A multi-functional guanine derivative for studying the DNA G-quadruplex structure†
Abstract
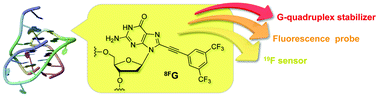
In the present study, we developed a multi-functional guanine derivative, 8FG, as a G-quadruplex stabilizer, a fluorescent probe for the detection of G-quadruplex formation, and a 19F sensor for the observation of the G-quadruplex. We demonstrate that the functional nucleoside bearing a 3,5-bis(trifluoromethyl)benzene group at the 8-position of guanine stabilizes the DNA G-quadruplex structure and fluoresces following the G-quadruplex formation. Furthermore, we show that the functional sensor can be used to directly observe DNA G-quadruplexes by 19F-NMR in living cells. To our knowledge, this is the first study showing that the nucleoside derivative simultaneously allows for three kinds of functions at a single G-quadruplex DNA. Our results suggest that the multi-functional nucleoside derivative can be broadly used for studying the G-quadruplex structure and serves as a powerful tool for examining the molecular basis of G-quadruplex formation in vitro and in living cells.


 Please wait while we load your content...
Please wait while we load your content...