Development of a high throughput (HT) Raman spectroscopy method for rapid screening of liquid blood plasma from prostate cancer patients
Dinesh K. R.
Medipally
 ab,
Adrian
Maguire
ab,
Jane
Bryant
b,
John
Armstrong
c,
Mary
Dunne
c,
Marie
Finn
c,
Fiona M.
Lyng†
ab,
Adrian
Maguire
ab,
Jane
Bryant
b,
John
Armstrong
c,
Mary
Dunne
c,
Marie
Finn
c,
Fiona M.
Lyng†
 *ab and
Aidan D.
Meade†
*ab and
Aidan D.
Meade†
 *ab
*ab
aSchool of Physics, Dublin Institute of Technology, Kevin Street, Dublin 8, Ireland. E-mail: fiona.lyng@dit.ie; aidan.meade@dit.ie
bDIT Centre for Radiation and Environmental Science, Focas Research Institute, Dublin Institute of Technology, Camden Row, Dublin 8, Ireland
cDepartment of Radiation Oncology, Saint Luke's Radiation Oncology Network, St Luke's Hospital, Dublin, Ireland
First published on 13th December 2016
Abstract
Extensive research has been undertaken on the examination of tissue biopsies using vibrational spectroscopic techniques. However, fewer studies have focused on less invasive and commonly acquired blood samples. Recent studies have shown the ability of Raman and Fourier transform infrared (FTIR) spectroscopy to discriminate between non-cancer controls and cancer cases using blood serum or plasma. Even though many studies have proposed Raman spectroscopy as a potential diagnostic tool in various cancers, the Raman spectroscopic technique has not been introduced as a routine clinical technology. This is due to multiple drawbacks with the application of the technique, including sample preparation, the requirement for expensive substrates and long acquisition times. The current study aims to overcome these limitations and focuses on the translation of Raman spectroscopy into a high throughput clinical diagnostic tool for prostate cancer. In this study, the effect of different instrumental and sample preparation parameters were investigated, with the aim of identifying a combination that would reduce the overall acquisition time for spectra from peripheral blood plasma, reduce the complexity of sample preparation and retain the classification accuracy from Raman spectroscopic diagnostics. A high throughput (HT) system was developed and Raman spectroscopic measurements were performed on plasma samples from 10 prostate cancer patients and 10 healthy volunteers. The spectra were pre-processed and classified by principal component analysis – linear discriminant analysis (PCA-LDA) in the R environment. Statistically significant differences were observed between Raman spectra of prostate cancer patients and non-cancer controls. The (HT) classification resulted in a sensitivity and specificity of 96.5% and 95% respectively. Overall, this study has overcome some of the limitations associated with clinical translation of Raman spectroscopy. The HT-Raman spectroscopy method developed in this study can be used for rapid and accurate diagnosis of prostate cancer using liquid plasma samples.
Introduction
In western countries, prostate cancer is the second most frequently diagnosed cancer and the third most common cause of death from cancer in men.1 Clinically there are several means of detecting prostate cancer including B-mode ultrasound, biopsy, and tumor marker screening, although each of these techniques possess some disadvantages. B-mode ultrasound images the formed solid tumor in the prostate gland and thus can only detect patients that have progressed through the early stages of cancer.2 Biopsy is the gold standard for detection of cancer, but it is invasive and impractical for a high-risk patient with multiple suspicious lesions, and particularly so for deep-seated cancers such as that of the prostate. Tumor marker screening, in particular the measurement of prostate specific antigen (PSA), has significantly improved early diagnosis. The PSA test has a good specificity (91%) but is limited in sensitivity (20.5%). PSA levels can be high in some subjects without prostate cancer leading to overdiagnosis; also PSA has poorer discriminating ability in men with symptomatic benign prostatic hyperplasia.3The potential of Raman and FTIR spectroscopy for diagnostic applications has been well investigated and demonstrated.4–7 The detailed information obtained from Raman spectroscopy provides information about the molecular structure and the composition of cells, tissues and biofluids, and ultimately promises an analysis of disease origin and progression. The analysis of biofluids can enable powerful minimally invasive diagnostics for many diseases.8
Infrared (IR) spectroscopy is based on the absorption of infrared radiation by the sample under study and the fact that molecules absorb specific frequencies of the incident light which are characteristic of their structure. FTIR spectroscopy is a non-destructive, label-free method for studying molecular composition and the structure of macromolecules, either in their isolated form9 or within complex biological systems like cells and tissues.10 It can in a single measurement detect spectral variations linked to various molecular constituents, such as nucleic acids, carbohydrates proteins or lipids, present in the sample, in a qualitative and quantitative way.11 The modality has been recently applied to biofluids for screening diseases, such as Alzheimer's disease, galactosemia, hepatic fibrosis and hepatocellular carcinoma with high specificity and sensitivity (>85%).7,11–13 However there are some disadvantages associated with FTIR spectroscopic analysis of biofluids. Biofluids must be measured in the dried state due to absorbance of water in the mid IR region. As glass has a spectral signature in the mid-IR region, glass slides cannot be used as substrates. In addition, because the FTIR method has constraints on sample thickness, uniformity and dilution to avoid saturation, some sample preparation is needed.14
Raman spectroscopy is based on inelastic scattering of monochromatic light, usually from a laser source. Inelastic scattering means that the frequency of photons in monochromatic light changes upon interaction with a sample.15 Like FTIR spectroscopy, Raman spectroscopy is a label free, non-destructive technique for studying molecular composition. Key advantages are that it requires little to no sample preparation and has no interference from water. This is due to polar molecules, such as water, producing relatively weak Raman signals.16 However, unlike FTIR spectroscopy, long spectral acquisition times are needed to achieve adequate SNR.17 Raman spectroscopy has recently been applied to biofluids (serum or plasma) for discriminating between non cancer controls and head and neck cancer patients,18 breast cancer patients19 and cervical cancer patients20 with sensitivity and specificity above 75%.
Despite its proven ability, Raman spectroscopy lags behind FTIR spectroscopy for the purposes of clinical diagnosis with biofluids due to certain limitations. Raman spectroscopic studies of biofluids to date have mainly been performed on dried samples deposited on spectroscopically neutral substrates such as CaF2. However, sample homogeneity and measurement reproducibility can be a problem.21,22 The CaF2 substrates are also very expensive and cannot be reused in clinical diagnosis. Due to the relatively low interaction cross-section for the Raman interaction, this also necessitates relatively long acquisition times in comparison with applications involving FTIR spectroscopy.
Therefore, the present study aims to overcome these limitations and contribute to the translation of Raman spectroscopy of biofluids to the clinic. In the study, a HT-Raman spectroscopy method was developed for rapid screening of prostate cancer patients. Different instrumental and sample preparation parameters were tested for the identification of a suitable parameter set for the HT-Raman spectroscopy method. This HT method was developed using low cost substrates and liquid plasma samples drawn from prostate cancer patients and healthy volunteers. The classification performance of this method provides an overall higher sensitivity (96.5%) and specificity (95%) when compared to the conventional PSA test. In addition this method can analyse plasma samples in a 96-well microplate in less than one hour. Overall, this HT-Raman spectroscopy approach has the potential to translate into clinics for HT screening of prostate cancer patients using samples of liquid plasma samples. It also has potential to be adaptable to any other human body fluid.
Materials and methods
Ethical approval
Blood samples were obtained from healthy volunteers after informed consent. Ethical approval has been obtained from the DIT Research Ethics Committee for this arm of the work. Blood samples from prostate cancer patients were obtained through an ongoing collaboration with Prof. John Armstrong, Consultant Radiation Oncologist, St Luke's Hospital (Dublin, Ireland). Ethical approval has been obtained from St Luke's Research Ethics Committee. The study is also sanctioned as part of an All-Ireland Cooperative Oncology Research Group (ICORG) translational study, ICORG 08-17.Patient and control demographics
The healthy controls were a cohort of both males and females, smokers and non-smokers, all between the ages of 21 and 64, while the patients were male with an age range from 58 to 85.Plasma separation
Whole blood was drawn into lithium-heparin tubes from 10 healthy volunteers and 10 prostate cancer patients. Plasma was isolated from these blood samples by centrifugation at 3500g for 5 minutes at 18 °C. The samples were subsequently stored at −80 °C prior to Raman acquisition.Raman spectroscopy
Optimisation study
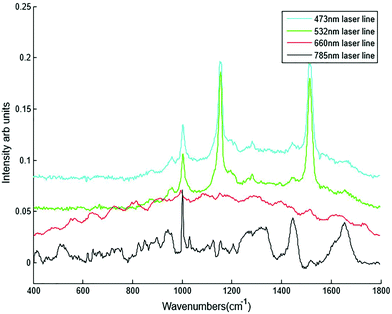
Extensive optimisation work has been carried out for the development of the HT Raman spectroscopy method. Plasma samples from healthy volunteers were used for the optimisation study. A Horiba Jobin Yvon LabRAM HR 800 was used to acquire Raman spectra from healthy plasma samples. The instrument was calibrated with the spectra using silicon (520.7 cm−1). A range of parameters (Table 1) were tested during the optimisation of HT-Raman spectroscopy.| Parameters | |
|---|---|
| Laser lines | 785 nm, 660 nm, 532 nm and 473 nm |
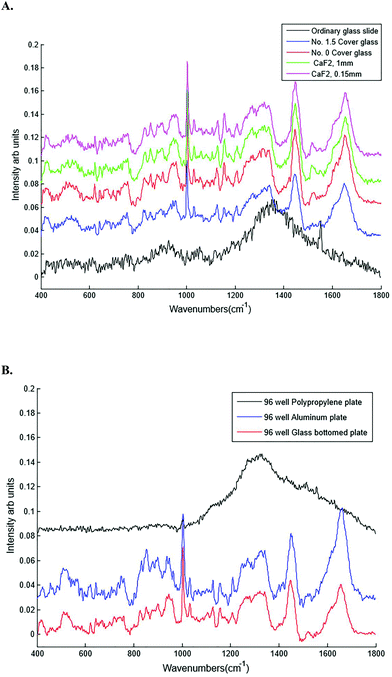
| Substrates | Glass, calcium fluoride (CaF2), aluminium and polypropylene |
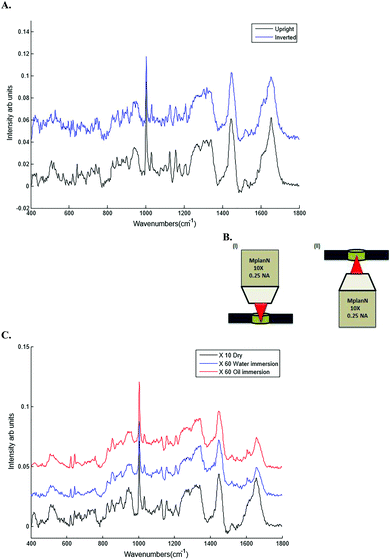
| Geometry | Upright and inverted |
| Microscope objectives | Dry (10×), water (60×) and oil (60×) immersion objectives. |
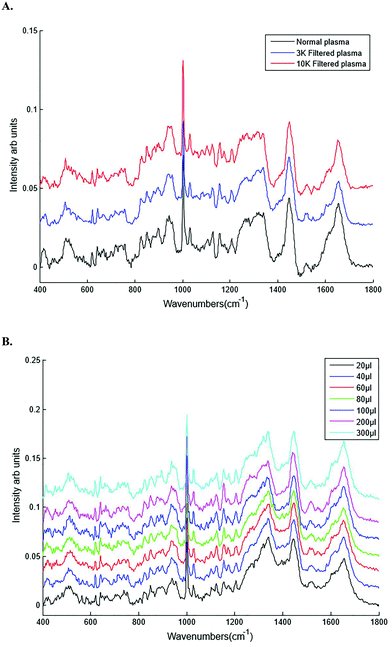
| Filtration | Unfiltered and filtered plasma (3 K and 10 K filtration) |
| Sample volume | 20, 40, 60, 80, 100, 200 and 300 μl |
| Gratings | 300 and 600 lines per mm |
| Confocal hole size | 100 to 400 μm |
| Acquisition times | 0.5, 3, 6, 10, 20, 40 and 60 seconds |
| Autoscan setup (for HT-Raman method) | Motorized XYZ sample stage was tested for the development of HT template |
HT-Raman spectroscopic analysis of blood plasma samples
Frozen plasma samples, after passive thawing, were subjected to Raman spectroscopy by placing 20 μl plasma on a cover glass bottomed 96 well plate and the spectra were recorded automatically from each well where the spectrometer was programmed using an in house developed high throughput macro template.Spectra of 1,4-bis (2-methylstyryl) benzene were acquired prior to each daily measurement as a further wavenumber calibration, and sample spectra from each day were wavenumber corrected to the standard spectrum of 1,4-bis (2-methylstyryl) benzene. The plasma samples were excited with the 785 nm, 660 nm, 532 nm and 473 nm laser focused through a 10× objective (N.A. 0.25). Spectra were recorded using a diffraction grating ruled with a grating of 300 lines per mm (for 785 nm and 660 nm) and 600 lines per mm (for 532 nm and 473 nm) giving a spectral resolution of ∼2.1 cm−1. Each spectrum was acquired in the region of 400–1800 cm−1. Multiple spectra were recorded from each individual sample using 785 nm and 532 nm laser lines at various acquisition times as shown in Table 2. Ten spectra per sample were recorded except for the acquisition time of 20 seconds and 2 accumulations using the 785 nm laser line whereby only 6 spectra per sample were recorded to avoid inconsistent spectra due to the drying of liquid plasma.
| Laser lines | Acquisition times | Spectra recorded/sample |
|---|---|---|
| 785 nm | 3 s × 2 | 10 |
| 5 s × 2 | 10 | |
| 10 s × 2 | 10 | |
| 20 s × 2 | 06 | |
| 532 nm | 0.25 s × 2 | 10 |
| 0.5 s × 2 | 10 | |
| 1s × 3 | 10 | |
| 3s × 3 | 10 |
Approximately 1–2 mg of lyophilized β-carotene was deposited on calcium fluoride slide and Raman spectra were recorded with both 532 and 785 nm laser lines. The laser was focused through a 50× objective (N.A. 0.55) using a diffraction grating ruled with a grating of 600 lines per mm for 532 nm and 300 lines per mm for 785 nm. Each spectrum was acquired in the region of 600–1800 cm−1. Five spectra per sample were recorded for the acquisition time of 5 seconds and 3 accumulations.
Data analysis
The raw Raman spectra were baseline-corrected (Rubber band), smoothed (Savitzky–Golay, Polynomial 5, Window 15) and vector-normalized. These processed spectra were used for spectral comparisons and for computing the difference spectra. The mean and difference spectra were also computed to illustrate the intra-group heterogeneity. The spectra were also subjected to principal component analysis–linear discriminant analysis (PCA–LDA).In brief, PCA is a routinely used method for multivariate data compression and visualization. It describes data variance by identifying a new set of orthogonal features, called principal components (PCs) or factors.23 In LDA, the discriminating plane is identified by maximising the ratio of between class variance and within class variance. LDA can be used in conjunction with PCA (PCA–LDA). PCA–LDA is a supervised method of using unsupervised PCA scores to define a linearly discriminating plane for classifying data sets using the feature space of the principal components. The advantage of doing this with Raman spectra is ultimately that the approach removes noise and covariance from the data and the generation of independent features that may be important for classification. PCA–LDA models were optimised using a leave-one-out-cross-validation (LOOCV), where the choice of the number of latent variables (principal components) was chosen from where the accuracy of classification reached the first maximum and where additional principal components did not contribute to the overall accuracy of the PCA–LDA performance.
LOOCV is a type of rotation estimation, a technique used for assessing the performance of a predictive model with a hypothetical validation set when an explicit validation set is not available. Leave-one-out involves using a single observation from the original sample as the validation data, and the remaining observations as training data. This is repeated such that each observation in the sample is used once as the validation data and all observations are used to form a confusion matrix. Though LOOCV may yield over optimistic results, in most cases it gives a reasonable first estimate of the model performance and is thus normally employed for small datasets.24 Since multiple spectra were acquired from each sample, data analysis is carried out using a spectrum-wise approach, where each spectrum is treated as an independent sample and all spectra are individually analysed by PC-LDA. Signal to noise ratio (SNR) and classification accuracies of the HT-Raman spectroscopy method were also calculated. Algorithms for these analyses were generated in-house implemented the R-based statistical software (version 0.99.896).25–27
Results and discussion
Optimisation results
Different dry and immersion objectives were tested for the HT-Raman method (Fig. 3). Immersion objectives gave slightly better quality spectra with higher SNR (43.6 dB) compared to dry objectives (43.3 dB) due to the increased numerical aperture. However, it was not possible to keep the drop of water or oil between the objective lens and each well consistently positioned across the 96 well plate due to smearing of the drop during movement of the stage from one well to the next. A low magnification 10× (N.A. 0.25) dry objective was selected for this study because it could more easily focus through the glass coverslip into the liquid plasma than a higher magnification objective lens. Use of an inverted geometry together with a low magnification objective lens allowed focussing of the laser into the liquid plasma and collection of the Raman scattered light without any interference from the glass coverslip.
Different plasma volumes of 20 to 300 μl (for increments refer to Table 1) were also tested. No significant difference in the spectral information or quality was observed in plasma samples excited with the 785 nm laser line in the inverted set up (Fig. 4B). This was also confirmed by SNR, all the sample volumes had the same SNR of 43.7 dB. As a result 20 μl of liquid plasma was selected for the development of the HT-Raman method to minimise the quantity of plasma required for the spectral acquisition.
 | ||
| Fig. 5 Schematic representation of HT-Raman spectroscopy method. (A) Top view (B) bottom view (these schematics are developed using Google Sketch up software). | ||
| Selected parameters for HT-Raman spectroscopy method | |
| Laser line | 785 and 532 nm |
| Substrate | 96 well plate (cover glass bottomed) |
| Objectives | 10× dry |
| Geometry | Inverted |
| Sample state | Unfiltered liquid plasma |
| Sample volume | 20 μl |
HT-Raman spectroscopy of liquid plasma samples from healthy volunteers and prostate cancer
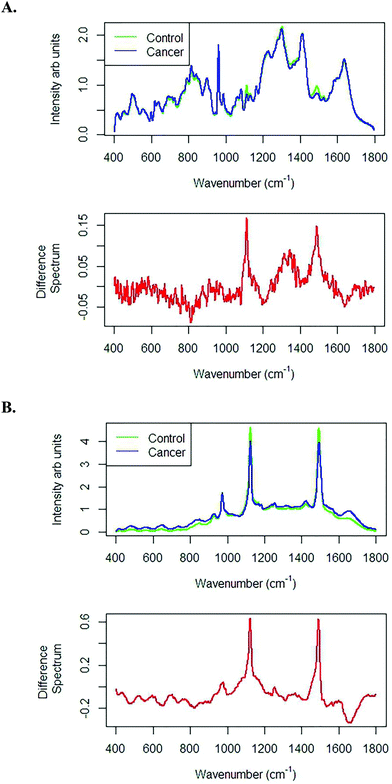
Fig. 6A shows the mean and difference spectra of liquid plasma samples from healthy volunteers and prostate cancer patients recorded with the 785 nm laser line using an integration time of 10 seconds averaged over 2 accumulations. The difference spectrum clearly shows highly intense bands at 1155 and 1523 cm−1, which are tentatively assigned to β-carotenoids.28Fig. 6B also shows the mean and difference spectra of liquid plasma samples from healthy volunteers and prostate cancer patients recorded with the 532 nm laser line with an integration time of 1 second averaged over 3 accumulations. Similar discriminating features were found with both the 785 and 532 nm laser lines and the discriminating features were resonant at 532 nm.
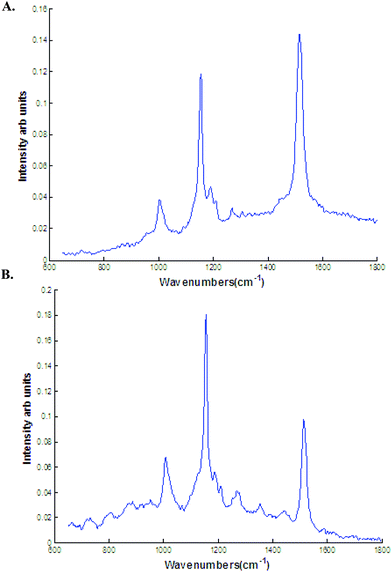
As mentioned earlier, the discriminating spectra can be assigned to β carotenoids. To interpret the discriminating spectra, β carotene spectra was recorded with both 532 and 785 nm laser line. The β carotene spectra resembles closely to the difference spectra with high intense bands in the regions 1155 and 1523 cm−1 (Fig. 7). The difference spectra in this study can be tentatively assigned to β carotenoids or any other related molecules.
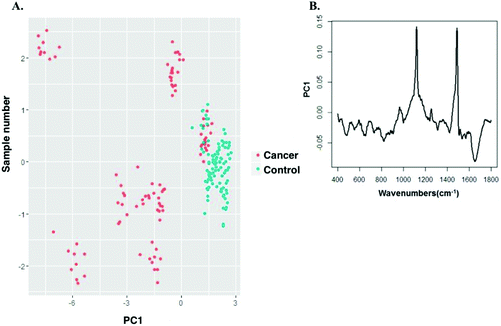
PCA was performed on the vector normalised spectra of cancer and control groups. Fig. 8 shows the scatter plot of the PCA classification of cancer and control group spectra acquired with the 532 nm laser line. The first two principal components (PC) were used to visualise the classification between the groups. Two minimally overlapping clusters for cancer and normal groups were observed. The PC's showed a high correlation to that of difference spectra shown in Fig. 6. PCA showed that two β carotene related peaks had differences between the control and cancer group. As mentioned earlier, these carotenoid levels are higher in control group compared to cancer.
Classification efficiency of HT-Raman using 532 and 785 nm laser lines
Blood plasma samples are colloidal dispersions, and can demonstrate heterogeneous behaviour in a micro-probing area, possibly because of the non-uniform distribution of the plasma constituents in the small volume under consideration. Thus, 10 spectra were recorded from each sample and the data were analysed using spectrum-wise approaches to investigate the uniformity of the distribution of the plasma analytes. The spectrum-wise approach, where each spectrum is treated as an independent entity, was adopted to explore if sample heterogeneity can influence the classification between control and cancer samples.The preprocessed spectra were subjected to supervised PCA–LDA, using the spectrum wise approach, and validated using leave-one-spectrum-out analysis (as described in Materials & methods). The results of PCA–LDA are depicted in the form of a scatter plot and a confusion matrix. The confusion matrix is generated to understand the discrimination between the groups obtained by accounting for the contribution of all factors selected for analysis.
The vector-normalized spectra from the control and cancer groups were imported into R software for processing using in-house developed algorithms and subsequent classification using PCA–LDA. Fig. 9A shows scatter plot of control (n = 10) and cancer (n = 10) plasma spectra recorded with 785 nm laser line. An LDA classifier was built using the first five principal components and the LDA scores from the classification are shown in the scatter plot. Two well differentiated clusters for control and cancer groups were observed. As shown in the confusion matrix in Table 4A, 58/60 cancer spectra and 57/60 control spectra were correctly classified with a sensitivity and specificity of 96.5% and 95% respectively.
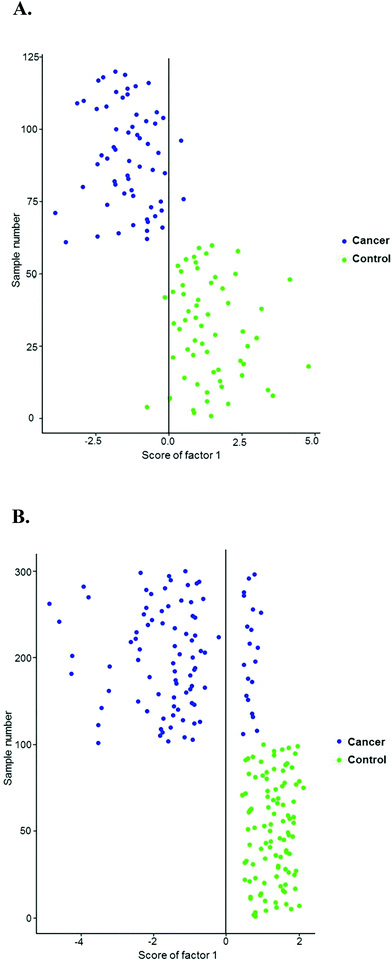 | ||
| Fig. 9 PCA–LDA for healthy control and prostate cancer plasma spectra recorded with (A) 785 nm laser line (B) 532 nm laser line. | ||
| Cancer | Control | Total | Efficiency | |
|---|---|---|---|---|
| A. | ||||
| Cancer | 58 | 2 | 60 | 96.5% (sensitivity) |
| Control | 3 | 57 | 60 | 95% (specificity) |
| B. | ||||
| Cancer | 80 | 20 | 100 | 80% (sensitivity) |
| Control | 0 | 100 | 100 | 100% (specificity) |
Fig. 9B shows the scatter plot of control (n = 10) and cancer (n = 10) plasma spectra recorded with 532 nm laser line. An LDA classifier was built using the first two principal components and the LDA scores from the classification are shown in the scatter plot. Two well differentiated clusters for control and cancer groups were observed. As shown in the confusion matrix in Table 4B, 80/100 cancer spectra and 100/100 control spectra were correctly classified with a sensitivity and specificity of 80% and 100% respectively.
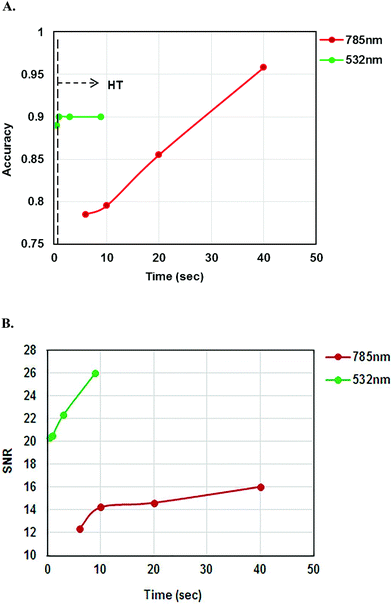
As multiple spectra were recorded with two different laser lines at various acquisition times, the classification accuracy of the two laser lines was compared in Fig. 10A. Classification accuracies were determined by the sum of the diagonal elements of the confusion matrix divided by total sum of all elements of the confusion matrix. The accuracy of the classification increased with increasing acquisition time for the 785 nm laser line but the classification accuracy remained the same after 1 second for the 532 nm laser line. The classification accuracies of 785 and 532 nm laser lines at different acquisition times are shown in Table 5. Fig. 10B shows the comparison of SNR values for the 785 and 532 nm laser lines. SNR increased with increasing acquisition time.
| Laser lines | Acquisition times | Classification accuracy |
|---|---|---|
| 785 nm | 3 s × 2 | 78.5% |
| 5 s × 2 | 79.5% | |
| 10 s × 2 | 85.5% | |
| 20 s × 2 | 95.8% | |
| 532 nm | 0.25 s × 2 | 89% |
| 0.5 s × 2 | 90% | |
| 1 s × 3 | 90% | |
| 3 s × 3 | 90% |
Conclusion
In the study, a high throughput methodology has been developed that can screen prostate cancer patients with a higher sensitivity and specificity than the conventional PSA test. Spectral differences are mainly observed in the region around 1155 and 1523 cm−1 across the analysed groups. These bands were highly expressed in healthy volunteers compared to prostate cancer patients. Control versus cancer cases could be classified with similar discriminating features using both 785 and 532 nm laser lines and the discriminating features were resonant at 532 nm. The discriminating feature in this study can be tentatively assigned to β carotenoids or any other related molecules. Carotenoid abundance has been shown to be depleted in the cancerous state of several organs including breast, lung, liver and colon,31 and to contribute to the discrimination between normal, benign and malignant breast tissues.32 It has been previously reported that the amount of β carotene present in blood decreases during cancer conditions. Reports suggest that β carotene levels are significantly reduced in blood plasma of women with histopathologically diagnosed cervical dysplasia and an inverse association between β carotene plasma levels and increasingly severe graded cervical histopathology.33 Epidemiological studies have suggested that high endogenous levels of pro-oxidants and deficiencies in levels of antioxidants are likely to be an important risk in the progression of pre-cancer to cancer.34 An increased breast cancer risk was also observed in subjects with lower levels of β carotene.19,35 Recent studies also reported lower levels of β carotene in the serum of oral cancers, buccal mucosa cancer, tongue cancer and cervical cancer as compared to healthy controls.20,36 The higher levels of β carotene in the healthy controls compared to cancer patients observed in this study are in concordance with the existing reports. However, further studies are required to confirm the molecule that differentiates control and cancer cohorts. PCA–LDA findings indicate the possibility of classifying the prostate cancer patients from healthy volunteers with a sensitivity of 96.5% (at 20 seconds and 2 accumulations) and 80% (at 0.5 seconds and 2 accumulations) using the 785 and 532 nm laser lines respectively.Classification accuracy and SNR increased with increasing acquisition times for the 785 nm laser line but classification accuracy remained the same after 1 second for the 532 nm laser line. A classification accuracy of 95.8% could be achieved with an acquisition time of 40 seconds (20 s with 2 integrations) using the 785 nm laser line but a classification accuracy of 90% could be achieved with an acquisition time of 1 second using the 532 nm laser line.
It is acknowledged that a limitation of the present study is that age matched controls were not used. However, the main aim of this study was to develop a HT Raman spectroscopy method for rapid screening of liquid plasma samples and the control and cancer samples were used to illustrate this method. Future work will further investigate the discrimination of non cancer controls and cancer cases using age matched samples. As the data set is small, LOOCV was used but for future validation a larger sample set would be required.
The cost of substrates is one of the factors limiting the clinical translation of Raman spectroscopy. Raman spectroscopy uses expensive substrates (such as CaF2) for analysis of biological samples making Raman based diagnostics more expensive than existing methods. Moreover these substrates cannot be reused in a clinical setting. Identifying suitable inexpensive substrates will help to overcome this limitation. This study has revealed that inexpensive cover glass substrates can be used rather than more expensive alternatives.
HT-Raman spectroscopy of blood plasma may be a more practical and patient friendly approach for screening of prostate cancer patients. This technology could produce a result within hours of patient admission as in total, 96 blood plasma samples could be analysed in less than one hour using this method. Future work will be based on validating the results of this HT-Raman study on a larger sample size with appropriate age matched controls.
Acknowledgements
This research was funded by the Dublin Institute of Technology Fiosraigh Fee and Materials Support Award and Science Foundation Ireland (11/RFP.1/BMT/3317).References
- J. Ferlay, I. Soerjomataram, R. Dikshit, S. Eser, C. Mathers, M. Rebelo, D. M. Parkin, D. Forman and F. Bray, Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012, Int. J. Cancer, 2015, 136, E359–E386 CrossRef CAS PubMed.
- S. Sarkar and S. Das, A Review of Imaging Methods for Prostate Cancer Detection, Biomed. Eng. Comput. Biol., 2016, 7, 1–15 CrossRef PubMed.
- M. Adhyam and A. K. Gupta, A Review on the Clinical Utility of PSA in Cancer Prostate, Indian J. Surg. Oncol., 2012, 3, 120–129 CrossRef PubMed.
- F. M. Lyng, D. Traynor, I. R. Ramos, F. Bonnier and H. J. Byrne, Raman spectroscopy for screening and diagnosis of cervical cancer, Anal. Bioanal. Chem., 2015, 407, 8279–8289 CrossRef CAS PubMed.
- M. Jermyn, K. Mok, J. Mercier, J. Desroches, J. Pichette, K. Saint-Arnaud, L. Bernstein, M. C. Guiot, K. Petrecca and F. Leblond, Intraoperative brain cancer detection with Raman spectroscopy in humans, Sci. Transl. Med., 2015, 7, 274ra19 CrossRef CAS PubMed.
- G. Theophilou, K. M. Lima, M. Briggs, P. L. Martin-Hirsch, H. F. Stringfellow and F. L. Martin, A biospectroscopic analysis of human prostate tissue obtained from different time periods points to a trans-generational alteration in spectral phenotype, Sci. Rep., 2015, 5, 13465 CrossRef CAS PubMed.
- C. Lacombe, V. Untereiner, C. Gobinet, M. Zater, G. D. Sockalingum and R. Garnotel, Rapid screening of classic galactosemia patients: a proof-of-concept study using high-throughput FTIR analysis of plasma, Analyst, 2015, 140, 2280–2286 RSC.
- D. M. Good, V. Thongboonkerd, J. Novak, J. L. Bascands, J. P. Schanstra, J. J. Coon, A. Dominiczak and H. Mischak, Body fluid proteomics for biomarker discovery: lessons from the past hold the key to success in the future, J. Proteome Res., 2007, 6, 4549–4555 CrossRef CAS PubMed.
- N. Mainreck, S. Brézillon, G. D. Sockalingum, F. X. Maquart, M. Manfait and Y. Wegrowski, Rapid characterization of glycosaminoglycans using a combined approach by infrared and Raman microspectroscopies, J. Pharm. Sci., 2011, 100, 441–450 CrossRef CAS PubMed.
- D. C. Fernandez, R. Bhargava, S. M. Hewitt and I. W. Levin, Infrared spectroscopic imaging for histopathologic recognition, Nat. Biotechnol., 2005, 23, 469–474 CrossRef CAS PubMed.
- X. Zhang, G. Thiéfin, C. Gobinet, V. Untereiner, I. Taleb, B. Bernard-Chabert, A. Heurgué, C. Truntzer, P. Ducoroy, P. Hillon and G. D. Sockalingum, Profiling serologic biomarkers in cirrhotic patients via high-throughput Fourier transform infrared spectroscopy: toward a new diagnostic tool of hepatocellular carcinoma, Transl. Res., 2013, 162, 279–286 CrossRef CAS PubMed.
- P. Carmona, M. Molina, M. Calero, F. Bermejo-Pareja, P. Martínez-Martín and A. Toledano, Discrimination analysis of blood plasma associated with Alzheimer's disease using vibrational spectroscopy, J. Alzheimers Dis., 2013, 34, 911–920 CAS.
- E. Scaglia, G. D. Sockalingum, J. Schmitt, C. Gobinet, N. Schneider, M. Manfait and G. Thiéfin, Noninvasive assessment of hepatic fibrosis in patients with chronic hepatitis C using serum Fourier transform infrared spectroscopy, Anal. Bioanal. Chem., 2011, 401, 2919–2925 CrossRef CAS PubMed.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood, K. A. Heys, C. Hughes, P. Lasch, P. L. Martin-Hirsch, B. Obinaju, G. D. Sockalingum, J. Sulé-Suso, R. J. Strong, M. J. Walsh, B. R. Wood, P. Gardner and F. L. Martin, Using Fourier transform IR spectroscopy to analyze biological materials, Nat. Protoc., 2014, 9, 1771–1791 CrossRef CAS PubMed.
- M. Keller, E. Kanter and A. Mahadevan, Raman spectroscopy for cancer diagnosis, Spectroscopy, 2006, 21, 33–41 Search PubMed.
- G. Clemens, J. R. Hands, K. M. Dorling and M. J. Baker, Vibrational spectroscopic methods for cytology and cellular research, Analyst, 2014, 139, 4411–4444 RSC.
- W. Huang, M. Li and R. Jarvis, Shining light on the microbial world the application of Raman microspectroscopy, Adv. Appl. Microbiol., 2010, 70, 153–186 CAS.
- A. Harris, A. Lungari and C. Needham, Potenial for Raman spectroscopy to provide cancer screening using peripheral blood sample, Head Neck Oncol., 2009, 1, 34 CrossRef PubMed.
- J. L. Pichardo, C. Frausto, O. Barbosa, R. Huerta, J. L. Gonzalez, C. A. Ramirez, G. Guitierrez and C. Medina, Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients, Laser Med. Sci., 2007, 22, 229–236 CrossRef PubMed.
- A. Gonzalez, L. Martinez, L. Torres, A. Aguilar-Lemarroy, L. A. Jave-Suarez and P. Palomares, Cervical cancer detection based on serum sample Raman spectroscopy, Lasers Med. Sci., 2014, 29, 979–985 CrossRef PubMed.
- K. W. Poon, F. M. Lyng, P. Knief, O. Howe, A. D. Meade, J. F. Curtin, H. J. Byrne and J. Vaughan, Quantitative reagent-free detection of fibrinogen levels in human blood plasma using Raman spectroscopy, Analyst, 2012, 137, 1807–1814 RSC.
- J. Filik and N. Stone, Analysis of human tear fluid by Raman spectroscopy, Anal. Chim. Acta, 2008, 616, 177–184 CrossRef CAS PubMed.
- C. Krafft, G. Steiner, C. Beleites and R. Salzer, Disease recognition by infrared and Raman spectroscopy, J. Biophotonics, 2009, 2, 13–28 CrossRef CAS PubMed.
- K. Varmuza and P. Filzmoser, Introductuion ti multivariate statiscal analysis in chemometrics, CRC press, Newyork, 2009 Search PubMed.
- W. Ripley, Modern applied statistics with S, 2002 Search PubMed.
- H. Wickham, Elegant graphics for data analysis (http://ggplot2.org), 2009.
- H. Borchers, Practical numerical math functions (http://cran.r-project.org/package=pracma), 2015.
- H. Abramczyk, I. Placek, B. Brozek-Pluska, K. Kurczewski, Z. Morawiec and M. Tazbir, Human breast tissue cancer diagnosis by Raman spectroscopy, Spectroscopy, 2008, 22, 113–121, DOI:10.3233/SPE-2008-0337.
- B. Brożek-Płuska, I. Placek, K. Kurczewski, Z. Morawiec, M. Tazbir and H. Abramczyk, Breast cancer diagnostics by Raman spectroscopy, J. Mol. Liq., 2008, 141, 145–148 CrossRef.
- F. Bonnier, F. Petitjean, M. J. Baker and H. J. Byrne, Improved protocols for vibrational spectroscopic analysis of body fluids, J. Biophotonics, 2014, 7, 167–179 CrossRef CAS PubMed.
- H. Abramczyk and B. Brozek-Pluska, New look inside human breast ducts with Raman imaging. Raman candidates as diagnostic markers for breast cancer prognosis: Mammaglobin, palmitic acid and sphingomyelin, Anal. Chim. Acta, 2016, 909, 91–100 CrossRef CAS PubMed.
- H. Abramczyk and B. Brozek-Pluska, Raman imaging in biochemical and biomedical applications. Diagnosis and treatment of breast cancer, Chem. Rev., 2013, 113(8), 5766–5781 CrossRef CAS PubMed.
- P. R. Palan, M. S. Mikhail, J. Basu and S. L Romney, Plasma levels of antioxidant beta-carotene and alpha-tocopherol in uterine cervix dysplasias and cancer, Nutr. Cancer, 1991, 15(1), 13–20 CrossRef CAS PubMed.
- L. Aravindh, P. Jagathesh, S. Shanmugam, S. Sarkar, P. Mahesh Kumar and S. Ramasubramanian, Estimation of plasma antioxidants beta carotene, vitamin C and vitamin E levels in patients with OSMF and Oral Cancer – Indian population, Int. J. Biol. Med. Res., 2012, 3(2), 1655–1657 Search PubMed.
- J. Surmacki, B. Brozek-Pluska, R. Kordek and H Abramczyk, The lipid-reactive oxygen species phenotype of breast cancer. Raman spectroscopy and mapping, PCA and PLSDA for invasive ductal carcinoma and invasive lobular carcinoma. Molecular tumorigenic mechanisms beyond Warburg effect, Analyst, 2015, 140(7), 2121–2133 RSC.
- A. Sahu, S. Sawant, H. Mamgain and C. M. Krishna, Raman spectroscopy of serum: an exploratory study for detection of oral cancers, Analyst, 2013, 138, 4161–4174 RSC.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |