multiplexing capability, QDs are widely utilized for long-term imaging.
Ding et al.84 synthesized uniform monodispersed C-dots on single-walled carbon nanotubes. Using the TTDDA-passivate method, the yellow fluorescence emitted by C-dots was transparent and stable, which showed almost no change after 2 h of continuous ultraviolet irradiation. Afterward, they labeled HeLa cells with luminescent C-dots to study the uptake process. In addition, extremely photostable CdSe/ZnS–QDs were reported by Chen and coworkers.85 They succeeded in visualizing the movement of the CdSe/ZnS-conjugated QDs from the cytoplasm to the nucleus, and observed the accumulation of the complex in the cell nucleus over a long time period. Experiments clearly showed that QDs remain stable during long-term intracellular biological visualization, which provides a nontoxic, long-term imaging platform for observing cellular mechanisms and processes.
However, there is still a lack of efficient techniques to deliver monodispersed QDs into the cytoplasm of living cells. In 2004, Derfus et al.86 used both biochemical and physical methods of delivering QDs into cells, and succeeded in improving the delivering scheme for intracellular tracking. For the biochemical methods, the authors compared different transfection reagents and found that cationic liposomes, among all methods tested, provided the highest efficiency of delivering QDs into living cells. For microinjection, they succeeded in targeting QDs to subcellular sites after direct cytoplasmic injection. The experiments showed good promise for effectively and safely delivering QDs into living cells.
3.2 Tracking and transporting
Benefiting from long-term imaging methods utilizing QDs, progress in intracellular tracking and transporting has also shown great improvements. The possibility of obtaining steady QD images motivated researchers to apply QDs in the tracing, tracking, or labeling of substances at the sub-cellular level, such as peptides,87 organelles,86 and RNAs,88,89 helping to further understand the delivery mechanism and intracellular structures.In contrast to dye-labeled organelles, Derfus et al.86 monitored QD-labeled mitochondria in live 3T3 fibroblast cells. No measurable loss of signal intensity was observed for over 8 min of continuous exposure, whereas the dye-labeled mitochondria rapidly showed photobleaching, and they could no longer be detected within less than 30 s. The results clearly showed that without photobleaching limitations, the QD conjugates could play a long-lasting role in the analysis of tracking and transporting.
Elucidating the delivery mechanism has also become a research hotspot in recent years. To trace the delivery paths, Ruan et al.87 combined Tat-peptide with QDs to examine the complex behavior of intracellular nanoparticle probes. To reveal the detailed delivery mechanism, labeling of QDs was employed to indicate where the bound Tat peptides were transferred. Consistent with general opinions, this method revealed that QDs internalized into living cells through a receptor-mediated endocytosis pathway tend to aggregate and be encapsulated in vesicles, endosomes, and lysosomes. Interestingly, the QD loaded vesicles are transported with the help of dyneins along microtubule tracks. The delivery ends at a final transportation of Tat–QDs to an asymmetric perinuclear region, the microtubule organizing center (MTOC). With the aid of conjugated Tat–QDs, a detailed mechanistic model for Tat peptide-mediated delivery was successfully established (Fig. 4).
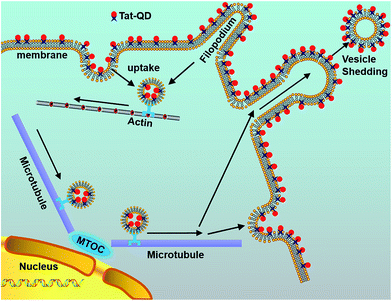 | ||
| Fig. 4 Schematic diagram illustrating the delivery mechanism of Tat–QDs inside living cells. The procedure of Tat–QD transportation includes the following steps: binding of Tat–QDs with cell membranes and filopodia, internalization by macropinocytosis, active transport of vesicles from the cell periphery to the microtubule organizing center (MTOC), and vesicle shedding in both manners of QD-membrane binding. Reproduced from ref. 87 with permission. Copyright © 2007, American Chemical Society. | ||
The high-efficiency tracking and tracing of QD-conjugated materials is also advantageous in medical science. Lee et al.88 reported an approach for the rapid analysis of intracellular small interfering RNA (siRNA) uptake, as well as the quantitative evaluation of the unpacking of siRNA from cationic QD carriers. Polyethyleneimine (PEI)-conjugated QD625, known as QD–PEI, was synthesized and nano-complexed with Cy5–siRNA, which together formed the cationic QD carriers. These two QD-based gene carriers, QD–PEI–Hph-1 and QD–PEI, were synthesized and used to monitor the intracellular trafficking of Cy5–siRNA in the experiment. Moreover, they used flow cytometry analysis to evaluate the quantity of Cy5–siRNA unpacking from the QD–PEI–Hph-1 and QD–PEI complexes.
To study the manipulation of key oncogenes, it is necessary to selectively, simultaneously, and efficiently deliver and monitor the siRNA molecules into tumor cells. Jung et al.89 demonstrated a method to synthesize and target-specifically deliver multifunctional siRNA–QD constructs. The reported constructs could selectively inhibit the expression of epidermal growth factor receptor (EGFR) variant III in the target cell line human U87 glioblastoma cells. Subsequently, the constructs could also be used to monitor the down-regulated signaling pathway with high efficiency.
Sun et al.90 reported the delivery ability of PEGylated nano-graphene oxide (NGO) sheets. They tested the intrinsic photoluminescence of NGO, and found that NGO can simply load doxorubicin through physisorption. Doxorubicin is a widely known anti-cancer drug that can be loaded onto NGO sheets, as it is capable of selectively killing cancer cells in vitro after functionalizing with antibodies. Taking advantage of its drug delivery capabilities, NGO is a promising material for biological and medical applications.
QD labels also enable researchers to trace viruses and understand the mechanism of their infection to mammalian cells. Li and colleagues91 utilized the molecular self-assembly method to encapsidate QDs in SV40 VLPs. It was observed that the formed SVLP-QDs endocytosed by cells, travelled along the microtubules and finally accumulated in endoplasmic reticulum. The procedure was just as what the wide-type SV40 do in the early stages of infection. Furthermore, they bound VLP-QDs with different nanoparticles such as AuNPs, and found clues to retain these structures robust.92 With the potential to be used for various purposes like tracing viruses and understanding the infection mechanism, the protein-based QD structures will gain more attention in medical and bioanalytical assays.
4. Intracellular analysis
Fluorescent probes are widely used to detect structures or substances in cell biology. Since the traditional organic fluorophore probes often suffer from fast photobleaching and asymmetric broad emission spectra, ordinary fluorescent dyes have many limitations in detecting and labeling intracellular substances. Therefore, the photostable and non-blinking QDs show great opportunities to be used as alternative intracellular probes.However, as mentioned above, QDs inside living cells are often captured and encapsulated in endosomes and lysosomes. It is crucial to develop efficient strategies to promote QDs escape into the cytoplasm to bind to the targets for subsequent intracellular analysis. Functionalized QDs can be degraded if they are transported from endosomes to lysosomes where biogenic thiols are accumulated actively.93 Therefore, it is important to deliver QDs into cytoplasm before QD loaded endosomes fuse with lysosomes.94 Duan et al.95 reported an endosomal escape method using a disrupting polymer PEI. QDs were coated with both PEG and PEI. The “proton-sponge effect” induced by plenty of amines on PEI macromolecules can enhance endosomal release of cargos and drugs. Xu et al.96 proposed a cell penetrating peptide (CPP) based methodology for QDs' delivery and release in live mammalian cells with high efficiency and minimal cytotoxicity. The probe (CPP–QDs) simply consists of CPP and QDs, and the cellular imaging of the probe provided insights into the mechanisms of CPP mediated QDs delivery and endosomal release. In general, the size, charge and surface coatings of QDs may play important roles in their delivery mechanisms and intracellular distributions.97 Microinjection of QDs into target cells is favorable to their subcellular distribution in cytoplasm. However, the crude microinjection procedure may cause severe damage to live cells. Therefore, efficient endosomal escape methods still need to be further studied.
4.1 Molecular localization
Owing to the robust properties of QDs, they have been extensively applied as fluorescent probes for locating tumor marker-related molecules or sub-cellular structures.Driven by the large need for fast, sensitive, and portable systems to detect biomarkers for clinical diagnosis, immunosensors coupled with QDs have been utilized to offer multiplexing capability for simultaneous measurements of multiple cancer biomarkers. Jie et al.98 reported an ultrasensitive immunosensor for CEA, one of the most commonly used tumor makers during clinical diagnosis. To form the Au–SiO2–CdSe–CdS–QDs nano-composites, they used CdSe–CdS QDs as the core and a dense monolayer of gold nanoparticles separated by a silica shell as the other combination material. Compared with pure QDs, the newly structured probe enhanced the chemiluminescence intensity by a magnitude of 17 orders and significantly improved the biocompatibility at the same time.
Besides tumor biomarkers, QDs have also been used to label and observe sub-cellular structures. Wu et al.39 conjugated QDs with immunoglobulin G, and then labeled the breast cancer marker HER2 with streptavidin on the surface of fixed cancer cells. They also used the processed probes to dye actin and microtubule fibers in the cytoplasm of the cells to detect the nuclear antigens. By means of the multiplexed labeling of cellular targets using QD-based probes, bright and specific signals were obtained for filamentous F-actin and nuclear antigens detection. The excellent properties of the QD-based probes during the detection of sub-cellular structures and substances make them excellent probes for molecular localization.
4.2 FRET
Combined with FRET technology, QDs conjugated with other fluorophores can form different probes and develop the ability to detect different substances or indices, including the intracellular pH value99 and physiological activity of biomacromolecules such as proteases.100,101QD-based FRET biosensors have unique features of quenching or increasing the photoluminescence with the addition of specific ions or functional groups, which suggests the feasibility of broad sensing applications. Medintz et al.102 designed and tested QD–protein assemblies that play the role of chemical sensors. They proved that β-cyclodextrin-QSY9 combined with probes resulted in quenching of the QD photoluminescence. Subsequently, maltose was added to displace the β-cyclodextrin-QSY9, leading to a steady increase in the QD photoluminescence.
Choosing QDs as excellent FRET donors while working with proximal organic acceptor dyes, Lee et al.88 conjugated QD625 with PEI to form cationic QD carriers. With flow cytometric analysis, they also studied the FRET interactions between Cy5–siRNA and cationic QD carriers. Dennis et al.99 reported QD–fluorescent protein FRET probes, which can be used to sense intracellular pH. They conjugated QDs either to mOrange, a protein that exhibits pH sensitivity, or to its homologue mOrange M163K, and found that the pH sensors based on QD–fluorescent protein FRET probes showed great sensitivity to intracellular pH values, which significantly varied by over 12-fold when the pH ranged from 6 to 8. The improved pH-sensitive, photostable probes show no environmental sensitivity. In addition, they were found to boost the visualization of acidification during succession in living cells. QD-based FRET probes show great potential in applications for intracellular pH imaging, which may not work using organic fluorophores or fluorescent proteins alone.
A FRET probe consisting of QD, nanogold, and EGF was designed for the long-term real-time monitoring of caspase-3 activity in HeLa cells.101 The newly developed EGF–QD–peptide–nanogold probe meets the demand of real-time monitoring of the prolonged protease activity in living cells, which could not be achieved with the commonly used dyes or fluorescent proteins. This probe could overcome the deficiencies of former probes, and improved the accuracy of reflecting the activity of proteases, as well as avoided the probes' susceptibility to photobleaching or low efficiency during cell penetration (Fig. 5). Such outstanding performance for detecting caspase-3 activity in living HeLa cells clearly demonstrated the major improvement of the high-quality probe over other methods developed for intracellular protease detection, representing a promising method for detecting enzyme activities, including DNA hydrolases and proteases.
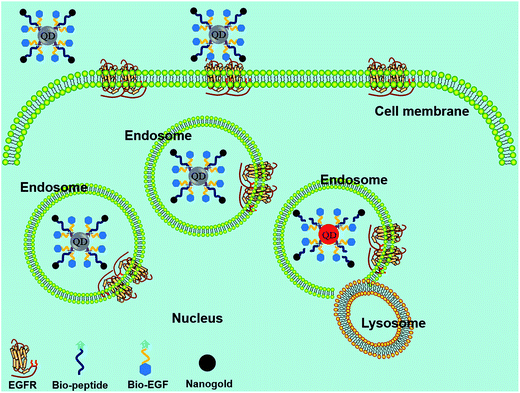 | ||
| Fig. 5 Schematic principle of protease detection inside living cells using the FRET probe. When the QD and nanogold are separated by enzyme digestion in the endosomes, fluorescence recovery of QD can be observed. Reproduced from ref. 101 with permission from The Royal Society of Chemistry. | ||
However, there are still limitations of QD-based FRET probes. One such limitation is due to donor–acceptor distance constraints. The overall dimensions of nanocrystals are larger than those of conventional organic dyes or other metal chelate energy donors, which may result in limitations to some of the FRET experiments.102
4.3 Metal ion detection
QD photoluminescence can be affected by the ionic environment. Ions such as cadmium(II) and zinc(II) can increase the luminescence of CdS nanocrystals, whereas copper(II) ions will quench the emission intensity.103,104 Moore et al.105 fluorometrically titrated aqueous Q-CdS samples against dozens of zinc and cadmium salts, and analyzed the improvement in the band-edge PL intensity of the Q-CdS. This unique feature of QDs suggests their feasibility in detecting heavy metal ions.Ruedas-Rama et al.106 reported the first zinc ion sensor, which uses QD nanoparticles in a host–guest and receptor-fluorophore (TACN, cyclen, and cyclam) system. They explained that as the zinc ion entered the azacrown, the energy level of electrons that was essential for the hole-transfer mechanism would no longer become available. This situation would switch on the QD emission and cause an increase in the detected fluorescence intensity (Fig. 6). The method allowed for the observation of low-concentrated zinc ions, with an observation limit lower than 2.4 μM, relative standard deviation (RSD) ∼3% (n = 10). Similarly, Wang et al.107 proposed an assay in which Pb2+ was detected based on the inhibition of FRET efficiency between QDs and gold nanoparticles by basement Pb2+ ions. The determination limit of Pb2+ was reported to be 30 ppb.
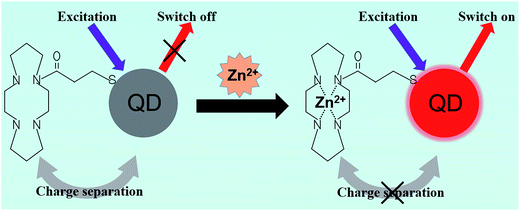 | ||
| Fig. 6 Schematic principle of zinc ion nanosensors. Conjugation of QDs and azamacrocycles causes charge separation and quenching of QDs. The complexation of Zn(II) with the ligand changes the energy level on the ligand, which prevents the charge separation and leads to the fluorescence recovery of QDs. Reproduced from ref. 106 with permission. Copyright © 2008, American Chemical Society. | ||
Carbonaceous QDs also play an important role in detecting heavy metal ions. Qu et al.108 developed an efficient method for selectively detecting Cu2+ with fluorescent C-dots conjugated with amino TPEA. After conjugation, they further applied C-dot–TPEA nanointegration to intracellular sensing and observations of Cu2+. During the experiments, low cytotoxicity and good cell-permeability of the fluorescent probe were observed, suggesting the application of the probe for the intracellular imaging of Cu2+. Analogously, Zhou et al.109 used carbon nanodots as newly developed environmentally friendly fluorescent probes for sensing Hg2+, which showed high sensitivity and selectivity.
Besides detecting heavy metal ions, QDs can also sense other types of ions. Wang et al.110 reported a Na–QD sensor for Na+ measurements. They conjugated 12-crown-4 to the QD surface to synthesize the nanosensor, which was applied for sensing the dynamic physiological responses of Na+ in human HEK-293F cells and primary rat cardiac myocytes. The fluorescence intensity of Na–QD clearly reflected the responses of Na+ when the membrane permeability of human HEK-293F cells and primary rat cardiac myocytes to Na+ were controllable with pharmacology methods. The experiment proved that this highly sensitive Na+ probe is a promising particle that could be further developed into assays for Na+ channel drug discovery.
4.4 Signaling pathways
QDs also contribute to the study of signaling pathways that are essential for cell–cell communication. Ting et al.111 reported targeting QDs to specific proteins on the cell membrane surface using Escherichia coli lipoic acid ligase and HaloTag as critical mediators. Combined with a previously reported QD targeting method using biotin-streptavidin E. coli ligase,112 they visualized a low-density lipoprotein receptor and EGFR orthogonally for two-color single-molecule imaging, as well as the transportation of neurexin1β and neuroligin1 in a dose-related manner.111 In this assay, QDs and the covalent linkage ensured the feasibility of the long-term tracking of single proteins, and introduction of a small peptide as the conjugation tag avoided the issue of conjugation of the large HaloTag–QD.Traditional biomedical applications and measurements are typically performed on bulk samples, which obscure heterogeneity of individual cells.113,114 In combination with a single-cells array,115 QD probes were further employed to study intracellular anti-EGFR functions as well as cell heterogeneity, which helped to gain an in-depth understanding of the EGF signaling pathway among the cells.116 EGF and the antibody against EGFR (anti-EGFR) were labeled with red and green QDs, respectively (Fig. 7). Based on the fluorescence intensity variations of the two-color QDs, the concentration of anti-EGFR, the amount of internalized EGFs, and the anti-EGFRs bounded to EGFRs on the cell surface were analyzed. The experimental results showed that the alterations of EGF engulfed by cells caused by the influence of anti-EGFR could be clearly detected by analyzing the QD fluorescence intensity. This method allows for multiplexed analysis at the single-cell level, which is of great significance for drug screening.
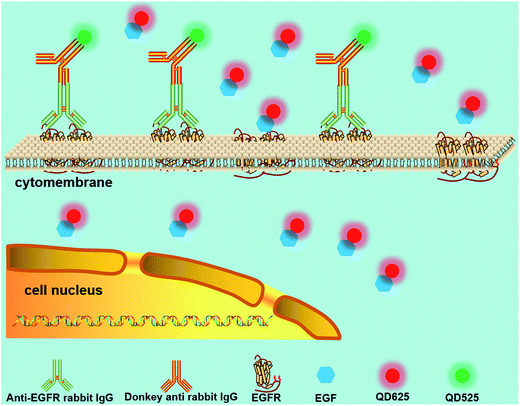 | ||
| Fig. 7 Schematic diagram of the testing assay of anti-EGFR effects. Multiplexed analysis of signaling pathways was achieved according to the fluorescence intensities of the two-color QDs. Reproduced from ref. 116 with permission. Copyright © 2016, American Chemical Society. | ||
GQDs were also utilized to investigate signaling pathways such as nerve growth factor receptor and insulin receptor-mediated signal transduction.117 Specific labeling and real-time tracking of insulin receptors with GQDs in 3T3-L1 adipocytes promulgated the dynamic cell functions of the molecular mechanisms at the single-cell level. Using total internal reflection microscopy, they observed four subpopulations during a 2 min imaging period based on the real-time tracking of individual insulin-GQD/insulin receptor clusters. Two cytokines, apelin and tumor necrosis factor-alpha, were proved to be the stimulator and the inhibitor of the insulin receptor dynamics, respectively. This study also revealed the great potential of GQDs in dynamic bioimaging and signaling pathway research.
5. In vivo probes
In vivo targeting and imaging is extremely important in medical diagnostics.119 QDs have been widely applied for high-sensitivity and high-specificity probes used in vivo owing to their excellent optical properties. In particular, the two-photon cross-section of QDs is much higher than that of organic fluorochromes,118 making them quite superior for the visualization of thick tissues. Originally, QDs containing cadmium or other toxic heavy metal elements were used for in vivo imaging and detection,120–123 and the potential cytotoxicity due to Cd2+ release over time was not studied in detail. Recently, C-dots and GQDs have attracted substantial attention and have been used for in vivo bioimaging because of their low toxicity.124,125 In addition, in vivo fluorescence imaging with QDs in the second near-infrared region (NIR-II, 1000–1350 nm) has increased in popularity owing to the greatly enhanced penetration depth and negligible tissue autofluorescence, enabling imaging of deeper tissues with high signal-to-noise ratios.126–1296. Conclusions
QDs have tremendous capacity in biological applications owing to their excellent properties, making them good substitutes for traditional fluorescent dyes. QD probes, especially in vivo probes, have shown great potential for cancer diagnosis and clinical therapy. However, there are still some challenges that must be further investigated. For example, the autofluorescence and fluorescence signal cross-talk of QDs need to be minimized in a multiplexed staining system. Moreover, methods for tuning the surface coatings and tagging strategies to effectively avoid nonspecific labeling and QD aggregation in vitro and in vivo remain to be studied in depth. In addition, it is imperative to understand the toxicity of QDs and other long-term effects during in vivo imaging, and the methodology for achieving timely and thorough QD clearance from the body needs to be further studied.Acknowledgements
This work is supported by the National Natural Science Foundation of China (No. 61071002), National Program for Significant Scientific Instruments Development of China (No. 2011YQ030134), the Funds for State Key Laboratory of China, the Scientific Research Foundation for Returned Overseas Chinese Scholars and the Fund for Beijing Laboratory of Biomedical Detection Technology and Instrument.References
- K. U. Hong, S. D. Reynolds, A. Giangreco, C. M. Hurley and B. R. Stripp, Am. J. Respir. Cell Mol. Biol., 2001, 24, 671–681 CrossRef CAS PubMed.
- X. Xie, R. Liu, Y. Xu, L. Wang, Z. Lan, W. Chen, H. Liu, Y. Lu and J. Cheng, RSC Adv., 2015, 5, 62007–62016 RSC.
- P. O. Krutzik and G. P. Nolan, Nat. Methods, 2006, 3, 361–368 CrossRef CAS PubMed.
- O. Shimomura, F. H. Johnson and Y. Saiga, J. Cell. Comp. Physiol., 1962, 59, 223–239 CrossRef CAS PubMed.
- R. F. Kubin and A. N. Fletcher, J. Lumin., 1983, 27, 455–462 CrossRef CAS.
- J. D. Marshall, W. C. Eveland and C. W. Smith, Exp. Biol. Med., 1958, 98, 898–900 CrossRef CAS.
- R. B. Mujumdar, L. A. Ernst, S. R. Mujumdar, C. J. Lewis and A. S. Waggoner, Bioconjugate Chem., 1993, 4, 105–111 CrossRef CAS PubMed.
- A. P. Alivisatos, W. Gu and C. Larabell, Annu. Rev. Biomed. Eng., 2005, 7, 55–76 CrossRef CAS PubMed.
- M. Stanisavljevic, S. Krizkova, M. Vaculovicova, R. Kizek and V. Adam, Biosens. Bioelectron., 2015, 74, 562–574 CrossRef CAS PubMed.
- M. Bruchez, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Science, 1998, 281, 2013–2016 CrossRef CAS PubMed.
- W. C. Chan and S. Nie, Science, 1998, 281, 2016–2018 CrossRef CAS PubMed.
- J. K. Jaiswal and S. M. Simon, Quantum Dots: Applications in Biology, 2007, pp. 93–104 Search PubMed.
- L. Brus, J. Phys. Chem., 1986, 90, 2555–2560 CrossRef CAS.
- R. E. Bailey and S. Nie, J. Am. Chem. Soc., 2003, 125, 7100–7106 CrossRef CAS PubMed.
- M. D. Regulacio and M. Y. Han, Acc. Chem. Res., 2010, 43, 621–630 CrossRef CAS PubMed.
- X. Michalet, F. F. Pinaud, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Sundaresan, A. M. Wu, S. S. Gambhir and S. Weiss, Science, 2005, 307, 538–544 CrossRef CAS PubMed.
- G. Rousserie, A. Sukhanova, K. Even-Desrumeaux, F. Fleury, P. Chames, D. Baty, V. Oleinikov, M. Pluot, J. H. M. Cohen and I. Nabiev, Crit. Rev. Oncol. Hematol., 2010, 74, 1–15 CrossRef PubMed.
- L. Li, G. J. Tian, Y. Luo, H. Brismar and Y. Fu, J. Phys. Chem. C, 2013, 117, 4844–4851 CAS.
- M. H. W. Stopel, J. C. Prangsma, C. Blum and V. Subramaniam, RSC Adv., 2013, 3, 17440–17445 RSC.
- S. M. Stuczynski, J. G. Brennan and M. L. Steigerwald, Inorg. Chem., 1989, 28, 4431–4432 CrossRef CAS.
- C. B. Murray, D. J. Norris and M. G. Bawendi, J. Am. Chem. Soc., 1993, 115, 8706–8715 CrossRef CAS.
- Y. Zhang, Y. Liu, C. Li, X. Chen and Q. Wang, J. Phys. Chem. C, 2014, 118, 4918–4923 CAS.
- Z. A. Peng and X. G. Peng, J. Am. Chem. Soc., 2001, 123, 183–184 CrossRef CAS PubMed.
- D. V. Talapin, I. Mekis, S. Gotzinger, A. Kornowski, O. Benson and H. Weller, J. Phys. Chem. B, 2004, 108, 18826–18831 CrossRef CAS.
- Z. Deng, L. Gao, F. Tang and B. Zou, J. Phys. Chem. B, 2005, 109, 16671–16675 CrossRef CAS PubMed.
- A. L. Rogach, L. Katsikas, A. Kornowski, D. Su, A. Eychmüller and H. Weller, Ber. Bunsen-Ges. Phys. Chem., 1996, 100, 1772–1778 CrossRef CAS.
- M. Gao, S. Kirstein, H. Möhwald, A. L. Rogach, A. Kornowski, A. Eychmüller and H. Weller, J. Phys. Chem. B, 1998, 102, 8360–8363 CrossRef CAS.
- H. Zhang, Z. Zhou, B. Yang and M. Gao, J. Phys. Chem. B, 2003, 107, 8–13 CrossRef CAS.
- Y. Cao, W. Geng, R. Shi, L. Shang, G. I. Waterhouse, L. Liu, L. Wu, C. Tung, Y. Yin and T. Zhang, Angew. Chem., Int. Ed., 2016, 55, 1–6 CrossRef.
- M. Ahmed, A. Guleria, M. C. Rath, A. K. Singh, S. Adhikari and S. K. Sarkar, J. Nanosci. Nanotechnol., 2014, 14, 5730–5742 CrossRef CAS PubMed.
- R. K. Beri and P. K. Khanna, J. Nanosci. Nanotechnol., 2011, 11, 5137–5142 CrossRef CAS PubMed.
- P. C. Huang, Q. Jiang, P. Yu, L. F. Yang and L. Q. Mao, ACS Appl. Mater. Interfaces, 2013, 5, 5239–5246 CAS.
- H. F. Bao, N. Hao, Y. X. Yang and D. Y. Zhao, Nano Res., 2010, 3, 481–489 CrossRef CAS.
- H. Q. Huang, M. X. He, W. X. Wang, J. L. Liu, C. C. Mi and S. K. Xu, Spectra Anal., 2012, 32, 1090–1093 CAS.
- S. R. Sturzenbaum, M. Hockner, A. Panneerselvam, J. Levitt, J. S. Bouillard, S. Taniguchi, L. A. Dailey, R. A. Khanbeigi, E. V. Rosca, M. Thanou, K. Suhling, A. V. Zayats and M. Green, Nat. Nanotechnol., 2013, 8, 57–60 CrossRef CAS PubMed.
- D. Ren, Y. Xia and Z. You, J. Biomed. Opt., 2013, 18(9), 096005 CrossRef PubMed.
- D. Gerion, F. Pinaud, S. C. Williams, W. J. Parak, D. Zanchet, S. Weiss and A. P. Alivisatos, J. Phys. Chem. B, 2001, 105, 8861–8871 CrossRef CAS.
- S. Kim and M. G. Bawendi, J. Am. Chem. Soc., 2003, 125, 14652–14653 CrossRef CAS PubMed.
- X. Wu, H. Liu, J. Liu, K. N. Haley, J. A. Treadway, J. P. Larson, N. Ge, F. Peale and M. P. Bruchez, Nat. Biotechnol., 2002, 21, 41–46 CrossRef PubMed.
- T. Pellegrino, L. Manna, S. Kudera, T. Liedl, D. Koktysh, A. L. Rogach, S. Keller, J. Rädler, G. Natileand and W. J. Parak, Nano Lett., 2004, 4, 703–707 CrossRef CAS.
- S. A. Díaz, G. O. Menendez, M. H. Etchehon, L. Giordano, T. M. Jovin and E. A. Jares-Erijman, ACS Nano, 2011, 5, 2795–2805 CrossRef PubMed.
- S. A. Díaz, L. Giordano, T. M. Jovin and E. A. Jares-Erijman, Nano Lett., 2012, 12, 3537–3544 CrossRef PubMed.
- H. S. Choi, W. Liu, P. Misra, E. Tanaka, J. P. Zimmer, B. I. Ipe, M. G. Bawendi and J. V. Frangioni, Nat. Biotechnol., 2007, 25, 1165–1170 CrossRef CAS PubMed.
- A. M. Smith and S. Nie, J. Am. Chem. Soc., 2008, 130, 11278–11279 CrossRef CAS PubMed.
- R. Gui, A. Wan, X. Liu, W. Yuan and H. Jin, Nanoscale, 2014, 6, 5467–5473 RSC.
- A. M. Derfus, W. C. Chan and S. N. Bhatia, Nano Lett., 2004, 4, 11–18 CrossRef CAS.
- C. Kirchner, T. Liedl, S. Kudera, T. Pellegrino, A. M. Javier and H. E. Gaub, Nano Lett., 2005, 5, 331–338 CrossRef CAS PubMed.
- C. Sealy, Nano Today, 2012, 7, 223–224 CrossRef.
- A. O. Choi, S. J. Cho, J. Desbarats, J. Lovrić and D. Maysinger, J. Nanobiotechnol., 2007, 5, 1 CrossRef PubMed.
- S. J. Soenen, J. Demeester, S. C. De Smedt and K. Braeckmans, Biomaterials, 2012, 33, 4882–4888 CrossRef CAS PubMed.
- S. J. Soenen, J. M. Montenegro, A. M. Abdelmonem, B. B. Manshian, S. H. Doak, W. J. Parak, S. C. De Smedt and K. Braeckmans, Acta Biomater., 2014, 10, 732–741 CrossRef CAS PubMed.
- B. B. Manshian, S. J. Soenen, A. Al-Ali, A. Brown, N. Hondow, J. Wills, G. J. S. Jenkins and S. H. Doak, Toxicol. Sci., 2015, 144, 246–258 CrossRef CAS PubMed.
- Y. Zhang, Y. Zhang, G. Hong, W. He, K. Zhou, K. Yang, F. Li, G. Chen, Z. Liu, H. Dai and Q. Wang, Biomaterials, 2013, 34, 3639–3646 CrossRef CAS PubMed.
- H. Mattoussi, J. M. Mauro, E. R. Goldman, G. P. Anderson, V. C. Sundar, F. V. Mikulec and M. G. Bawendi, J. Am. Chem. Soc., 2000, 122, 12142–12150 CrossRef CAS.
- H. Mattoussi, J. M. Mauro, E. R. Goldman, T. M. Green, G. P. Anderson, V. C. Sundar and M. G. Bawendi, Phys. Status Solidi B, 2001, 224, 277–283 CrossRef CAS.
- A. P. Alivisatos, Science, 1996, 271, 933 CAS.
- B. Ballou, B. C. Lagerholm, L. A. Ernst, M. P. Bruchez and A. S. Waggoner, Bioconjugate Chem., 2004, 15, 79–86 CrossRef CAS PubMed.
- K. D. Wegner, P. T. Lanh, T. Jennings, E. Oh, V. Jain, S. M. Fairclough, J. M. Smith, E. Giovanelli, N. Lequeux, T. Pons and N. Hildebrandt, ACS Appl. Mater. Interfaces, 2013, 5, 2881–2892 CAS.
- M. Han, X. Gao, J. Z. Su and S. Nie, Nat. Biotechnol., 2001, 19, 631–635 CrossRef CAS PubMed.
- W. C. Chan, D. J. Maxwell, X. Gao, R. E. Bailey, M. Han and S. Nie, Curr. Opin. Biotechnol., 2002, 13, 40–46 CrossRef CAS PubMed.
- A. M. Smith, S. Dave, S. Nie, L. True and X. Gao, Expert Rev. Mol. Diagn., 2006, 6, 231–244 CrossRef CAS PubMed.
- E. L. Bentzen, I. D. Tomlinson, J. Mason, P. Gresch, M. R. Warnement, D. Wright, E. Sanders-Bush, R. Blakely and S. J. Rosenthal, Bioconjugate Chem., 2005, 16, 1488–1494 CrossRef CAS PubMed.
- B. A. Kairdolf, M. C. Mancini, A. M. Smith and S. Nie, Anal. Chem., 2008, 80, 3029–3034 CrossRef CAS PubMed.
- J. Shen, Y. Zhu, X. Yang and C. Li, Chem. Commun., 2012, 48, 3686–3699 RSC.
- L. Lin, M. Rong, F. Luo, D. Chen, Y. Wang and X. Chen, TrAC, Trends Anal. Chem., 2014, 54, 83–102 CrossRef CAS.
- P. Alivisatos, Nat. Biotechnol., 2004, 22, 47–52 CrossRef CAS PubMed.
- I. L. Medintz, H. T. Uyeda, E. R. Goldman and H. Mattoussi, Nat. Mater., 2005, 4, 435–446 CrossRef CAS PubMed.
- F. Pinaud, X. Michalet, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Iyer and S. Weiss, Biomaterials, 2006, 27, 1679–1687 CrossRef CAS PubMed.
- P. Zrazhevskiy, M. Sena and X. H. Gao, Chem. Soc. Rev., 2010, 39, 4326–4354 RSC.
- A. Valizadeh, H. Mikaeili, M. Samiei, S. M. Farkhani, N. Zarghami, A. Akbarzadeh and S. Davaran, Nanoscale Res. Lett., 2012, 7, 480 CrossRef PubMed.
- B. A. Kairdolf, A. M. Smith, T. H. Stokes, M. D. Wang, A. N. Young and S. Nie, Annu. Rev. Anal. Chem., 2013, 6, 143–162 CrossRef CAS PubMed.
- E. Petryayeva, W. R. Algar and I. L. Medintz, Appl. Spectrosc., 2013, 67, 215–252 CrossRef CAS PubMed.
- J. Li and J. J. Zhu, Analyst, 2013, 138, 2506–2515 RSC.
- A. Nagy, K. B. Gemmill, J. B. Delehanty, I. L. Medintz and K. E. Sapsford, IEEE J. Sel. Top. Quantum Electron., 2014, 3, 1–12 Search PubMed.
- W. J. Kang, J. R. Chae, Y. L. Cho, J. D. Lee and S. Kim, Small, 2009, 5, 2519–2522 CrossRef CAS PubMed.
- S. Wu, G. Han, D. J. Milliron, S. Aloni, V. Altoe, D. V. Talapin, B. E. Cohen and P. J. Schuck, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10917–10921 CrossRef CAS PubMed.
- H. Qin, Y. Niu, R. Meng, X. Lin, R. Lai, W. Fang, X. Peng, H. Han, D. Fukumura, R. K. Jain and M. G. Bawendi, J. Am. Chem. Soc., 2014, 136, 179–187 CrossRef CAS PubMed.
- O. Chen, J. Zhao, V. P. Chauhan, J. Cui, C. Wong, D. K. Harris and H. Wei, Nat. Mater., 2013, 12, 445–452 CrossRef CAS PubMed.
- X. Y. Xu, R. Ray, Y. L. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed.
- L. Cao, X. Wang, M. J. Meziani, F. Lu, H. Wang, P. G. Luo, Y. Lin, B. A. Harruff, L. M. Veca, D. Murray, S. Xie and Y. Sun, J. Am. Chem. Soc., 2007, 129, 11318–11319 CrossRef CAS PubMed.
- P. Zrazhevskiy and X. Gao, Nat. Commun., 2013, 4, 1619 CrossRef PubMed.
- Y. He, Y. Zhong, F. Peng, X. Wei, Y. Su, Y. Lu, S. Su, W. Gu, L. Liao and S. Lee, J. Am. Chem. Soc., 2011, 133, 14192–14195 CrossRef CAS PubMed.
- Y. Zhong, F. Peng, F. Bao, S. Wang, X. Ji, L. Yang, Y. Su, S. Lee and Y. He, J. Am. Chem. Soc., 2013, 135, 8350–8356 CrossRef CAS PubMed.
- H. Ding, L. Cheng, Y. Ma, J. Kong and H. Xiong, New J. Chem., 2013, 37, 2515–2520 RSC.
- F. Chen and D. Gerion, Nano Lett., 2004, 4, 1827–1832 CrossRef CAS.
- A. M. Derfus, W. C. W. Chan and S. N. Bhatia, Adv. Mater., 2004, 16, 961–966 CrossRef CAS.
- G. Ruan, A. Agrawal, A. I. Marcus and S. Nie, J. Am. Chem. Soc., 2007, 129, 14759–14766 CrossRef CAS PubMed.
- H. Lee, I. Kim and T. Gwan Park, Bioconjugate Chem., 2010, 21, 289–295 CrossRef CAS PubMed.
- J. Jung, A. Solanki, K. A. Memoli, K. Kamei, H. Kim, M. A. Drahl, L. J. Williams, H. Tseng and K. Lee, Angew. Chem., Int. Ed., 2010, 49, 103–107 CrossRef CAS PubMed.
- X. Sun, Z. Liu, K. Welsher, J. T. Robinson, A. Goodwin, S. Zaric and H. Dai, Nano Res., 2008, 1, 203–212 CrossRef CAS PubMed.
- F. Li, Z. P. Zhang, J. Peng, Z. Q. Cui, D. W. Pang, K. Li, H. P. Wei, Y. F. Zhou, J. K. Wen and X. E. Zhang, Small, 2009, 5, 718–726 CrossRef CAS PubMed.
- F. Li, H. Chen, L. Ma, K. Zhou, Z. P. Zhang, C. Meng, X. E. Zhang and Q. Wang, Small, 2014, 10, 536–543 CrossRef CAS PubMed.
- Z. J. Zhu, Y. C. Yeh, R. Tang, B. Yan, J. Tamayo, R. W. Vachet and V. M. Rotello, Nat. Chem., 2011, 3, 963–968 CrossRef CAS PubMed.
- M. A. Yessine and J. C. Leroux, Adv. Drug Delivery Rev., 2004, 56, 999–1021 CrossRef CAS PubMed.
- H. Duan and S. Nie, J. Am. Chem. Soc., 2007, 129, 3333–3338 CrossRef CAS PubMed.
- J. Xu, Y. Yu, H. C. Lee, Q. Fan, J. Winter and G. Yang, 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 2014, pp. 4260–4263 Search PubMed.
- D. Maysinger, J. Lovrić, A. Eisenberg and R. Savić, Eur. J. Pharm. Biopharm., 2007, 65, 270–281 CrossRef CAS PubMed.
- G. Jie, P. Liu and S. Zhang, Chem. Commun., 2010, 46, 1323–1325 RSC.
- A. M. Dennis, W. J. Rhee, D. Sotto, S. N. Dublin and G. Bao, ACS Nano, 2012, 6, 2917–2924 CrossRef CAS PubMed.
- D. Ren, J. Wang, B. Wang and Z. You, Biosens. Bioelectron., 2016, 79, 802–809 CrossRef CAS PubMed.
- D. Ren, J. Wang and Z. You, RSC Adv., 2014, 4, 54907–54918 RSC.
- I. L. Medintz, A. R. Clapp, H. Mattoussi, E. R. Goldman, B. Fisher and J. M. Mauro, Nat. Mater., 2003, 2, 630–638 CrossRef CAS PubMed.
- Y. F. Chen and Z. Rosenzweig, Anal. Chem., 2002, 74, 5132–5238 CrossRef CAS PubMed.
- J. Chen, A. Zheng, Y. Gao, C. He, G. Wu, Y. Chen, X. Kai and C. Zhu, Spectrochim. Acta, Part A, 2008, 69, 1044–1052 CrossRef PubMed.
- D. E. Moore and K. Patel, Langmuir, 2001, 17, 2541–2544 CrossRef CAS.
- M. J. Ruedas-Rama and E. A. H. Hall, Anal. Chem., 2008, 80, 8260–8268 CrossRef CAS PubMed.
- X. Wang and X. Guo, Analyst, 2009, 134, 1348–1354 RSC.
- Q. Qu, A. Zhu, X. Shao, G. Shi and Y. Tian, Chem. Commun., 2012, 48, 5473–5475 RSC.
- L. Zhou, Y. Lin, Z. Huang, J. Ren and X. Qu, Chem. Commun., 2012, 48, 1147–1149 RSC.
- Y. Wang, H. Mao and L. B. Wong, Talanta, 2011, 85, 694–700 CrossRef CAS PubMed.
- D. S. Liu, W. S. Phipps, K. H. Loh, M. Howarth and A. Y. Ting, ACS Nano, 2012, 6, 11080–11087 CAS.
- M. Howarth, K. Takao, Y. Hayashi and A. Y. Ting, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 7583–7588 CrossRef CAS PubMed.
- H. C. Fan, G. K. Fu and S. P. Fodor, Science, 2015, 347, 1258367 CrossRef PubMed.
- A. M. Klein, L. Mazutis, I. Akartuna, N. Tallapragada, A. Veres, V. Li, L. Peshkin, D. A. Weitz and M. W. Kirschner, Cell, 2015, 161, 1187–1201 CrossRef CAS PubMed.
- D. Ren, Y. Xia, J. Wang and Z. You, Sens. Actuators, B, 2013, 188, 340–346 CrossRef CAS.
- D. Ren, Y. Xia, B. Wang and Z. You, Anal. Chem., 2016, 88, 4318–4327 CrossRef CAS PubMed.
- X. T. Zheng, A. Than, A. Ananthanaraya, D. H. Kim and P. Chen, ACS Nano, 2013, 7, 6278–6286 CrossRef CAS PubMed.
- X. Gao and S. Nie, Trends Biotechnol., 2003, 21, 371–373 CrossRef CAS PubMed.
- R. Liu, F. Song, C. Jin and J. Liu, Int. J. Nanomed., 2012, 8, 33–38 Search PubMed.
- B. Dubertret, P. Skourides, D. J. Norris, V. Noireaux, A. H. Brivanlou and A. Libchaber, Science, 2002, 298, 1759–1762 CrossRef CAS PubMed.
- D. R. Larson, W. R. Zipfel, R. M. Williams, S. W. Clark, M. P. Bruchez, F. W. Wise and W. W. Webb, Science, 2003, 300, 1434–1436 CrossRef CAS PubMed.
- X. Gao, Y. Cui, R. M. Levenson, L. W. Chung and S. Nie, Nat. Biotechnol., 2004, 22, 969–976 CrossRef CAS PubMed.
- S. Kim, Y. T. Lim, E. G. Soltesz, A. M. De Grand, J. Lee, A. Nakayama, J. A. Parker, T. Mihaljevic, R. G. Laurence, D. M. Dor, L. H. Cohn, M. G. Bawendi and J. V. Frangioni, Nat. Biotechnol., 2004, 22, 93–97 CrossRef CAS PubMed.
- M. Nurunnabi, Z. Khatun, K. M. Huh, S. Y. Park, D. Y. Lee, K. J. Cho and Y. K. Lee, ACS Nano, 2013, 7, 6858–6867 CrossRef CAS PubMed.
- C. Ding, A. Zhu and Y. Tian, Acc. Chem. Res., 2014, 47, 20–30 CrossRef CAS PubMed.
- A. M. Smith, M. C. Mancini and S. Nie, Nat. Nanotechnol., 2009, 4, 710 CrossRef CAS PubMed.
- Y. Zhang, G. Hong, Y. Zhang, G. Chen, F. Li, H. Dai and Q. Wang, ACS Nano, 2012, 6, 3695–3702 CrossRef CAS PubMed.
- C. Li, Y. Zhang, M. Wang, Y. Zhang, G. Chen, L. Li, D. Wu and Q. Wang, Biomaterials, 2014, 35, 393–400 CrossRef CAS PubMed.
- Y. Du, B. Xu, T. Fu, M. Cai, F. Li, Y. Zhang and Q. Wang, J. Am. Chem. Soc., 2010, 132, 1470–1471 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |