Open Access Article
Open Access ArticleNanomolar small-molecule detection using a genetically encoded 129Xe NMR contrast agent†
B. W.
Roose
 ,
S. D.
Zemerov
and
I. J.
Dmochowski
,
S. D.
Zemerov
and
I. J.
Dmochowski
 *
*
Department of Chemistry, University of Pennsylvania, 231 South 34th St., Philadelphia, PA 19104-6323, USA. E-mail: ivandmo@sas.upenn.edu
First published on 20th September 2017
Abstract
Genetically encoded magnetic resonance imaging (MRI) contrast agents enable non-invasive detection of specific biomarkers in vivo. Here, we employed the hyper-CEST 129Xe NMR technique to quantify maltose (32 nM to 1 mM) through its modulation of conformational change and xenon exchange in maltose binding protein (MBP). Remarkably, no hyper-CEST signal was observed for MBP in the absence of maltose, making MBP an ultrasensitive “smart” contrast agent. The resonance frequency of 129Xe bound to MBP was greatly downfield-shifted (Δδ = 95 ppm) from the 129Xe(aq) peak, which facilitated detection in E. coli as well as multiplexing with TEM-1 β-lactamase. Finally, a Val to Ala mutation at the MBP–Xe binding site yielded 34% more contrast than WT, with 129Xe resonance frequency shifted 59 ppm upfield from WT. We conclude that engineered MBPs constitute a new class of genetically encoded, analyte-sensitive molecular imaging agents detectable by 129Xe NMR/MRI.
Introduction
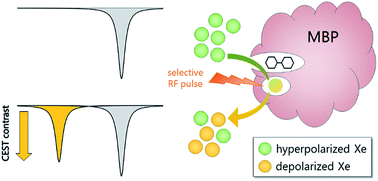
The development of molecular probes for quantifying the in vivo distribution of biologically active species (e.g., metabolites, neurotransmitters) is critical to understanding both normal physiology and disease pathologies. Magnetic resonance imaging (MRI) offers excellent spatiotemporal resolution and penetration depth without the use of ionizing radiation. Functional MRI (fMRI) has spurred the development of “smart” contrast agents that are responsive to specific physiological triggers1 such as metal ions,2 metabolites,3 and enzymes.4 Genetically encoded (GE) contrast agents can be expressed directly in the tissue of interest and tailored to bind specific analytes through biomolecular engineering techniques such as directed evolution.5,6 In pioneering work, Shapiro and coworkers evolved bacterial cytochrome P450-BM3 to bind dopamine near the paramagnetic heme, which was designed to decrease 1H MRI signal: 89 μM BM3 enabled detection of 75 μM dopamine in live rat brain.6 An alternative contrast approach is 1H chemical exchange saturation transfer (1H-CEST), which uses selective radiation to depolarize solute-bound protons, then measures loss of bulk solvent proton signal as protons exchange from solute to solvent (i.e., magnetization transfer).7 By this method, Oskolkov and coworkers observed MR contrast from 733 μM human protamine-1, an arginine-rich nuclear protein that was sensitive to pH, phosphorylation state, nucleotides, and heparin.8 Notably, the low sensitivity of 1H MRI has limited the development of GE T1, T2, and 1H-CEST contrast agents for molecular imaging.9Nuclear hyperpolarization provides a unique strategy to overcome natural limitations in NMR sensitivity.10,11 For example, the spin-½ nucleus of 129Xe can be hyperpolarized (hp) through spin-exchange optical pumping (SEOP) to increase net magnetization by several orders of magnitude.12,13 Synthetic xenon biosensors14 have been developed by targeting a cryptophane-A or curburbit[6]uril host to report on proteins,15–18 glycans,19 oligonucleotides,20 metal ions,21 free thiols,22 and pH.23 A technique known as hyper-CEST can be employed to detect the hp 129Xe-host interaction at nM–fM concentrations by applying saturation pulses at the 129Xe-host chemical shift and monitoring magnetization transfer to the large population of 129Xe nuclear spins in aqueous solution.24 This saturation transfer process accelerates the loss of hp 129Xe(aq) signal relative to the natural depolarization rate, 1/T1, which provides an ultrasensitive mechanism for generating MR contrast.25–28 Shapiro et al. demonstrated the first GE hyper-CEST agents via pM detection of proteinaceous nano-to-micron-scale gas vesicles.29 It remains challenging, however, to develop monomeric protein reporters, as Xe typically binds proteins weakly (Ka ≪ 200 M−1) and with fast exchange (koff > 105 s−1).30 Recently, our laboratory identified TEM-1 β-lactamase (Bla) as a monomeric protein reporter, with sub-micromolar Bla producing hyper-CEST signal in mammalian cells.31 Here, we developed maltose binding protein (MBP) as a small-molecule-responsive, GE xenon biosensor capable of detecting nanomolar concentrations of maltose using the hyper-CEST 129Xe NMR technique (Scheme 1).
Xe binding to MBP has been well-characterized by NMR, which showed that 129Xe chemical shift depends on MBP conformation.32 X-ray crystallography identified a single Xe-binding site near the maltose binding cleft of MBP (Fig. 1 and S1†).33 MBP is a periplasmic protein encoded by the malE gene that serves as an initial receptor in the maltose/maltodextrin transport systems of Gram-negative bacteria.34 WT MBP binds maltose with good affinity (Kd ≈ 1 μM),34 as well as other maltodextrins, between two nearly symmetrical lobes that transition from an “open” (MBPopen) to “closed” (MBPclosed) conformation upon ligand binding.35 This structural response to ligand binding has led to the utilization of MBP as a versatile platform for biosensing applications.36 Small-molecule detection with MBP, as well as other periplasmic binding proteins (PBPs),37 has been achieved through a variety of signal transduction modalities, including fluorescence,38 fluorescence resonance energy transfer (FRET),39 and electrochemical response.40 Moreover, MBP has been engineered to increase ligand affinity41,42 and to bind non-maltodextrin ligands such as sucrose43 and zinc.44 Thus, we set out to evaluate the hyper-CEST NMR contrast generated by MBP as a function of maltose binding.
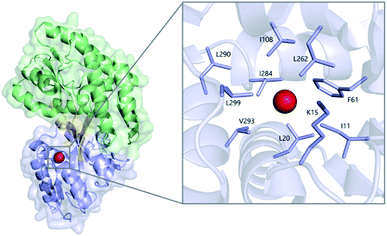 | ||
| Fig. 1 Xe (red sphere) bound to MBPopen (PDB ID 1LLS), with the N-terminal domain colored blue, C-terminal domain colored green, and linking segments colored orange. (Inset) detailed view of the Xe-binding cavity. | ||
Results and discussion
Maltose detection by MBP
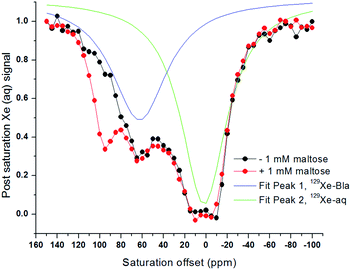
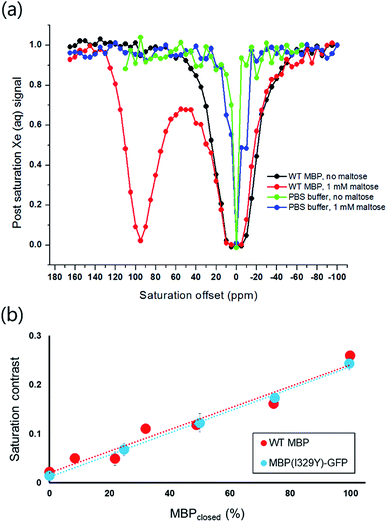
Xenon hyper-CEST z-spectra were acquired from recombinant MBP in both the presence and absence of maltose to assess the magnitude and frequency of NMR saturation contrast (Fig. 2a). Multiple selective d-SNOB saturation pulses were scanned over the chemical shift range of −100 to 180 ppm in 5 ppm steps, and the 129Xe(aq) signal was measured as a function of saturation pulse offset. MBP in the absence of maltose (MBPopen) showed a single saturation response corresponding to free 129Xe in solution centered at 0 ppm. In contrast, MBP in the presence of maltose (MBPclosed) showed a pronounced saturation response 95 ppm downfield of the Xe(aq) peak, corresponding to Xe@MBPclosed. This peak is 35 ppm further downfield than Xe@Bla,31 suggesting that Xe@MBPclosed experiences a more hydrophobic environment. Also, the width of the Xe@MBPclosed peak (35 ppm) is narrower than Xe@Bla (60 ppm), indicating slower Xe exchange with MBP (Table S1†).To assess the detection sensitivity of MBP, time-dependent saturation transfer experiments were performed by measuring Xe(aq) polarization as a function of saturation time (Fig. S2†). Saturation frequencies of d-SNOB pulses were positioned +95 ppm and −95 ppm, referenced to the Xe(aq) peak, for on- and off-resonance, respectively. The normalized difference between on- and off-resonance saturation transfer was measured as on-resonance hyper-CEST contrast. By this method, 100 nM MBP with 1 mM maltose reported 0.26 ± 0.01 saturation contrast; by comparison, 0.23 ± 0.02 saturation contrast was observed for 100 nM Bla.31 To evaluate the responsiveness of MBP MR contrast to maltose, additional saturation contrast measurements were taken for 100 nM MBP with decreasing maltose. The lowest maltose concentration that could be readily detected by MBP was 100 nM, with an observed saturation contrast of 0.050 ± 0.007. For each maltose concentration, the amount of MBPclosed (i.e., contrast “ON”) was calculated using a Kd of 1 μM. The observed saturation contrast was linearly proportional (R2 = 0.953) to the percentage of MBP in the maltose-bound closed conformation, which provides a measure of maltose concentration (Fig. 2b).
Mutagenesis to modulate maltose detection sensitivity
To extend the detection threshold of MBP, Ile-329 was mutated to Tyr to greatly increase maltose affinity (Kd = 22 nM).41 This mutation alters the conformational dynamics of MBP to disfavor the unliganded, open conformation, thereby promoting maltose binding through conformational coupling.41,42 Importantly, Ile-329 is located in a “hinge region” opposite the maltose-binding cleft, far enough away from the Xe-binding site to reasonably assume that mutations at this position should not affect Xe exchange. The I329Y mutation was introduced to a MBP–GFP fusion construct prepared to facilitate protein quantitation during cell studies (vide infra). The Xe hyper-CEST z-spectrum of MBP(I329Y)–GFP in the presence of 1 mM maltose is comparable to WT MBP (Fig. S3†), though the saturation response of Xe@MBP(I329Y)–GFP is shifted 5 ppm downfield and is slightly attenuated compared to WT. Saturation contrast measurements for 100 nM MBP(I329Y)–GFP followed the same procedure used for WT MBP, but with the saturation frequencies of d-SNOB pulses positioned +100 ppm and −100 ppm (Fig. S4†). As observed for WT MBP, saturation contrast was linearly proportional (R2 = 0.997) to the percentage of MBP in the maltose-bound closed conformation (Fig. 2b). The detection threshold for the I329Y mutant was 32 nM maltose, which gave rise to saturation contrast of 0.07 ± 0.01. These saturation contrast data demonstrate that MBP can be “tuned” through mutagenesis to detect maltose across varying concentration ranges. High-affinity mutants such as I329Y can be employed for nM-to-low μM maltose detection, whereas WT MBP can be used for low μM-to-mM maltose detection.Hyper-CEST of MBP in E. coli
To evaluate the hyper-CEST contrast detectable from MBP in a cellular environment, MBP with a C-terminal GFP tag (MBP–GFP) was expressed in BL21 (DE3) E. coli cells and time-dependent saturation transfer measurements were taken following the same protocol used for purified WT MBP (Fig. S5†). Using the GFP tag, MBP concentration was quantified in real-time by fluorescence intensity at 510 nm (λex = 489 nm). E. coli growths not induced with IPTG served as controls to measure background contrast. Cells were washed with PBS and then transferred to an NMR tube for data collection. Control E. coli reported saturation contrasts of 0.11 ± 0.01 and 0.09 ± 0.01 with and without 1 mM maltose, respectively. Background contrast at 95 ppm downfield from Xe(aq) likely arises from Xe exchanging with the hydrophobic interior of cellular membranes. E. coli expressing MBP in the presence of maltose reported 0.25 ± 0.02 saturation contrast (Table 1), nearly five-fold higher than E. coli expressing MBP in the absence of maltose (0.14 ± 0.01), after subtraction of background (0.11 ± 0.01) from both. This highlights a mechanism for designing xenon-based MRI molecular imaging agents capable of detecting a specific analyte in cellular milieu.| E. coli sample | T 1on (s) | T 1off (s) | Saturation contrast | [MBP–GFP]a (μM) |
|---|---|---|---|---|
| a MBP–GFP concentration measured by fluorescence. | ||||
| Non-induced, no maltose | 19.1 ± 0.8 | 26 ± 2 | 0.09 ± 0.01 | <0.001 |
| Non-induced, 1 mM maltose | 18.9 ± 0.8 | 28 ± 2 | 0.11 ± 0.01 | <0.001 |
| Induced, no maltose | 17.9 ± 0.7 | 28.5 ± 0.8 | 0.14 ± 0.01 | 1 |
| Induced, 1 mM maltose | 13.3 ± 0.6 | 8 ± 2 | 0.25 ± 0.02 | 1 |
Multiplexed MR contrast
The saturation frequency of Xe@MBP is sufficiently downfield of Xe@Bla that we hypothesized that the two proteins could be detected sequentially and very sensitively with minimal crosstalk using hyper-CEST in the same solution. The hyper-CEST z-spectrum of a mixture of MBP and Bla was acquired, where the ratio of MBP to Bla was lowered to approximately equalize the magnitude of contrast produced by the two proteins (Fig. 3). The z-spectrum of 27 μM MBP and 80 μM Bla in the absence of maltose showed two peaks: Xe(aq) at 0 ppm, and Xe@Bla at 60 ppm. In the presence of 1 mM maltose, three peaks were observed: Xe(aq) at 0 ppm, Xe@Bla at 60 ppm, and Xe@MBP at 95 ppm. Critically, the magnitude of saturation contrast at 60 ppm for Xe@Bla was not affected by maltose or whether MBP contrast was “on” or “off”. We envision that the ability to multiplex the responsive hyper-CEST agent, MBP, with a non-responsive hyper-CEST agent such as Bla, should enable the in vivo quantitation of maltose via ratiometric analysis. Indeed, ratiometric approaches employing fluorescent small molecules and proteins have been widely applied for detection of ions and biomolecules in solution and in cellular studies.45CEST contrast dependence on MBP conformation
To ascertain whether hyper-CEST contrast with MBP results from the closed conformation induced by maltose binding or from sugar binding alone, the z-spectrum of MBP in the presence of β-cyclodextrin (βCD) was obtained (Fig. S6†). βCD binds MBP with good affinity (Kd = 1.8 μM) in the same cleft as maltose, but its larger size prevents MBP from adopting a closed conformation.46 The z-spectrum of MBP with βCD shows no downfield saturation response, indicating that the maltose-bound closed conformation is required for producing hyper-CEST contrast. The lack of saturation contrast with MBP + βCD was unexpected given that Xe has higher affinity for MBP + βCD than MBP + maltose. Previous 1H–15N HSQC NMR experiments measured a Ka of 20 ± 10 M−1 for Xe binding to MBP + βCD and concluded that Ka was too low to measure for Xe binding to MBP + maltose: the addition of Xe to MBP + maltose produced no measurable changes in the 1H–15N HSQC resonances.33 Indeed, the fact that the Xe(aq) peak is broadened more by βCD (FWHM = 60 ppm) than by maltose (47 ppm) reveals a greater degree of Xe exchange with MBP + βCD than MBP + maltose (Table S1†). The structural basis for a difference in Xe affinity is not obvious, however, as the contours of the Xe-binding cavity are largely conserved among several MBP complexes (Fig. S7†). It therefore appears that the primary determinant of hyper-CEST contrast in MBP is rate of Xe exchange, not Xe affinity. Xe exchange is likely too fast (∼MHz frequency) with MBPopen (with or without βCD), but the binding of maltose and the conformational change to MBPclosed slows the rate of Xe exchange to resolve a separate peak in the z-spectrum.Mutagenesis to identify origin of hyper-CEST contrast
We performed mutagenesis experiments to further probe the mechanism of Xe CEST with MBP. Val-293 was chosen as the site for single-point mutations due to its proximity to bound Xe identified in the maltose-free MBP structure (Fig. 1). The methyl carbons of Val-293 are only 4.0 and 4.2 Å from Xe, respectively, thus it was expected that mutations at this position would affect Xe binding affinity and/or kinetics. Val-293 was mutated to Leu to reduce the cavity volume, and to Ala to enlarge the cavity. CD and fluorescence spectroscopy, along with thermal stability assays, confirmed that the V293L and V293A mutants retained the same global structure and maltose-binding properties as WT MBP (Fig. S8 and S9, Table S2†).Hyper-CEST z-spectra of V293L and V293A were obtained following the same protocol used for WT MBP (Fig. S10†). V293L with and without maltose showed only a saturation response for 129Xe(aq), suggesting that substitution of valine for the bulkier leucine sidechain effectively blocks Xe from occupying the MBP cavity. This result helps to confirm that the crystallographically determined Xe site for MBPopen is also the site of hyper-CEST with MBPclosed. The 129Xe(aq) peaks for V293L are narrower (19.1 ppm without maltose, 21.5 ppm with maltose) than WT MBP (41.8 ppm without maltose, 47 ppm with maltose) (Table S1†), further confirming the overall reduction of Xe exchange with V293L relative to WT MBP. However, the 129Xe(aq) peaks for V293L are broader than PBS in the absence of protein, likely due to non-specific Xe–protein interactions elsewhere on the surface of the protein.47
The z-spectrum of 80 μM V293A without maltose showed only the 129Xe(aq) peak, though in the presence of maltose a plateau of saturation response was observed between 50 and −10 ppm. Lowering the V293A concentration to 10 μM resolved this broad saturation response into two peaks – one at 0 ppm for 129Xe(aq), the other at 36 ppm for Xe@V293Aclosed. This large change in chemical shift from 95 ppm observed for WT follows a trend observed in T4 lysozyme,33 clathrate cages,48 and zeolites,49 where it has been noted that larger cavities produce smaller downfield 129Xe chemical shifts, and vice versa. Notably, this mutation increased the magnitude of CEST saturation contrast with 100 nM MBP from 0.26 ± 0.01 (WT) to 0.35 ± 0.02 for V293A (Fig. S11†). The molecular features of this signal enhancement and chemical shift change are under investigation.
Conclusions
In summary, we have demonstrated that 100 nM MBP generates significant saturation contrast in vitro and that observed contrast is proportional to % MBPclosed, thereby characterizing MBP as a “smart” analyte-sensitive biosensor. The I329Y MBP sensor at 100 nM concentration detected maltose in the range 32 nM to 5 μM, whereas 100 nM WT MBP detected maltose in the range 100 nM to 1 mM. Notably, the large (+95 ppm) 129Xe NMR chemical shift was generated within the GE MBP molecule and did not require post-translational modification or cofactor such as a lanthanide or other paramagnetic shift agent. Additionally, WT MBP appended with GFP was readily detected at 1 μM via hyper-CEST NMR when expressed in E. coli. The large downfield shift of WT MBP (δ = 95 ppm) makes it compatible with Bla (δ = 60 ppm) for multiplexing applications and ratiometric analysis. Notably, Xe–Bla was cleanly detected in the multiplexing experiment with minimal crosstalk from Xe–MBP, by withholding maltose until Xe–MBP signal was desired. Experiments with I329Y MBP confirm 3–4 orders-of-magnitude higher small-molecule sensitivity than that achieved with available GE T1 or 1H-CEST contrast agents. Indeed, nM maltose detection via MBP hyper-CEST NMR rivals the small-molecule detection sensitivity of many GE fluorescent sensors.The increased contrast generated by V293A (and loss of contrast observed for V293L) confirms that MBP–CEST efficiency can be enhanced with mutations to the xenon binding site: coupling this with mutations promoting the maltose-bound, MBP-closed conformation41,42 should yield a superior biosensor. Furthermore, the modulated saturation frequency of V293A (δ = 36 ppm, shifted 59 ppm upfield from WT MBP) suggests that rational mutagenesis will yield MBP variants with a broad range of 129Xe NMR chemical shifts, akin to the palette of fluorescent proteins such as GFP and mCherry commonly used for multiplexed cellular imaging.50 Similar attempts to engineer Bla through site-directed mutagenesis have so far failed to improve or modulate its CEST signal (unpublished data), which makes MBP a particularly versatile protein system for elucidating the hyper-CEST mechanism. Finally, MBP variants that have been engineered previously to bind different ligands43,44 highlight the exciting potential for employing MBP-enhanced 129Xe NMR/MRI to detect bioactive molecules present in mammalian cells.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by NIH R01-GM097478 to IJD. We thank UPenn Chemistry NMR facility for spectrometer time.Notes and references
- G.-L. Davies, I. Kramberger and J. J. Davis, Chem. Commun., 2013, 49, 9704–9721 RSC.
- A. C. Esqueda, J. A. López, G. Andreu-de-Riquer, J. C. Alvarado-Monzón, J. Ratnakar, A. J. M. Lubag, A. D. Sherry and L. M. De León-Rodríguez, J. Am. Chem. Soc., 2009, 131, 11387–11391 CrossRef CAS PubMed.
- S. Zhang, R. Trokowski and A. D. Sherry, J. Am. Chem. Soc., 2003, 125, 15288–15289 CrossRef CAS PubMed.
- A. Y. Louie, M. M. Hüber, E. T. Ahrens, U. Rothbächer, R. Moats, R. E. Jacobs, S. E. Fraser and T. J. Meade, Nat. Biotechnol., 2000, 18, 321–325 CrossRef CAS PubMed.
- G. Angelovski and É. Tóth, Chem. Soc. Rev., 2017, 46, 324–336 RSC.
- M. G. Shapiro, G. G. Westmeyer, P. A. Romero, J. O. Szablowski, B. Küster, A. Shah, C. R. Otey, R. Langer, F. H. Arnold and A. Jasanoff, Nat. Biotechnol., 2010, 28, 264–270 CrossRef CAS PubMed.
- M. T. McMahon and A. A. Gilad, Top. Magn. Reson. Imag., 2016, 25, 197–204 CrossRef PubMed.
- N. Oskolkov, A. Bar-Shir, K. W. Y. Chan, X. Song, P. C. M. van Zijl, J. W. M. Bulte, A. A. Gilad and M. T. McMahon, ACS Macro Lett., 2015, 4, 34–38 CrossRef CAS PubMed.
- G. Angelovski, Angew. Chem., Int. Ed., 2016, 55, 7038–7046 CrossRef CAS PubMed.
- M. S. Albert, G. D. Cates, B. Driehuys, W. Happer, B. Saam, C. S. Springer and A. Wishnia, Nature, 1994, 370, 199–201 CrossRef CAS PubMed.
- P. Berthault, G. Huber and H. Desvaux, Prog. Nucl. Magn. Reson. Spectrosc., 2009, 55, 35–60 CrossRef CAS.
- M. G. Mortuza, S. Anala, G. E. Pavlovskaya, T. J. Dieken and T. Meersmann, J. Chem. Phys., 2003, 118, 1581–1584 CrossRef CAS.
- D. A. Barskiy, A. M. Coffey, P. Nikolaou, D. M. Mikhaylov, B. M. Goodson, R. T. Branca, G. J. Lu, M. G. Shapiro, V. Telkki, V. V. Zhivonitko, I. V. Koptyug, O. G. Salnikov, K. V. Kovtunov, V. I. Bukhtiyarov, M. S. Rosen, M. J. Barlow, S. Safavi, I. P. Hall, L. Schröder and E. Y. Chekmenev, Chemistry, 2017, 23, 725–751 CrossRef CAS PubMed.
- Y. Wang and I. J. Dmochowski, Acc. Chem. Res., 2016, 49, 2179–2187 CrossRef CAS PubMed.
- M. M. Spence, S. M. Rubin, I. E. Dimitrov, E. J. Ruiz, D. E. Wemmer, A. Pines, S. Q. Yao, F. Tian and P. G. Schultz, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 10654–10657 CrossRef CAS PubMed.
- Q. Wei, G. K. Seward, P. A. Hill, B. Patton, I. E. Dimitrov, N. N. Kuzma and I. J. Dmochowski, J. Am. Chem. Soc., 2006, 128, 13274–13283 CrossRef CAS PubMed.
- J. M. Chambers, P. A. Hill, J. A. Aaron, Z. Han, D. W. Christianson, N. N. Kuzma and I. J. Dmochowski, J. Am. Chem. Soc., 2009, 131, 563–569 CrossRef CAS PubMed.
- Y. Wang, B. W. Roose, J. P. Philbin, J. L. Doman and I. J. Dmochowski, Angew. Chem., Int. Ed., 2016, 55, 1733–1736 CrossRef CAS PubMed.
- C. Witte, V. Martos, H. M. Rose, S. Reinke, S. Klippel, L. Schröder and C. P. R. Hackenberger, Angew. Chem., Int. Ed., 2015, 54, 2806–2810 CrossRef CAS PubMed.
- V. Roy, T. Brotin, J.-P. Dutasta, M.-H. Charles, T. Delair, F. Mallet, G. Huber, H. Desvaux, Y. Boulard and P. Berthault, ChemPhysChem, 2007, 8, 2082–2085 CrossRef CAS PubMed.
- K. Jeong, C. C. Slack, C. C. Vassiliou, P. Dao, M. D. Gomes, D. J. Kennedy, A. E. Truxal, L. J. Sperling, M. B. Francis, D. E. Wemmer and A. Pines, ChemPhysChem, 2015, 16, 3573–3577 CrossRef CAS PubMed.
- S. Yang, W. Jiang, L. Ren, Y. Yuan, B. Zhang, Q. Luo, Q. Guo, L.-S. Bouchard, M. Liu and X. Zhou, Anal. Chem., 2016, 88, 5835–5840 CrossRef CAS PubMed.
- B. A. Riggle, Y. Wang and I. J. Dmochowski, J. Am. Chem. Soc., 2015, 137, 5542–5548 CrossRef CAS PubMed.
- L. Schröder, T. J. Lowery, C. Hilty, D. E. Wemmer and A. Pines, Science, 2006, 314, 446–449 CrossRef PubMed.
- Y. Wang and I. J. Dmochowski, Chem. Commun., 2015, 51, 8982–8985 RSC.
- S. Korchak, W. Kilian, L. Schröder and L. Mitschang, J. Magn. Reson., 2016, 265, 139–145 CrossRef CAS PubMed.
- Y. Bai, P. A. Hill and I. J. Dmochowski, Anal. Chem., 2012, 84, 9935–9941 CrossRef CAS PubMed.
- Y. Bai, Y. Wang, M. Goulian, A. Driks and I. J. Dmochowski, Chem. Sci., 2014, 5, 3197–3203 RSC.
- M. G. Shapiro, R. M. Ramirez, L. J. Sperling, G. Sun, J. Sun, A. Pines, D. V. Schaffer and V. S. Bajaj, Nat. Chem., 2014, 6, 629–634 CrossRef CAS PubMed.
- R. F. Tilton and I. D. Kuntz, Biochemistry, 1982, 21, 6850–6857 CrossRef CAS PubMed.
- Y. Wang, B. W. Roose, E. J. Palovcak, V. Carnevale and I. J. Dmochowski, Angew. Chem., Int. Ed., 2016, 55, 8984–8987 CrossRef CAS PubMed.
- S. M. Rubin, M. M. Spence, I. E. Dimitrov, E. J. Ruiz, A. Pines and D. E. Wemmer, J. Am. Chem. Soc., 2001, 123, 8616–8617 CrossRef CAS PubMed.
- S. M. Rubin, S. Y. Lee, E. J. Ruiz, A. Pines and D. E. Wemmer, J. Mol. Biol., 2002, 322, 425–440 CrossRef CAS PubMed.
- W. Boos and H. Shuman, Microbiol. Mol. Biol. Rev., 1998, 62, 204–229 CAS.
- J. Evenäs, V. Tugarinov, N. R. Skrynnikov, N. K. Goto, R. Muhandiram and L. E. Kay, J. Mol. Biol., 2001, 309, 961–974 CrossRef PubMed.
- I. L. Medintz and J. R. Deschamps, Curr. Opin. Biotechnol., 2006, 17, 17–27 CrossRef CAS PubMed.
- F. S. Grünewald, in Advances in Chemical Bioanalysis, ed. F.-M. Matysik, Springer, Cham, 2013, vol. 6, pp. 205–235 Search PubMed.
- J. S. Marvin, E. R. Schreiter, I. M. Echevarría and L. L. Looger, Proteins, 2011, 79, 3025–3036 CrossRef CAS PubMed.
- M. Fehr, W. B. Frommer and S. Lalonde, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9846–9851 CrossRef CAS PubMed.
- D. E. Benson, D. W. Conrad, R. M. de Lorimier, S. A. Trammell and H. W. Hellinga, Science, 2001, 293, 1641–1644 CrossRef CAS PubMed.
- M.-H. Seo, J. Park, E. Kim, S. Hohng and H.-S. Kim, Nat. Commun., 2014, 5, 3724 CAS.
- J. S. Marvin and H. W. Hellinga, Nat. Struct. Biol., 2001, 8, 795–798 CrossRef CAS PubMed.
- G. Guntas, T. J. Mansell, J. R. Kim and M. Ostermeier, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11224–11229 CrossRef CAS PubMed.
- J. S. Marvin and H. W. Hellinga, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 4955–4960 CrossRef CAS PubMed.
- M. H. Lee, J. S. Kim and J. L. Sessler, Chem. Soc. Rev., 2015, 44, 4185–4191 RSC.
- A. J. Sharff, L. E. Rodseth and F. A. Quiocho, Biochemistry, 1993, 32, 10553–10559 CrossRef CAS PubMed.
- S. M. Rubin, M. M. Spence, A. Pines and D. E. Wemmer, J. Magn. Reson., 2001, 152, 79–86 CrossRef CAS PubMed.
- J. A. Ripmeester, C. I. Ratcliffe and J. S. Tse, J. Chem. Soc., Faraday Trans. 1, 1988, 84, 3731–3745 RSC.
- J.-L. L. Bonardet, J. Fraissard, A. Gedeon and M.-A. A. Springuel-Huet, Catal. Rev.: Sci. Eng., 1999, 41, 115–225 CAS.
- N. C. Shaner, P. A. Steinbach and R. Y. Tsien, Nat. Methods, 2005, 2, 905–909 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc03601a |
| This journal is © The Royal Society of Chemistry 2017 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.