Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Influence of washing and quenching in profiling the metabolome of adherent mammalian cells: a case study with the metastatic breast cancer cell line MDA-MB-231†
Rahul Vijay
Kapoore
 a,
Rachael
Coyle
ab,
Carolyn A.
Staton
b,
Nicola J.
Brown
a,
Rachael
Coyle
ab,
Carolyn A.
Staton
b,
Nicola J.
Brown
 b and
Seetharaman
Vaidyanathan
b and
Seetharaman
Vaidyanathan
 *a
*a
aChELSI Institute, Advanced Biomanufacturing Centre, Department of Chemical and Biological Engineering, The University of Sheffield, Mappin Street, Sheffield, S1 3JD, UK. E-mail: s.vaidyanathan@sheffield.ac.uk
bDepartment of Oncology and Metabolism, Faculty of Dentistry, Medicine and Health, The University of Sheffield, Sheffield, S10 2RX, UK
First published on 5th May 2017
Abstract
Metabolome characterisation is a powerful tool in oncology. To obtain a valid description of the intracellular metabolome, two of the preparatory steps are crucial, namely washing and quenching. Washing must effectively remove the extracellular media components and quenching should stop the metabolic activities within the cell, without altering the membrane integrity of the cell. Therefore, it is important to evaluate the efficiency of the washing and quenching solvents. In this study, we employed two previously optimised protocols for simultaneous quenching and extraction, and investigated the effects of a number of washing steps/solvents and quenching solvent additives, on metabolite leakage from the adherent metastatic breast cancer cell line MDA-MB-231. We explored five washing protocols and five quenching protocols (including a control for each), and assessed for effectiveness by detecting ATP in the medium and cell morphology changes through scanning electron microscopy (SEM) analyses. Furthermore, we studied the overall recovery of eleven different metabolite classes using the GC-MS technique and compared the results with those obtained from the ATP assay and SEM analysis. Our data demonstrate that a single washing step with PBS and quenching with 60% methanol supplemented with 70 mM HEPES (−50 °C) results in minimum leakage of intracellular metabolites. Little or no interference of PBS (used in washing) and methanol/HEPES (used in quenching) on the subsequent GC-MS analysis step was noted. Together, these findings provide for the first time a systematic study into the washing and quenching steps of the metabolomics workflow for studying adherent mammalian cells, which we believe will improve reliability in the application of metabolomics technology to study adherent mammalian cell metabolism.
Introduction
Mammalian metabolomics has received increased attention in recent years mainly because of its potential in the prognosis and diagnosis of cancer and for assessing treatment efficacy by analysing cells, fluids or tissues for specific biomarkers in experimental, translational and clinical studies. Adherent mammalian cell-lines are used extensively in oncology research as preclinical models but to date there is insufficient information on the application of metabolomics for the analysis of cultured mammalian cell lines.1,2 To obtain reliable metabolomics data for adherent cells, an optimised workflow must be identified which will provide maximum coverage for all classes of metabolites with minimum leakage.Breast cancer (BC) is a highly heterogeneous cancer for which morphologically and clinically distinct subgroups of cell lines have been established.3 Triple negative breast cancer (TNBC) is characterised by the absence of estrogen receptor (ER), progesterone receptor (PR) and lack of overexpression of human epidermal growth factor receptor (HER-2). TNBC represents approximately 20% of all BC and is typically associated with poor prognosis. Moreover, due to its aggressive phenotype, TNBC only partially responds to chemotherapy. At present, there is a lack of clinically established targeted therapies and a resultant increase in patient fatalities.4–6 In order to sustain growth and proliferation of tumour cells, they constantly require supplements of macromolecular precursors and an exhibition of altered metabolism compared to quiescent cells. Several researchers have employed the metabolomics approach to catalogue these changes and have focused on the classification approach where healthy cells are compared against tumour cells.6 Nevertheless, while doing so it is important to obtain reliable metabolomics data using an optimised workflow which will provide maximum coverage for all classes of metabolites with minimum leakage.7 To our knowledge this has not been done so far for the breast cancer studies involving metabolomics approach, which we address here.
The major bottlenecks associated with the metabolomics sample preparation workflow are efficient sampling, quenching and extraction of metabolites that do not alter the internal metabolite signatures.8 The high turnover rates of most of the intracellular metabolites require rapid sampling and instantaneous quenching of enzyme activity for reliably capturing metabolite levels. The ideal quenching solvent should instantly arrest cellular metabolic activity without causing significant damage to the cell envelop and thus preventing leakage of intracellular metabolites from the cells.9 Quenching with 60% v/v cold methanol at −40 or −50 °C was originally pioneered in Saccharomyces cerevisiae and has been used widely for microbial metabolomics.10 However, quenching with cold methanol in prokaryotic cells results in leakage of intracellular metabolites caused by cold shock. As a result buffer additives have been employed to reduce metabolite leakage.11 The commonly employed buffer additives are HEPES,10,12 AMBIC,13,14 tricine,10,12,15 or NaCl.16 Although the influence of these buffer additives in preserving the membrane integrity and therefore in minimising metabolite leakage is well studied for bacteria,11,12,17–19 fungi20 and yeast,10,15,21,22 they have not been as rigorously studied in mammalian cells.
Due to basic differences in the cell envelope structure, the sampling methods cannot simply be adapted for mammalian cells. Briefly, mammalian cells lack a cell wall, and instead have a cell membrane that makes it more prone to metabolite leakage due to the delicate and fragile nature of the cell envelope compared to that of prokaryotes. This has been clearly demonstrated for suspension mammalian cultures (CHO cells)13,16,23,24 where researchers have proposed various washing solvents and additives for quenching solvents that maintain the ionic strength and avoid osmotic shock, to aid in preserving the membrane integrity of these delicate cells. In the case of CHO cells, methanol/water (buffered/un-buffered) was compared with cold isotonic saline (0.9% w/v NaCl),16 and it was shown that quenching with isotonic saline did not damage the cells and resulted in proper metabolic arrest, as evidenced by the halted conversion of ATP to ADP and AMP. However, in contrast, another report on CHO cells13 showed little or no improvement in metabolite recoveries with the use of 0.9% NaCl compared to that of 60% aqueous methanol. Consequently, the use of methanol supplemented AMBIC has been proposed, which appears to preserve the membrane integrity, as evidenced by minimal leakage of intracellular ATP.13,25 A fluorescent marker such as green fluorescence protein (GFP) has been used as a visualisation marker to estimate the rate of metabolite efflux following membrane damage calculated by numerical modelling from CHO cells.24 The authors reported that 90% of small metabolites would be lost within ≤1 s when 5% of membrane damage is caused by washing, quenching solvents or harsh sampling techniques. This clearly underlines the requirement for rapid sampling methods with suitable quenching or washing solvents to avoid metabolite leakage.
The sampling and quenching of adherently grown cells is much more complicated than of suspension cultures, as the former involves detachment of adhered cells from a culture flask. In the case of simultaneous quenching methods, direct quenching and extraction using methanol of human breast cancer cells has been postulated.26 A similar approach using liquid nitrogen (LN2) was recently reported where washing with water, direct LN2 quenching and rapid single step extraction for LC-MS based metabolomics of adherent cultures was recommended.27 Our previous work demonstrated that selection of the sampling methods largely depends on the cell type, culture conditions (adherent or non-adherent), analytes of interest (exo-metabolome or endo-metabolome), and that different washing steps/solvent and various quenching additives will likely influence the analysis.9
There have been few rigorous studies demonstrating effective washing and quenching methods for adherent cell cultures. The use of buffer additives such as AMBIC, HEPES and NaCl have been evaluated with methanol/AMBIC identified as an effective quenching solvent for OVCAR-8 cells (ovarian cancer cells).28 Another study, explored three different quenching protocols (flash freezing (−80 °C), cold methanol (−20 °C) and cold methanol (−40 °C)) without any buffer additives to quench NSC-34 cell metabolism, and reported methanol quenching at −40 °C to be reliable.29 A further study evaluated the washing protocols with the use of phosphate buffered saline (PBS), water and no washing step for the colorectal cancer cell line HCT116 and reported extraction of metabolites with cold methanol without any washing to be satisfactory.30 It is important to note that the composition of media used for mammalian cultures are very rich in nutrients such as amino acids, vitamins, sugars, which are also present inside the cells as intracellular metabolites. Hence subsequent extraction of cells followed by quenching, without any washing step might result in false higher recoveries for specific intracellular metabolites that are found to be present in the medium as well. Moreover, these media carryovers might also interfere in the post extraction analytical protocols. Inclusion of a washing step for adherent cultures would therefore be advantageous as it can be performed rapidly prior to quenching, does not prolong the quenching time, and thus might improve the validity of the intracellular metabolite measurements. However, this needs to be investigated further, as there is little quantitative information on the effectiveness of the washing step (with respect to the solvents used and the number of steps) on metabolite leakage with adherent cells.
The major aim of the current study was to determine quantitatively, whether significant leakage occurs in adherent cultures during the washing and quenching steps, and if so whether this can be minimised. We determined the metabolite levels in individual fractions to establish any differences between fractions and to identify where improvements can be made to minimise metabolite leakage. We further aimed to determine the properties of the quenching solvent with different buffer additives, the number of washing steps required, and interferences with the analytical technique (GC-MS). We believe this to be the first extensive report on metabolite leakage of adherent mammalian cells that points the way forward in dealing with these cell types for reliable metabolomics analyses.
Experimental
Chemicals and reagents
The RPMI-1640 medium was obtained from Lonza (Gibco-BRL, Paisley, UK). All other reagents and consumables were obtained from Sigma-Aldrich (Dorset, UK), unless stated otherwise.Cell line, cell culture and growth assessment
The MDA-MB-231 epithelial breast cancer cell line was obtained from American Type Culture Collection (ATCC), (http://www.atcc.org/). The cells were cultured at 37 °C with 5% CO2 in 100 mm Nunclon dish (Nunclon™, Thermo Scientific) with 2 × 105 cells per dish with 10 ml RPMI media containing 10% fetal bovine serum (originated from the same batch), 1% penicillin/streptomycin (GIBCO®) and 1% glutamine. Growth curves were produced by seeding cells in 24 well plates with 500 μL media at a seeding density of 5 × 104 cells per well. Viable cell counts were obtained at 24, 36, 48 and 72 hours using a Beckman Coulter Vi-Cell XR cell viability analyser (Beckman Coulter, Germany) after trypsinization. Control flasks containing media but no cells were used to determine background chemical (metabolite) signals from the culture medium.ATP assay
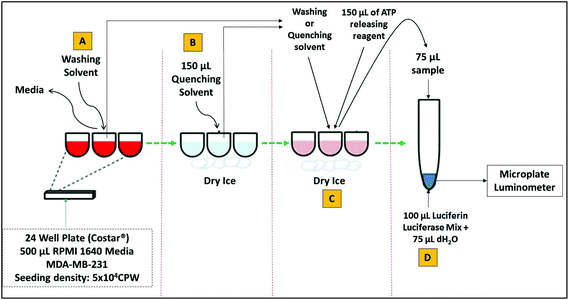
For all ATP assays, assay vials, glassware and pipette tips were soaked in 1 N HCl overnight followed by washing 3 times with ultrapure water before drying in an oven at 40 °C for 1 hour. MDA-MB-231 cells were seeded in triplicate (5 × 104 per well) in 24 well plate (Corstar®). The cells were incubated in 500 μL of RPMI 1640 media containing 1% fungazone, 10% fetal bovine serum (originated from the same batch), 1% penicillin/streptomycin (GIBCO®) and 1% glutamine at 37 °C with 5% CO2. After 36 hours of incubation, the culture medium was removed from each well and cells were washed and/or quenched using different reagents, including direct quenching with liquid nitrogen (LN2). The level of ATP released into the medium was then determined using ATP bioluminescent somatic cell assay kit (FLASC) (Sigma-Aldrich, Dorset, UK). See Table S1† for the assay parameters. Briefly, 100 μL of ATP assay mix was added to the assay vials and incubated for at least 3 minutes at room temperature in order to hydrolyse any ATP in the vial. Media and solvents after washing and quenching steps in each well were added to 150 μL of ATP releasing agent (ARR) which includes: p-tertiary-octylphenoxy polyethyl alcohol and edetic acid. Finally 75 μL of sterile distilled water was added to the vial containing luciferase assay mix, followed by the addition of 75 μL of the sample as shown in Fig. 1. The amount of light emitted from each reaction was then measured immediately in luminescence units (RLU) using a microplate luminometer (Centro LB 960, Berthold, Germany). The concentration of ATP in nM in each sample was determined from the log–log plot of ATP standards (0 to 1.5 nM) against relative luminescence units (RLU). To normalise for differences in cell number between various treatments, the concentration levels of free ATP were corrected for the level of protein (in micrograms) present in the same cell extracts prepared for the ATP assay. Determination of total protein content is described in detail in section ‘Protein assay as a normalisation to ATP content’. The nM amounts of ATP per mg of protein produced by MDA-MB-231 cells, after each treatment was determined in triplicates (n = 3). See Fig. S1† for the standard curves generated for both luciferase bioluminescence ATP assay (a) and BCA protein assay (b).Protein assay as a normalisation to ATP content
The total intracellular protein content was determined using the detergent-compatible bicinchoninic acid (BCA) protein assay using the MicroBCA Protein Assay Kit, (Thermo Scientific UK). Briefly, seven diluted albumin (BSA) standards in mg mL−1 (1, 0.8, 0.6, 0.4, 0.2, 0.1 and 0 mg mL−1) were prepared initially from a BSA stock solution (2 mg mL−1) using ARR as a diluent. The total volume of Micro BCA working reagent (WR) required was determined and prepared according to the manufacturer's instructions (Micro BCA Protein Assay Kit, Pierce/Thermo Scientific, Rockford, IL, USA) by mixing 25 parts of Micro BCA reagent MA and 24 parts of reagent MB with 1 part of reagent MC (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, reagent MA
1, reagent MA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MB
MB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MC). Finally, 75 μL of cell extract (MDA-MB-231) was added to the 96 well plate (Corstar®) followed by addition of 75 μL of BCA WR. The components were gently mixed for 30 s and incubated at 37 °C for 2 hours wrapped in aluminium foil. The plates were cooled to room temperature for 5 minutes after incubation and absorbance was measured at 570 nm on a plate reader (Centro LB 960, Berthold, Germany). The average absorbance reading of the blank standard was subtracted from the absorbance reading of all individual standards. Average values of all replicates samples (blank-corrected) were determined and used to generate a BCA standard curve by plotting the mean standard absorbance values against the respective concentrations. A best-fit polynomial equation was used for the standard curve and to estimate the amount of total protein content (mg).
MC). Finally, 75 μL of cell extract (MDA-MB-231) was added to the 96 well plate (Corstar®) followed by addition of 75 μL of BCA WR. The components were gently mixed for 30 s and incubated at 37 °C for 2 hours wrapped in aluminium foil. The plates were cooled to room temperature for 5 minutes after incubation and absorbance was measured at 570 nm on a plate reader (Centro LB 960, Berthold, Germany). The average absorbance reading of the blank standard was subtracted from the absorbance reading of all individual standards. Average values of all replicates samples (blank-corrected) were determined and used to generate a BCA standard curve by plotting the mean standard absorbance values against the respective concentrations. A best-fit polynomial equation was used for the standard curve and to estimate the amount of total protein content (mg).
Scanning electron microscopy (SEM) analysis
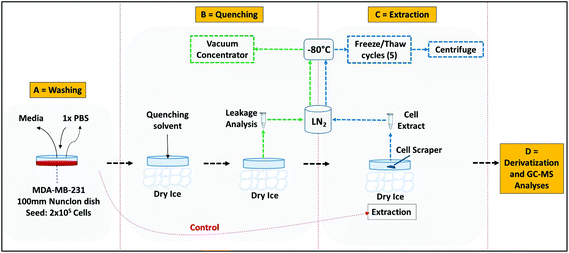
SEM analysis was performed on samples of control (non-quenched cells) and on MDA-MB-231 cells quenched with four different quenching reagents. The cells were grown in 12 well plate under similar conditions as described in section ‘Cell line, cell culture and growth assessment’. However, for ease of SEM analysis we used silicon wafers for growing adherent mammalian cells (Fig. S2†). Briefly, the specimens were fixed in 3% glutaraldehyde in 0.1 M phosphate buffer at 4 °C. The specimens were then washed thrice in 0.1 M phosphate buffer with 30 min intervals at 4 °C. Secondary fixation was carried out in 1% osmium tetroxide aqueous for 1 h at room temperature. Dehydration was carried out through a graded series of ethanol concentrations in the following order: 75% ethanol for 15 min, 95% ethanol for 15 min, 100% ethanol for 15 min, 100% ethanol for 15 min, 100% ethanol dried over anhydrous copper sulphate for 15 min. All the above dehydration steps were carried out at room temperature. The specimens were then placed in a 50/50 mixture of 100% ethanol and 100% hexamethyldisilazane for 30 min followed by 30 minutes in 100% hexamethyldisilazane. The specimens were then allowed to air dry overnight at room temperature before mounting on 12.5 mm aluminium stubs using carbon-sticky tabs and then coated in an Edwards S150B sputter coater with approximately 25 nm of gold. The specimens were examined in a Philips XL-20 Scanning Electron Microscope at an accelerating voltage of 20 kV.Cell quenching
The cells were grown to mid-“exponential” phase (36 h) and then rapidly quenched before extraction. Initially, the culture medium was removed and cells were quickly washed once with 3 mL of optimised washing solvent (∼60 seconds). The residual PBS was removed by suction. The cells were rapidly quenched with 3 mL of pre-chilled (−50 °C) quenching solvent (buffered or non-buffered): (i) 100% methanol alone, (ii) 60% aqueous methanol, (iii) 60% aqueous methanol supplemented with 0.85% (w/v) ammonium bicarbonate (AMBIC, pH 7.4) or (iv) 60% aqueous methanol supplemented with 70 mM HEPES. Addition of cells to the quenching solvent increased the temperature by no more than 15 °C. After 60 s, the quenching solution was removed, and an aliquot (2 mL) was transferred to a 2 mL pre-chilled Eppendorf, and then snap frozen in LN2 and stored at −80 °C prior to determining the leakage of the internal metabolites.Metabolite extraction
Quenching and extractions were performed using the methods illustrated in Fig. 2. The adherent cells were first quenched using all the quenching solvents as described in section ‘Cell quenching’, followed by extraction with 1 mL of pre-chilled 100% methanol (−40 °C). The cells were then suspended in extraction solvent on dry ice after cell lifting with a pre-chilled rubber tipped cell scraper. The cell suspension was then transferred to a 2 mL pre-chilled Eppendorf and stored on dry ice. A further 1 mL aliquot of 100% methanol was added to the culture flask and the same procedure was repeated. The first and second aliquot were combined and the metabolites extracted by performing freeze–thaw cycles as suggested elsewhere.31 Briefly the aliquot was snap frozen in liquid nitrogen (LN2) for 3 minutes, followed by thawing on dry ice and vortexed. The freeze–thaw cycle was repeated five times for complete cell disruption and followed by centrifugation at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at −9 °C for 5 minutes. The pellet was re-extracted with 500 μL of 100% methanol. The first and second aliquots were then combined and the extract was lyophilized overnight prior to derivatization.
000g at −9 °C for 5 minutes. The pellet was re-extracted with 500 μL of 100% methanol. The first and second aliquots were then combined and the extract was lyophilized overnight prior to derivatization.
Simultaneous quenching and extraction with liquid nitrogen
Recent protocols for simultaneous quenching and extraction,3,9,27,32 were adopted with a few modifications. Briefly, after removal of the culture medium and washings with PBS, the cells were rapidly quenched by directly adding about 10 mL LN2. The adherent cells were then immediately subjected to extraction on dry ice by addition of 1 mL of pre-chilled 100% methanol followed by lifting the cells with a pre-chilled rubber tipped cell scraper. The cells were re-extracted and further steps were performed as described in section ‘Metabolite extraction’.Metabolite derivatization
Metabolite derivatization was performed as described elsewhere.31 Briefly, to the lyophilized extract, 40 μL of 20 mg mL−1 methoxyamine hydrochloride in pyridine was added and samples were shaken for 80 min at 37 °C. The samples were then derivatized by trimethylsilylation of acidic protons by addition of 40 μL MSTFA (N-methyl-N-trimethylsilyltrifluoroacetamide) with further incubations in shaking conditions at 40 °C for 80 min. A retention index solution was added for the chromatographic alignment prior to analysis by GC-MS (20 μL, 0.6 mg mL−1 C10, C12, C15, C19, C22 n-alkanes).GC-MS analysis
Metabolite data was acquired on a Thermo Finnigan TRACE DSQ GC-MS System (Thermo Scientific, Hertfordshire, UK) operating in EI mode onto a TRACE TR-5MS capillary column (30 m × 0.25 mm × 0.25 μm). The derivatized sample volume of 1 μL was injected in splitless mode at 230 °C, with the transfer line temperature maintained at 250 °C. The GC was operated at a constant flow of 1 mL min−1 helium. The temperature program was started at 80 °C for 6 min, followed by temperature ramping at 6 °C min−1 to a final temperature of 290 °C and held constant at 310 °C for 5 min. Data acquisition was performed on a DSQ MS system with a mass range of 50 to 650.Metabolite identification
The metabolites were identified as trimethylsilyl (TMSi) derivatives by comparing their mass spectral and RI index with online databases (the GOLM Metabolome database: http://csbdb.mpimp-golm.mpg.de/ and NIST 05 database). The acquired spectra were deconvoluted by AMDIS (Automated Mass Spectral Deconvolution and Identification System), before comparing with the database. Spectra of individual components were further transferred to the NIST mass spectral search system and matched with NIST main library, RI index library and the GMD (GOLM metabolome database). The metabolite identification adopted here could be classified as Metabolomics Standard Initiative (MSI), level 2.Data analysis
GC-MS analysis yields complex data sets (time × mass × intensity) which require deconvolution, as fragment ions may be shared between two co-eluting compounds. All GC-MS chromatograms were processed using freely available AMDIS 2.70 software. The peaks were deconvoluted and the retention indices (RIs) were automatically calculated according to the retention time of the alkane mixture by exporting the RI calibration file into AMDIS. AMDIS deconvolution parameters used are as follows: resolution was set to high, sensitivity was high, shape requirement was medium, and component width was 12 (validated with 12 metabolite standard mixture).9 For identification, the minimum match factor was kept at 60, resolution: high; sensitivity: high; shape requirement: medium. Finally, a report was generated in *.xls format and the first hit considered. Compounds identified in at least two out of three biological replicates were considered true hits. Data for retention time, S/N ratio, peak tailing, m/z value and peak area was collected manually by exporting to MS Excel 2013. For intracellular metabolites the GC-MS data was normalised to viable cell count and the sum of peak areas, as suggested elsewhere.3Results and discussion
We have evaluated metabolite leakage in the adherent metastatic breast cancer cell line MDA-MB-231 using a range of washing and quenching solvents (buffered/non-buffered). Results from the different treatments are evaluated and compared using the ATP assay, SEM analysis (for visual observation of membrane integrity) and GC-MS based untargeted metabolomics.ATP assay in response to washing and quenching protocols
ATP is a key central metabolite of all live cells, has a high turnover rate (1.5 mM s−1) and constant concentration across each cell.16 Unlike other metabolites, ATP is never secreted into the extracellular environment by cells under normal conditions. However, when cells are subjected to environmental stress or membrane damage, rapid changes in ATP concentration occurs because of the high turnover rate, with subsequent leakage into the extracellular medium.33,34 Quenching protocols in metabolomics studies are well known to cause severe leakage of metabolites from cells due to cold shock. Therefore, measuring the amount of ATP released by the cells, in response to various washing and quenching solvent, will provide an indication of the extent of metabolite leakage and quenching efficiency of the solvents.11,13,25Based on the above rationale, the first objective of this investigation was to evaluate the extent of metabolite leakage in response to different washing and quenching solvents (buffered/non-buffered) using the ATP assay. The ATP content released in the washing/quenching supernatant was determined using a luciferase bioluminescence assay and normalised to total protein content of the cell. This approach has been successfully applied to bacterial18,35,36 and mammalian cell suspensions,11,13,16,25 although there is little or no information in the published literature on the application of the approach to adherent cell cultures. Hence prior to application of these assays to actual experiments on adherent cultures, standard curves were generated for both ATP and protein assay using standard solutions, an additional test standard solution and the MDA-MB-231 cell extract (n = 3) (Fig. S1†).
In adherent cultures, a similar workflow to that used in suspension cultures cannot be applied since lifting of cells by trypsinization or cell scraping is likely to result in more mechanical damage with subsequent increased leakage than would be caused by quenching or washing steps alone. Hence, it is essential to quench the adherent cultures in the adhered state after removal of the medium. It is known that usage of large volumes of quenching solvents removes the majority of media components and extracellular metabolites potentially reducing the need for a washing step. However, in cases where the media components are more concentrated than the intracellular metabolites, the inclusion of a washing step to remove media components and extracellular metabolites could be advantageous. That said, a key concern is whether the use of washing solvents, and the number of washing steps employed, influence metabolite leakage from adherent cultures. In order to evaluate the effects of washing, the levels of ATP released into the washing solvents were determined after the washing step (Table S2†).
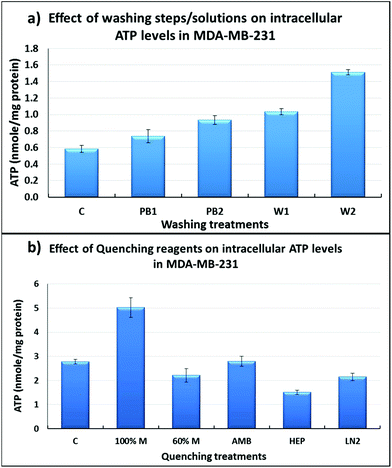
As can be seen in Fig. 3a, washing with de-ionised water (W1 & W2) resulted in increased leakage of ATP when compared to washing with PBS (PB1 & PB2) (P < 0.05), and with two washes there was increased leakage of ATP (W1 vs. W2; PB1 vs. PB2). Washing once with PBS (PB1) resulted in similar results to the control sample (C) suggesting that one PBS wash is less damaging than a two-step PBS wash. Mammalian cells are delicate and to preserve their membrane integrity, it is essential that the ionic strength of the medium is kept highly compatible. The extremely low ionic strength of water might be responsible for more leakage of intracellular ATP into the extracellular environment, likely caused by osmotic shock. The effect of more than two washes with PBS was not investigated due to an increase in the processing time which will result in decreased ATP levels due to the conversion of ATP to ADP and AMP, consequently the estimation of metabolite leakage will not be reliable. For subsequent experiments, a single wash with PBS prior to quenching was selected as the optimal washing step.
Scanning electron microscopy (SEM) analysis in response to quenching protocols
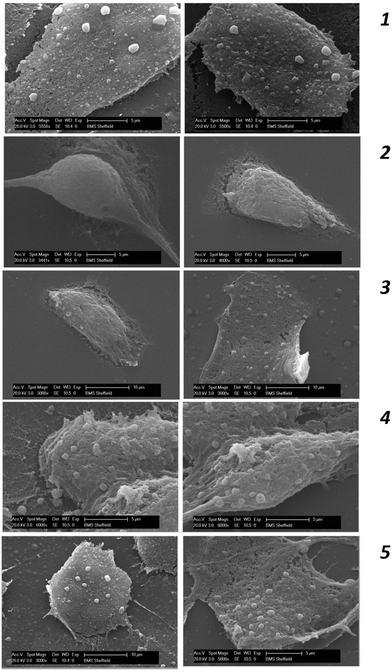
In addition to the ATP assay, the cell membrane integrity of MDA-MB-231 was further studied using SEM analysis following the different quenching protocols which are summarised in Fig. 4. In the case of non-quenched cells (control) (Fig. 4-1), it can be clearly seen that MDA-MB-231 cells adopt a polygonal and a flat morphology in the adhered/fixed state, where the patterned network and vascular channels are clearly visible. However, when the cells are quenched with 100% methanol (Fig. 4-2), detrimental effects on cellular morphology can be clearly seen where patterned network, vascular channels and pores have been obliterated. The use of 60% aqueous methanol (Fig. 4-3) also has a similar effect, however there is evidence of some membrane integrity preservation compared to that of 100% methanol. The use of additives such as HEPES and AMBIC preserved the membrane integrity to a greater extent than when alcohol was used alone, however there is some damage to the membrane when compared to the control (Fig. 4-4). Overall, quenching with methanol supplemented with 70 mM HEPES (Fig. 4-5) appears to be the most appropriate quenching solvent and compares well with findings from the ATP assay.GC-MS based overall recovery of metabolites
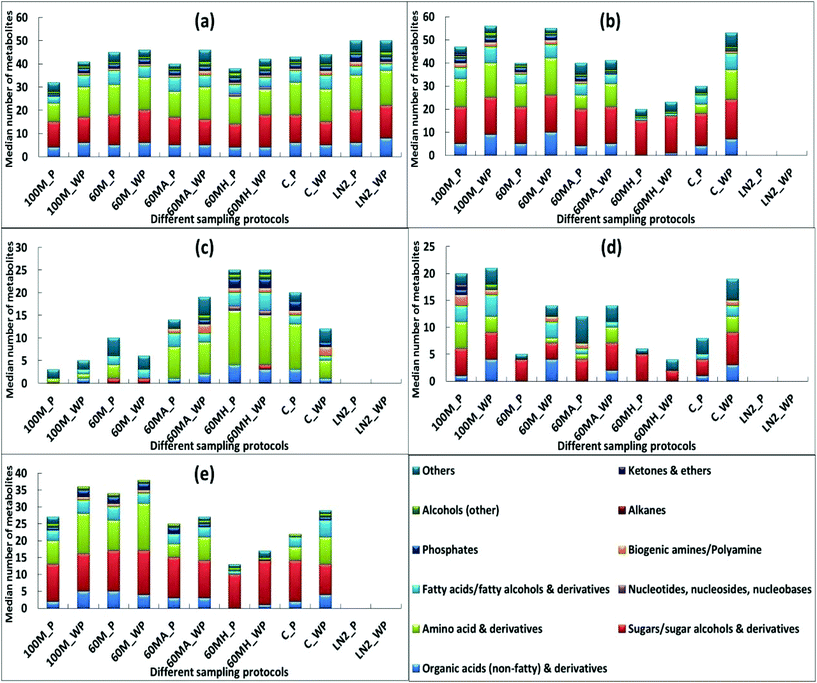
In order to obtain confirmation of the findings from the ATP assay and SEM analysis, GC-MS sample analysis was undertaken following the different quenching protocols (buffered or non-buffered). In total, 112 intracellular metabolites were identified after correction for leaked metabolites (Table S3†) across all treatments. However, since the study objective is to maximise metabolome coverage, characterisation of leakage with respect to metabolite class is required. For this, it is essential to first identify and classify the metabolites detected based on their physicochemical properties, thus generating a metabolite matrix that can be utilized for interpretations. Hence, all the identified metabolites with the different sample treatment approaches were initially classified into eleven different metabolite classes based on their physicochemical properties.9As can be seen from Fig. 5c, 60% aqueous methanol supplemented with HEPES yielded similar recoveries of metabolites unique to the cells with or without a washing step (60MH_P and 60MH_WP) and demonstrated the highest recovery compared to other treatments (except that of LN2). Correspondingly, metabolites common to both the cells and supernatant (Fig. 5e) are the lowest (with slightly higher recovery when the washing step was excluded in 60% methanol supplemented with HEPES (60MH_WP)). The number of metabolites detected in the supernatant is also the least compared to other treatments (Fig. 5d). Among other treatments, exclusion of washing step with 100% methanol (100M_WP) and 60% methanol supplemented with AMBIC (60MA_WP) resulted in overall higher recovery of metabolites unique to the cells compared to those with inclusion of the PBS wash (100M_P and 60MA_P) (Fig. 5c). In contrast, higher recoveries were obtained with 60% methanol and unquenched sample involving washing step (60M_P and C_P), compared to that when the washing step was excluded (60M_WP and C_WP). However, recoveries of metabolites common to both the cells and supernatant (Fig. 5e) were higher when the washing step was excluded. One possible reason might be carry-over of media components along with the intracellular metabolites contributing to the higher recoveries with all the treatments when the washing step was not included. Only the use of HEPES, and direct quenching with LN2 (with or without a PBS wash) showed negligible variations in the recovery of metabolites in cell extracts (Fig. 5a).
Our findings for adherent cells are in contrast to the previously published report for suspension CHO cells,13 where the authors suggested that no washing step is required based on an ATP assay. The authors reported loss of ATP with all the evaluated treatments (methanol alone and methanol supplemented with 70 mM HEPES, 0.85% AMBIC and 0.9% NaCl) when compared to no washing. However it is important to note that the washing step was performed with the quenching reagents which may further compromise membrane integrity leading to elevated metabolite leakage, due to cold shock phenomenon and extended time periods for quenching. In another report,25 a similar strategy was adopted for CHO cells, where the authors investigated the effect of additional washing steps (one, two and three) against no washing, where PBS was used for both washing and quenching steps. There was severe leakage with all the washing steps, which may be due to the longer protocol duration accelerating the conversion of ATP to ADP and AMP. We have demonstrated minimal variation in ATP leakage between non-washed (control) and cells washed once with PBS, in adherent mammalian cultures, which was confirmed by GC-MS analysis, and demonstrated minimal variations in recovery of intracellular metabolites (Fig. 5c) across all the applied treatments.
In contrast to both buffered and non-buffered methanol, direct quenching with LN2 yielded the highest recoveries for all the metabolite classes (Fig. 5a). However, there were problems in using LN2 for simultaneous quenching and extraction of metabolites. The procedure involved pouring of LN2 directly into T75 culture flasks (small neck diameter) or well plates, which may result in the loss of sample and also has operator health and safety issues (this could result in frostbite or cryogenic burns, and possible asphyxiation), when working with small volumes, as is often the case.
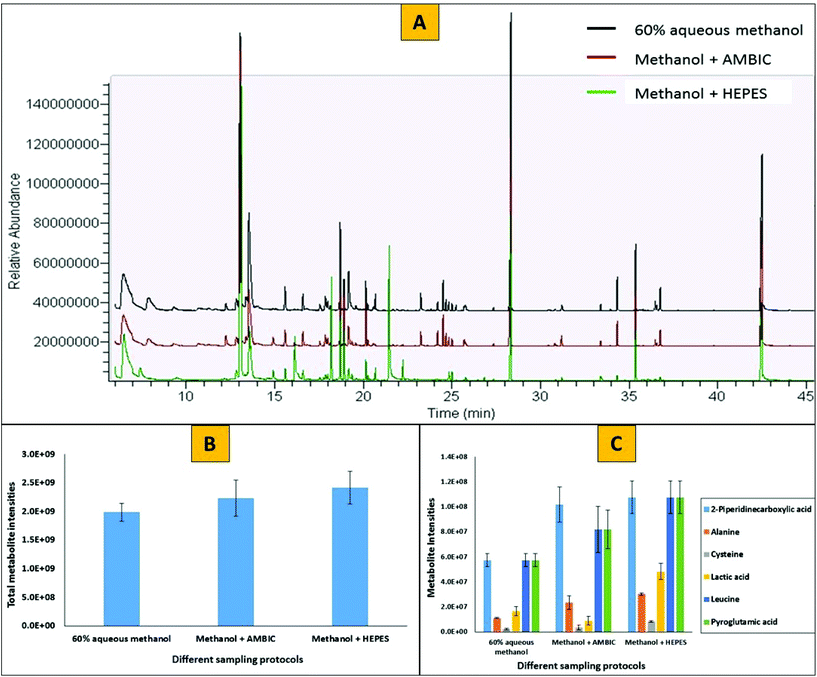
To evaluate the effects of a PBS washing step and the use of quenching additives (HEPES or AMBIC) on two step derivatization protocols (methoximation followed by silylation) in GC-MS based analysis, we have profiled the metabolome and compared the chromatograms of methanol supplemented with HEPES to that of methanol supplemented with AMBIC and methanol with no additive. Fig. 6 represents the GC-MS profiles of intracellular metabolites extracted from adherent MDA-MB-231 mammalian cells. Cells were washed once with PBS followed by quenching with either 60% aqueous methanol alone (black), or 60% aqueous methanol supplemented with 0.85% AMBIC (red) or 60% aqueous methanol supplemented with 70 mM HEPES (green) prior to extraction of metabolites using cold methanol. For clarity, the Y-axis for 60% aqueous methanol and 60% aqueous methanol + 0.85% AMBIC has been offset. Comparison between three different sample types with respect to total numbers and amounts of metabolites recovered showed no major differences. In total 41 ± 3 unique metabolites were identified with each of the three different sample types. However, slightly higher relative peak areas for all the metabolites were obtained with methanol supplemented with HEPES treatment compared to other two treatments (20% higher compared to aqueous methanol alone and 12% higher compared to methanol supplemented with AMBIC). This analysis clearly reveals that the presence of HEPES and AMBIC does not interfere with the derivatization reactions and GC-MS analysis. Our results for inclusion of AMBIC were in agreement with that of Sellick and co-workers,13 however in addition to that we note no apparent interference of HEPES with the derivatization reactions and the GC-MS analysis.
Conclusions
Estimation of the intracellular ATP levels in response to various washing steps/solvents results in minimal ATP leakage between non-quenched (control) cells and cells washed once with PBS, indicating a single washing step with PBS maintains the membrane integrity of adherent mammalian cultures. Moreover, GC-MS analysis confirms this finding, where similar numbers of intracellular metabolites are recovered with all the applied treatments. Estimation of extracellular ATP levels in response to various quenching solvents results in less leakage of ATP using methanol supplemented with 70 mM HEPES, and this treatment appears to preserve the membrane integrity of MDA-MB-231 cells compared to the other quenching solvents evaluated. We obtained similar findings using GC-MS based metabolomics, where analysis based on recoveries of eleven different metabolite classes clearly demonstrated severe leakage of nearly all metabolite classes with the use of non-buffered methanol quenching treatments compared to that of cells quenched with buffered methanol or direct quenching with LN2. Among buffered quenching solvents containing 60% aqueous methanol, HEPES supplemented methanol results in the highest recoveries of organic acids, amino acids, nucleotides and phosphates compared to that of AMBIC supplemented methanol. Furthermore, SEM analysis in response to various quenching solvents yields similar findings to that of the ATP assay and GC-MS based analyses. In addition, we have demonstrated no interferences on the derivatization reactions in GC-MS analysis when PBS and HEPES are employed in the protocol. The findings from different analytical platforms compare well and clearly indicate that a single washing step with PBS and quenching with methanol supplemented with 70 mM HEPES (−50 °C) results in minimum leakage of intracellular metabolites from an adherent mammalian cell-line. We believe this to be the first report of a systematic study into the washing and quenching steps of the metabolomics workflow for studying adherent mammalian cells, which we believe will improve reliability in the application of metabolomics technology to study adherent mammalian cell metabolism.Acknowledgements
Funding support from Chemical Engineering at the Life Science Interface (ChELSI) (EPSRC: EP/E036252/1) for part funding of a PhD studentship to Rahul Vijay Kapoore and the BBSRC-DTA: BB/F016840/1 studentship funding to Rachael Coyle are gratefully acknowledged.References
- M. Cuperlovic-Culf, D. A. Barnett, A. S. Culf and I. Chute, Drug Discovery Today, 2010, 15, 610–621 CrossRef CAS PubMed.
- S. H. G. Khoo and M. Al-Rubeai, Biotechnol. Appl. Biochem., 2007, 47, 71–84 CrossRef CAS PubMed.
- A. Hutschenreuther, A. Kiontke, G. Birkenmeier and C. Birkemeyer, Anal. Methods, 2012, 4, 1953–1963 RSC.
- C. Criscitiello, H. A. Azim, Jr., P. C. Schouten, S. C. Linn and C. Sotiriou, Ann. Oncol., 2012, 23, 13–18 CrossRef PubMed.
- O. Gluz, C. Liedtke, N. Gottschalk, L. Pusztai, U. Nitz and N. Harbeck, Ann. Oncol., 2009, 20, 1913–1927 CrossRef CAS PubMed.
- F. Podo, L. M. C. Buydens, H. Degani, R. Hilhorst, E. Klipp, I. S. Gribbestad, S. Van Huffel, H. W. M. van Laarhoven, J. Luts, D. Monleon, G. J. Postma, N. Schneiderhan-Marra, F. Santoro, H. Wouters, H. G. Russnes, T. Sorlie, E. Tagliabue, A.-L. Borresen-Dale and F. Consortium, Mol. Oncol., 2010, 4, 209–229 CrossRef CAS PubMed.
- R. V. Kapoore and S. Vaidyanathan, Philos. Trans. R. Soc., A, 2016, 374, 20150363 CrossRef PubMed.
- R. V. Kapoore, Mass spectrometry based hyphenated techniques for microalgal and mammalian metabolomics, University of Sheffield, 2014 Search PubMed.
- R. V. Kapoore, R. Coyle, C. A. Staton, N. J. Brown and S. Vaidyanathan, Metabolomics, 2015, 11, 1743–1755 CrossRef CAS.
- A. B. Canelas, C. Ras, A. ten Pierick, J. C. van Dam, J. J. Heijnen and W. M. Van Gulik, Metabolomics, 2008, 4, 226–239 CrossRef CAS.
- M. Faijes, A. E. Mars and E. J. Smid, Microb. Cell Fact., 2007, 6, 27 CrossRef PubMed.
- H. Taymaz-Nikerel, M. de Mey, C. Ras, A. ten Pierick, R. M. Seifar, J. C. Van Dam, J. J. Heijnen and W. M. Van Glilik, Anal. Biochem., 2009, 386, 9–19 CrossRef CAS PubMed.
- C. A. Sellick, R. Hansen, A. R. Maqsood, W. B. Dunn, G. M. Stephens, R. Goodacre and A. J. Dickson, Anal. Chem., 2009, 81, 174–183 CrossRef CAS PubMed.
- G. D. Tredwell, B. Edwards-Jones, D. J. Leak and J. G. Bundy, PLoS One, 2011, 6, e16286 CAS.
- J. I. Castrillo, A. Hayes, S. Mohammed, S. J. Gaskell and S. G. Oliver, Phytochemistry, 2003, 62, 929–937 CrossRef CAS PubMed.
- S. Dietmair, N. E. Timmins, P. P. Gray, L. K. Nielsen and J. O. Kroemer, Anal. Biochem., 2010, 404, 155–164 CrossRef CAS PubMed.
- C. J. Bolten, P. Kiefer, F. Letisse, J.-C. Portais and C. Wittmann, Anal. Chem., 2007, 79, 3843–3849 CrossRef CAS PubMed.
- H. Link, B. Anselment and D. Weuster-Botz, Metabolomics, 2008, 4, 240–247 CrossRef CAS.
- R. Marcinowska, J. Trygg, H. Wolf-Watz, T. Mortiz and I. Surowiec, J. Microbiol. Methods, 2011, 87, 24–31 CrossRef CAS PubMed.
- H. Hajjaj, P. J. Blanc, G. Goma and J. Francois, FEMS Microbiol. Lett., 1998, 164, 195–200 CrossRef CAS.
- S. Kim, D. Y. Lee, G. Wohlgemuth, H. S. Park, O. Fiehn and K. H. Kim, Anal. Chem., 2013, 85, 2169–2176 CrossRef CAS PubMed.
- J. Spura, L. C. Reimer, P. Wieloch, K. Schreiber, S. Buchinger and D. Schomburg, Anal. Biochem., 2009, 394, 192–201 CrossRef CAS PubMed.
- M. Volmer, J. Gettmann, S. Scholz, H. Buntemeyer and T. Noll, BMC Proc., 2011, 5(Suppl 8), P93–P93 CrossRef PubMed.
- M. Wurm and A.-P. Zeng, Metabolomics, 2012, 8, 1081–1089 CrossRef CAS.
- J. Kronthaler, G. Gstraunthaler and C. Heel, OMICS, 2012, 16, 90–97 CrossRef CAS PubMed.
- Q. Teng, W. Huang, T. W. Collette, D. R. Ekman and C. Tan, Metabolomics, 2009, 5, 199–208 CrossRef CAS.
- M. A. Lorenz, C. F. Burant and R. T. Kennedy, Anal. Chem., 2011, 83, 3406–3414 CrossRef CAS PubMed.
- P. Purwaha, P. L. Lorenzi, L. P. Silva, D. H. Hawke and J. N. Weinstein, Metabolomics, 2014, 10, 909–919 CrossRef CAS PubMed.
- B. M. Hounoum, H. Blasco, L. Nadal-Desbarats, B. Dieme, F. Montigny, C. R. Andres, P. Emond and S. Mavel, Anal. Bioanal. Chem., 2015, 407, 8861–8872 CrossRef PubMed.
- Z. Ser, X. Liu, T. Ngoc Nu and J. W. Locasale, Anal. Biochem., 2015, 475, 22–28 CrossRef CAS PubMed.
- C. L. Winder, W. B. Dunn, S. Schuler, D. Broadhurst, R. Jarvis, G. M. Stephens and R. Goodacre, Anal. Chem., 2008, 80, 2939–2948 CrossRef CAS PubMed.
- H. Bi, K. W. Krausz, S. K. Manna, F. Li, C. H. Johnson and F. J. Gonzalez, Anal. Bioanal. Chem., 2013, 405, 5279–5289 CrossRef CAS PubMed.
- D. Y. Lee and O. Fiehn, Plant Methods, 2008, 4, 7 CrossRef PubMed.
- E. M. Schwiebert and A. Zsembery, Biochim. Biophys. Acta, Biomembr., 2003, 1615, 7–32 CrossRef CAS.
- U. Nasution, W. M. Van Gulik, R. J. Kleijn, W. A. Van Winden, A. Proell and J. J. Heiinen, Biotechnol. Bioeng., 2006, 94, 159–166 CrossRef CAS PubMed.
- F. Schaedel, F. David and E. Franco-Lara, Appl. Microbiol. Biotechnol., 2011, 92, 1261–1274 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7an00207f |
| This journal is © The Royal Society of Chemistry 2017 |