Open Access Article
Open Access ArticleSelective inhibition of cancer cells by enzyme-induced gain of function of phosphorylated melittin analogues†
Qian-Qian
Li
a,
Pu-Guang
Chen
a,
Zhi-Wen
Hu
a,
Yuan
Cao
a,
Liang-Xiao
Chen
a,
Yong-Xiang
Chen
a,
Yu-Fen
Zhao
a and
Yan-Mei
Li
 *ab
*ab
aKey Lab of Bioorganic Phosphorus Chemistry & Chemical Biology, Department of Chemistry, Tsinghua University, Beijing 100084, P. R. China. E-mail: liym@mail.tsinghua.edu.cn; Fax: +86-10-62781695; Tel: +86-10-62796197
bBeijing Institute for Brain Disorders, Beijing 100069, P. R. China
First published on 12th September 2017
Abstract
The selective killing of cancer cells and the avoidance of drug resistance are still difficult challenges in cancer therapy. Here, we report a new strategy that uses enzyme-induced gain of function (EIGF) to regulate the structure and function of phosphorylated melittin analogues (MelAs). Original MelAs have the capacity to disrupt plasma membranes and induce cell death without selectivity. However, phosphorylation of Thr23 on one of the MelAs (MelA2-P) efficiently ameliorated the membrane lysis potency as well as the cytotoxicity for normal mammalian cells. After treatment with alkaline phosphatase (ALP), which is more active in cancer cells than normal cells, MelA2-P restored the pore-forming function around the cancer cells and induced cancer cell death selectively. This mechanism was independent of the receptor proteins and the cell uptake process, which may partially bypass the development of drug resistance in cancer cells.
Introduction
Phosphorylation is a ubiquitous modification method of biomolecules to regulate their structure, function, and activity.1 Combined with kinases and phosphatases, nature can manipulate protein–protein interactions, enzyme activity, and cell signal pathways on a spatiotemporal scale.2 Although enzyme-instructed phosphorylation and dephosphorylation are common regulatory mechanisms in nature, the utilization of this strategy to regulate the function of artificial chemical molecules is still in the early stage. For example, switching phosphorylation and dephosphorylation was used to manipulate the assembly process of silk protein,3 the morphology of micellar nanoparticles,4 and microstructure formation of polymer–peptide conjugates in vitro.5 Recently, Xu and others have succeeded in developing an enzyme-instructed self-assembly (EISA) method to regulate the aggregation process of phosphorylated compounds at the cellular level.6 After dephosphorylation, the phosphorylated compounds, which were originally soluble, could self-assemble into hydrogels around the cancer cells and kill the cancer cells selectively.6 Here, we expand a new strategy by using enzyme-induced gain of function (EIGF) to manipulate the anticancer activity of the designed compounds. Based on the differences in enzyme activity between cancer cells and normal cells, EIGF of cancer chemotherapeutic compounds may achieve selective cytotoxicity for cancer cells, which could reduce the side effects of chemotherapy.Chemotherapy is a principal method to treat malignant cancers. However, drug resistance and selectivity are two major obstacles for current chemotherapeutic molecules.7 Clinical chemotherapeutic drugs mostly act on intracellular targets, so cancer cells can resist drugs by decreasing the expression of receptor proteins or by excreting drugs out of cells.8 Recently, antimicrobial peptides have attracted intense attention for their potential application as anticancer peptides.9 Cationic amphiphilic peptides (CAPs), a type of anticancer peptide, could form an α helix conformation upon interacting with the cell membrane.10 CAPs cause membrane lysis by different mechanistic models, such as the barrel-stave model and the toroidal model.11 Because CAPs act on the cell membrane directly, but not on intracellular targets, these peptides may bypass many drug resistance mechanisms.12 However, the majority of CAPs have no cell selectivity, killing both cancer cells and normal cells. We hypothesized that using EIGF to regulate the anticancer activity of CAPs may achieve selective cytotoxicity for cancer cells. As a proof of concept, we chose one of the well-studied CAPs, melittin, as a model peptide.
Melittin, derived from bee venom, is a 26 amino acid peptide.13 After binding with lipid membrane, melittin forms two independent α helixes separated by a helix-breaking proline residue, and shows potent pore forming activity.14 Meanwhile, the C terminus of melittin is unstructured.15 Melittin has been widely used to kill cancer cells,16 like human gastric cancer17 and human ovarian cancer cells.18 Moreover, the detailed and precise structure–function relationships of melittin analogues (MelAs) have been well elucidated.19 According to the results of single amino acid mutation on MelAs, extending the α helix of the C terminus could increase the membrane lytic capacity,20 and, conversely, reducing the α helix conformation caused a loss of this function.21 Although MelAs are highly poisonous to cancer cells, they are also indistinguishably toxic to normal mammalian cells,22 which is not beneficial for cancer chemotherapy. Inspired by the widely important correlation between phosphorylation induced structural variations and functions in nature, we hypothesized that phosphorylation of MelAs at a specific site might influence their secondary structure and cause further loss of the membrane lytic function. Moreover, dephosphorylation of phosphorylated MelAs may restore the membrane disruption function, especially around the cancer cells bearing high phosphatase activity, which could result in selective cytotoxicity for cancer cells.
Results and discussion
Design, synthesis and screening of the phosphorylated MelAs with reduced membrane lysis capacity
To prove the concept of EIGF, we chose the reported MelA peptide Mel-P5, with the sequence of GIGAVLKVLATGLPALISWIKAAQQL, as an initial peptide for the study, and we designated it as MelA1 (Table 1). Through high-throughput screening of a melittin-based peptide library, MelA1 was reported to have the best potency with respect to membrane leakage and accessibility.20 Further investigations proved that MelA1 could assemble into more stable and larger pores in the lipid membrane than native melittin.23 In MelA1, two residues, Thr11 and Ser18, can be phosphorylated. So, we firstly synthesized two peptides, MelA1-P11 and MelA1-P18 (Table 1), phosphorylated at Thr11 and Ser18 of MelA1, respectively. The membrane lysis capacity was evaluated through measuring calcein leakage of two different calcein-entrapped synthetic lipid vesicles POPC (made of 100% POPC lipid) and POPG (made of 90% POPC lipid and 10% POPG) after treatment with the peptides. However, neither MelA1-P11 nor MelA1-P18 reduced the calcein leakage amount compared to MelA1 (Fig. S1†). On the contrary, MelA1-P11 exhibited slightly stronger membrane leakage with respect to the POPG vesicles than MelA1 (Fig. S1B†).| Peptide | Sequence |
|---|---|
| MelA1 | GIGAVLKVLATGLPALISWIKAAQQL |
| MelA1-P11 | GIGAVLKVLAT(p)GLPALISWIKAAQQL |
| MelA1-P18 | GIGAVLKVLATGLPALIS(p)WIKAAQQL |
| MelA2 | GIGAVLKVLATGLPALISWIKATQQL |
| MelA2-P | GIGAVLKVLATGLPALISWIKAT(p)QQL |
It has been reported that the replacement of the 23rd site of melittin with Ala, Arg and Gln residues could retain the membrane leaking activity,18 which suggested that the 23rd residue was much more compatible. Therefore, we substituted Ala23 with Thr23 in MelA1 to construct peptide MelA2 (Table 1) and measured the secondary structure by Circular Dichroism (CD) and calcein leakage of both peptides. Compared to MelA1, MelA2 had a similar α helix structure and a membrane lysis capacity when interacted with the two synthetic POPC and POPG lipid vesicles (Fig. S2†). The conservation of the structure and function made it possible to use MelA2 for further phosphorylated modifications.
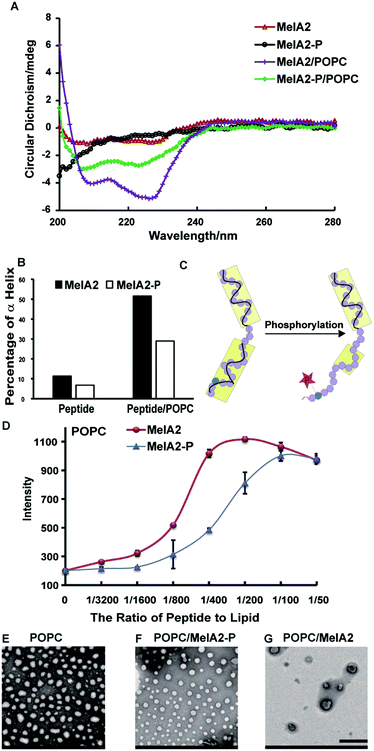
To explore the influence of phosphorylation, we synthesized the MelA2-P peptide (Table 1), which was phosphorylated at the Thr23 site on MelA2. According to the secondary structures measured by CD, MelA2 formed little of the α helix conformation and MelA2-P was almost completely unstructured. Meanwhile, the addition of empty POPC or POPG vesicles extremely enhanced the α helix conformation in both of the peptides (Fig. 1A and S3A†). This result matched the previous reports that liposomes could induce MelAs to fold into amphiphilic α helix structures.14 However, it was obvious that MelA2-P formed less of the α helix structure than MelA2, either with or without the liposomes. Quantification by CDNN software demonstrated that the percentage of α helix in MelA2-P (29%) was only half of that in MelA2 (51.5%) when the peptides interacted with the POPC vesicles (Fig. 1B). Moreover, similar results were observed when POPG vesicles were used (Fig. S3B†). So, it is reasonable to suppose that phosphorylation on the Thr23 site of MelA2 could reduce the amount of lipid-induced α helix conformation (Fig. 1C).
It was reported that the reduction of the α helix on melittin impaired the membrane lysis potency.21 To investigate the effect of phosphorylation at Thr23 on the membrane lytic function, MelA2-P and MelA2 were incubated with calcein-entrapped POPC and POPG vesicles, respectively. The quantified results of calcein leakage indicated that MelA2-P triggered less calcein leakage than MelA2 from both of the POPC and POPG vesicles (Fig. 1D and S3C–E†), which indicated that MelA2-P was less harmful than MelA2 toward the two types of liposome. Furthermore, the morphology and integrity of the POPC and POPG vesicles were monitored by transmission electron microscopy (TEM). The untreated synthetic POPC and POPG vesicles were uniformly round in shape with a diameter of about 100 nm (Fig. 1E and S3F†). However, after treatment with MelA2, the majority of the vesicles were destroyed and became stacked with each other (Fig. 1G and S3H†). In contrast, MelA2-P had almost no effect on the integrity of the POPC and POPG vesicles, as most of the vesicles kept their round shape (Fig. 1F and S3G†). Based on the different effects of MelA2-P and MelA2 on the vesicles, it was quite apparent that phosphorylation at Thr23 on MelA2 could partially reduce the α helix conformation and membrane lysis function.
ALP could dephosphorylate MelA2-P and restore its membrane disruption potency
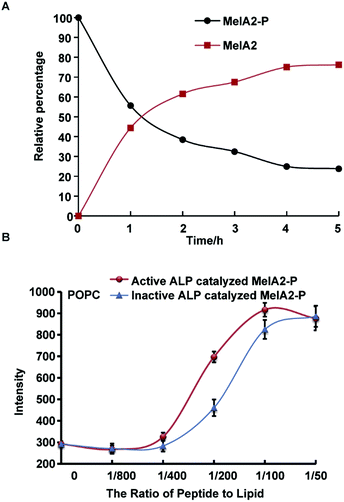
The mammalian alkaline phosphatase (ALP) is expressed in many tissues as a kind of ectoenzyme to remove phosphate groups from proteins, nucleotides, and alkaloids,24 which indicates the broad substrate range of ALP. In many types of cancer, including osteoblastic bone tumors and leukemia, ALP is overexpressed and highly active.25 Investigations using ALP as a target for killing cancer cells have shown positive results.26 For example, ALP-mediated EISA is toxic to drug resistant uterine cancer cells.6d Here, ALP was chosen to dephosphorylate MelA2-P and further verify the feasibility of the ALP-mediated EIGF strategy. Firstly, the dephosphorylation efficiency of MelA2-P by ALP was verified by incubation of MelA2-P with ALP. The amount of MelA2-P and dephosphorylation-resulted MelA2 was monitored and traced by analytical RP-HPLC after ALP treatment for various times (Fig. 2A). We found that more than 50% of MelA2-P was dephosphorylated into MelA2 within 1 h and that the dephosphorylation reaction activity reached a plateau within 5 h where about 80% of MelA2-P was hydrolyzed, which indicated that ALP could effectively catalyze dephosphorylation on MelA2-P. Furthermore, we incubated ALP-hydrolyzed MelA2-P with calcein-entrapped POPC and POPG vesicles and discovered that ALP treated MelA2-P recovered its membrane lysis potency compared with control groups for which the same amount of inactive ALP was used to treat MelA2-P in experiments (Fig. 2B and S4A†). Thus, ALP was proven to be an effective phosphatase to dephosphorylate MelA2-P, allowing it to regain its membrane disrupting function, which implied that ALP-mediated EIGF was able to regulate MelA2-P activity in vitro.MelA2-P could selectively kill cancer cells with higher ALP activity
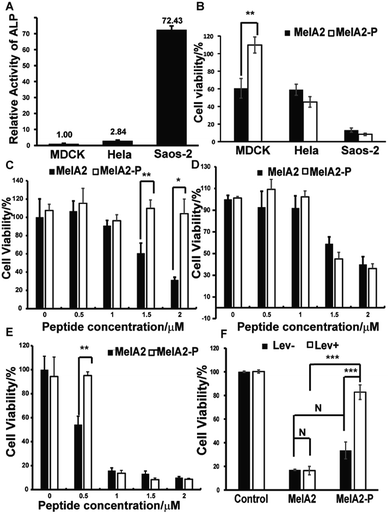
To test whether this ALP-mediated EIGF mechanism functions at the cellular level, the normal mammalian cell line MDCK, cervical carcinoma cell line Hela, and human osteosarcoma cell line Saos-2 were chosen because of their differing ALP activities.6f We found that the ALP activity, as quantified by an ALP activity kit, was about three times higher in Hela and seventy times higher in Saos-2 than that in the MDCK control cells (Fig. 3A). MelAs, which have the pore forming tendency, have been proven to disrupt the plasma membrane and induce cell death in mammalian cells. Using the MTT assay, we demonstrated that the MelA2 peptide was cytotoxic to Hela and Saos-2, as well as to MDCK (Fig. 3B). Furthermore, the cytotoxicity of MelA2 for MDCK, Hela and Saos-2 was dose dependent (Fig. 3D and E). Considering that MelA2-P induced less membrane lysis in the two synthetic vesicles, MelA2-P should be less toxic to normal cells. The MTT assay indicated that MelA2-P almost doubled the viability of normal MDCK cells (Fig. 3B). Based on our hypothesis, MelA2-P should gain the function of membrane lysis capacity and trigger cell death after dephosphorylation by ALP, which is highly active around Hela and Saos-2 cells. As expected, the cell viability of Hela after incubation with MelA2-P was as low as that after MelA2 treatment (Fig. 3B). A similar result was obtained in Saos-2 cells (Fig. 3B). Moreover, the cytotoxicity of MelA2-P for Hela and Saos-2 was concentration dependent (Fig. 3D and E). These results demonstrate that MelA2 could kill all of the three cell lines, but MelA2-P only had significant cytotoxicity toward the high ALP active cells Hela and Saos-2. Furthermore, during the dosage testing, MelA2-P showed almost no toxicity for MDCK (Fig. 3C). Therefore, the results of the MTT assays implied that MelA2-P could distinguish cancer cells from normal cells by differences in the ALP activity and could selectively kill cancer cells with higher ALP activity. To verify the crucial role of ALP in MelA2-P induced cancer cell death, we measured the effect of levamisole, which is a widely used ALP inhibitor, on the cell viability of Saos-2 and MelA2 or MelA2-P treated Saos-2. The MTT assay results indicated that there was no difference in the cytotoxicity of MelA2 treated cells with or without ALP inhibitor treatment (Fig. 3F). But levamisole could significantly reduce the MelA2-P induced cell death. Moreover, the levamisole had no obvious effect on the cell viability alone. These results demonstrated that the process of dephosphorylation by ALP was vital for the MelA2-P induced cell death of Saos-2 (Fig. 3F). Besides, to further investigate the effect of dephosphorylation on the MelA2-P induced cell death, the phosphorylated Thr residue in MelA2-P was mutated to glutamate (E), which is widely used to mimic phosphorylation modification, to give MelA2-T23E. The MelA2-T23E peptide is resistant to phosphatase. We tested the cytotoxicity of MelA2-T23E and MelA2-P by MTT assay. The results showed that the MelA2-T23E treatment induced less cell death than MelA2-P treatment in Saos-2 cells (Fig. S4B†). This suggested that the cytotoxicity of MelA2-P was dependent on the ALP mediated dephosphorylation of MelA2-P, which indicated that the ALP mediated EIGF strategy could work on the cellular level as the proposed mechanism. Therefore, MelA2-P could selectively kill cancer cells with higher ALP activity through an ALP-mediated EIGF strategy.Experimental
Peptide synthesis
All of the peptides were manually synthesized by the standard Fmoc-based solid-phase peptide synthesis (Fmoc SPPS). Rink amide-MBHA resin and Fmoc-protected amino acids were purchased from GL Biochem (Shanghai) Ltd. Other chemicals were purchased from Sigma-Aldrich. For all of the peptides, amide-MBHA resin was used. Crude peptides were further purified by RP-HPLC. The identification of the pure peptides was confirmed by ESI-MS and analytical RP-HPLC.Empty vesicle preparation
For the POPG vesicles, 337.5 μl of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) stock solution (10 mg ml−1 in chloroform) and 37.5 μl of POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol) stock solution (10 mg ml−1 in chloroform) were mixed and dried under nitrogen gas. For the POPC vesicles, 375 μl POPC was dried. The dried lipids were dissolved in Tris buffer (10 mM Tris, 150 mM NaCl, pH 7.4), separately. Vesicles with a size of 100 nm were constructed with an Avanti Mini Extruder. The final lipid concentration was 5 mM.Calcein entrapped vesicle preparation
For the POPG vesicles, 675 μl of POPC stock solution (10 mg ml−1 in chloroform) and 75 μl of POPG stock solution (10 mg ml−1 in chloroform) was mixed and dried under nitrogen gas. For the POPC vesicles, 750 μl of POPC was used. The dried lipids were dissolved in calcein buffer (10 mM Tris, 90 mM NaCl, 60 mM calcein). Vesicles with a size of 100 nm were constructed with an Avanti Mini Extruder. To remove the free calcein molecules, vesicles were purified with a PD-10 column (GE Healthcare Life Sciences). The final lipid concentration was determined by the previous method.27Membrane leakage
Calcein entrapped vesicles were diluted into 25 μM or 100 μM with Tris buffer (10 mM Tris, 150 mM NaCl, pH 7.4). Different doses of peptides were added into 25 μM or 100 μM of the POPC or POPG vesicles to achieve ratios of the peptide and lipid of 0, 1/3200, 1/1600, 1/800, 1/400, 1/200, 1/100, and 1/50. The calcein leakage was measured by a microplate reader (BioTek). Also, the calcein fluorescence was excited at 485 and emitted at 535 nm.Circular dichroism (CD)
Peptides (10 μM) were used for the CD measurements. Peptide (10 μM) was cocultured with the 500 μM vesicles for 1.5 h before the CD measurements for the mixtures of peptides and vesicles. The CD spectra were collected from 200 nm to 280 nm in wavelength in the CD spectropolarimeter purchased from Applied Photophysics.Cell culture
MDCK was propagated in a RPMI 1640 medium with 10% FBS (fetal bovine serum) and 1% penicillin–streptomycin solution (penicillin 100 U ml−1 and streptomycin 100 μg ml−1). Hela was propagated in a DMEM medium containing 10% FBS and 1% penicillin–streptomycin solution. The Saos-2 cell line was purchased from China Infrastructure of Cell Line Resources and propagated in a McCoy’ 5A medium with 20% FBS and 1% penicillin–streptomycin solution. All of the media and the FBS were purchased from Gibco. The cells were cultured in a sterile incubator containing 5% CO2 at 37 °C.MTT assay
The cells in the exponential growth phase were harvested and seeded in a 96-well plate at a concentration of 6000 cells per well. The cells were cultured for 24 h at 37 °C. Then the medium was removed and 200 μl of opti MEM containing MelA2 and MelA2-P peptides of gradient concentrations (0, 0.5, 1, 1.5, and 2 μM) was added into each well. After 24 h of incubation, 20 μl of 5 mg ml−1 MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was introduced into each well and the plate was cultured at 37 °C for another 4 h. After that, the medium was removed and 150 μl of DMSO was added to each well to dissolve the purple precipitation. The absorption at 490 or 570 nm of the solution was measured in a microplate reader.ALP inhibitor levamisole treatment
For the experimental groups, the Saos-2 cells were treated with 0.5 mM levamisole for 3 h before the addition of the mixture of 1 μM peptides (MelA2 or MelA2-P) and 0.5 mM levamisole for 24 h of culturing. For the blank control, the Saos-2 cells were treated with equivalent water and DMSO compared to the experimental group. For the levamisole control, the Saos-2 cells were treated with 0.5 mM levamisole for 3 h before the addition of the mixture of equivalent DMSO and 0.5 mM levamisole for 24 h of culturing. For the peptide treatment groups (MelA2 or MelA2-P), the cells were treated with equivalent water for 3 h compared to the experimental control before the addition of the mixture of 1 μM peptides (MelA2 or MelA2-P) and equivalent water for 24 h of culturing. Levamisole was dissolved in water to prepare the 100 mM stock buffer. MelA2-P and MelA2 were dissolved in DMSO to prepare the 2 mM peptide stock solutions. After 24 h of cell culturing, the MTT assay was measured. n = 3.ALP activity assay
The Hela, Saos-2 and MDCK cells were plated in a 96-well plate with 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well, respectively, and the cells were allowed to attach for 4 h. The medium was removed and the cells were washed with PBS buffer 3 times. 25 μl of dd H2O was added into each well and the plate was incubated at 37 °C for 1 h. Then the plate was moved into a refrigerator set at −80 °C and stored for 1 h. After that, the plate was placed on ice and allowed to defrost. The ALP activity in each well was measured with an Alkaline Phosphatase Assay Kit purchased from Beyotime Biotechnology.
000 cells per well, respectively, and the cells were allowed to attach for 4 h. The medium was removed and the cells were washed with PBS buffer 3 times. 25 μl of dd H2O was added into each well and the plate was incubated at 37 °C for 1 h. Then the plate was moved into a refrigerator set at −80 °C and stored for 1 h. After that, the plate was placed on ice and allowed to defrost. The ALP activity in each well was measured with an Alkaline Phosphatase Assay Kit purchased from Beyotime Biotechnology.
Dephosphorylation of MelA2-P by ALP
MelA2-P (100 μM) was incubated with 100 U ml−1 of ALP (Alkaline Phosphatase, Calf Intestinal, New England Biolabs) in Tris buffer (10 mM Tris, 150 mM NaCl, pH 7.4) at 37 °C for 5 h. Every hour, 50 μl of the reaction solution was used to run an analytical RP-HPLC measurement, and the dephosphorization ratio was quantified by the peak area integration and normalization. For the calcein leakage experiment, MelA2-P (100 μM) was incubated with 100 U ml−1 active and inactive ALP (Alkaline Phosphatase, Calf Intestinal, New England Biolabs) in Tris buffer (10 mM Tris, 150 mM NaCl, pH 7.4) at 37 °C for 5 h and then all samples were heated at 100 °C for 30 min to stop catalytic reactions. The leakage intensity of the 100 μM (lipid concentration) calcein-entrapped vesicles at a peptide-to-lipid ratio from 1/800 to 1/50 was measured for 20 min and the amount of ALP enzyme was kept the same in each well to avoid the influence of the ALP on the calcein leakage.Transmission electron microscopy (TEM) analysis
For the mixtures of the peptides and vesicles, the peptide (10 μM) was cocultured with the 500 μM vesicles for 10 h before TEM. All of the samples (8 μl) were loaded onto a copper grid with carbon support films 3 times and stained with tungstophosphoric acid. The samples were imaged on a Hitachi H-7650B TEM after being dried overnight.Conclusions
In conclusion, we have verified that phosphorylation at Thr23 of MelA2 could reduce the membrane lysis capacity and cytotoxicity of the studied CAP toward normal mammalian cells. After ALP-mediated dephosphorylation, MelA2-P gained the function of disrupting the membranes not only for the synthetic vesicles but also for the mammalian cell membranes, resulting in further cell death. Moreover, through the ALP mediated EIGF of MelA2-P, we achieved selective killing of cancer cells with high ALP activity, and these direct membrane lysis mechanisms might partially reduce the drug resistance responses of cancer cells. Furthermore, this ALP-mediated EIGF method should be applicable to regulate the function and activity of other CAPs as well as different types of bioactive molecule, which provides new strategies for the design and discovery of prodrugs and cancer-targeted drugs.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21332006, 21672126).Notes and references
- (a) B. Mayr and M. Montminy, Nat. Rev. Mol. Cell Biol., 2001, 2, 599 CrossRef CAS PubMed; (b) J. Lilien and J. Balsamo, Curr. Opin. Cell Biol., 2005, 17, 459 CrossRef CAS PubMed; (c) J. T. Du, Y. M. Li, W. Wei, G. S. Wu, Y. F. Zhao, K. Kanazawa, T. Nemoto and H. Nakanishi, J. Am. Chem. Soc., 2005, 127, 16350 CrossRef CAS PubMed; (d) M. R. Ma, Z. W. Hu, Y. F. Zhao, Y. X. Chen and Y. M. Li, Sci. Rep., 2016, 6, 37130 CrossRef CAS PubMed; (e) Z. W. Hu, M. R. Ma, Y. X. Chen, Y. F. Zhao, W. Qiang and Y. M. Li, J. Biol. Chem., 2016, 292, 2611 CrossRef PubMed.
- (a) S. Y. Shieh, M. Ikeda, Y. Taya and C. Prives, Cell, 1997, 91, 325 CrossRef CAS PubMed; (b) C. Prives, Cell, 1998, 95, 5 CrossRef CAS PubMed.
- S. Winkler, D. Wilson and D. L. Kaplan, Biochemistry, 2000, 39, 14002 CrossRef CAS PubMed.
- T. H. Ku, M. P. Chien, M. P. Thompson, R. S. Sinkovits, N. H. Olson, T. S. Baker and N. C. Gianneschi, J. Am. Chem. Soc., 2011, 133, 8392 CrossRef CAS PubMed.
- H. Kuhnle and H. G. Borner, Angew. Chem., Int. Ed. Engl., 2009, 48, 6431 CrossRef PubMed.
- (a) C. Ren, H. Wang, D. Mao, X. Zhang, Q. Fengzhao, Y. Shi, D. Ding, D. Kong, L. Wang and Z. Yang, Angew. Chem., Int. Ed. Engl., 2015, 54, 4823 ( Angew. Chem. , 2015 , 127 , 4905 ) CrossRef CAS PubMed; (b) B. Mei, Q. Miao, A. Tang and G. Liang, Nanoscale, 2015, 7, 15605 RSC; (c) R. A. Pires, Y. M. Abul-Haija, D. S. Costa, R. Novoa-Carballal, R. L. Reis, R. V. Ulijn and I. Pashkuleva, J. Am. Chem. Soc., 2015, 137, 576 CrossRef CAS PubMed; (d) Y. Kuang, J. Shi, J. Li, D. Yuan, K. A. Alberti, Q. Xu and B. Xu, Angew. Chem., Int. Ed. Engl., 2014, 53, 8104 ( Angew. Chem. , 2014 , 126 , 8242 ) CrossRef CAS PubMed; (e) J. Zhou, X. Du and B. Xu, Angew. Chem., Int. Ed. Engl., 2016, 55, 5770 ( Angew. Chem. , 2016 , 128 , 5864 ) CrossRef CAS PubMed; (f) J. Zhou, X. Du, N. Yamagata and B. Xu, J. Am. Chem. Soc., 2016, 138, 3813 CrossRef CAS PubMed; (g) H. Wang, Z. Feng, Y. Wang, R. Zhou, Z. Yang and B. Xu, J. Am. Chem. Soc., 2016, 138, 16046 CrossRef CAS PubMed; (h) H. Wang, Z. Feng, D. Wu, K. J. Fritzsching, M. Rigney, J. Zhou, Y. Jiang, K. Schmidt-Rohr and B. Xu, J. Am. Chem. Soc., 2016, 138, 10758 CrossRef CAS PubMed.
- (a) R. Liang, S. You, L. Ma, C. Li, R. Tian, M. Wei, D. Yan, M. Yin, W. Yang, D. G. Evans and X. Duan, Chem. Sci., 2015, 6, 5511 RSC; (b) G. Yu, J. Zhou, J. Shen, G. Tang and F. Huang, Chem. Sci., 2016, 7, 4073 RSC.
- (a) S. T. Pan, Z. L. Li, Z. X. He, J. X. Qiu and S. F. Zhou, Clin. Exp. Pharmacol. Physiol., 2016, 43, 723 CrossRef CAS PubMed; (b) R. R. Gordon and P. S. Nelson, Drug Resist. Updates, 2012, 15, 123 CrossRef CAS PubMed.
- (a) I. A. Al-Rayahi and R. H. Sanyi, Front. Immunol., 2015, 6, 2 Search PubMed; (b) D. W. Hoskin and A. Ramamoorthy, Biochim. Biophys. Acta, 2008, 1778, 357 CrossRef CAS PubMed; (c) N. I. Kalinovskaya, L. A. Romanenko, A. I. Kalinovsky, P. S. Dmitrenok and S. A. Dyshlovoy, Nat. Prod. Commun., 2013, 8, 381 CAS.
- C. Wang, Y. Zhou, S. Li, H. Li, L. Tian, H. Wang and D. Shang, Life Sci., 2013, 92, 1004 CrossRef CAS PubMed.
- R. E. Hancock and A. Rozek, FEMS Microbiol. Lett., 2002, 206, 143 CrossRef CAS PubMed.
- (a) J. S. Mader and D. W. Hoskin, Expert Opin. Invest. Drugs, 2006, 15, 933 CrossRef CAS PubMed; (b) F. Schweizer, Eur. J. Pharmacol., 2009, 625, 190 CrossRef CAS PubMed.
- N. Oršolić, Cancer Metastasis Rev., 2011, 31, 173 CrossRef PubMed.
- (a) M. T. Lee, T. L. Sun, W. C. Hung and H. W. Huang, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 14243 CrossRef CAS PubMed; (b) D. Gimenez, O. L. Sanchez-Munoz and J. Salgado, Langmuir, 2015, 31, 3146 CrossRef CAS PubMed; (c) R. Smith, F. Separovic, T. J. Milne, A. Whittaker, F. M. Bennett, B. A. Cornell and A. Makriyannis, J. Mol. Biol., 1994, 241, 456 CrossRef CAS PubMed.
- (a) S. Akashi and K. Takio, J. Am. Soc. Mass Spectrom., 2001, 12, 1247 CrossRef CAS PubMed; (b) T. C. Terwilliger and D. Eisenberg, J. Biol. Chem., 1982, 257, 6010 CAS; (c) T. C. Terwilliger and D. Eisenberg, J. Biol. Chem., 1982, 257, 6016 CAS.
- N. R. Soman, S. L. Baldwin, G. Hu, J. N. Marsh, G. M. Lanza, J. E. Heuser, J. M. Arbeit, S. A. Wickline and P. H. Schlesinger, J. Clin. Invest., 2009, 119, 2830 CrossRef CAS PubMed.
- G. M. Kong, W. H. Tao, Y. L. Diao, P. H. Fang, J. J. Wang, P. Bo and F. Qian, World J. Gastroenterol., 2016, 22, 3186 CrossRef CAS PubMed.
- (a) M. Liu, J. Zong, Z. Liu, L. Li, X. Zheng, B. Wang and G. Sun, Cancer Immunol. Immunother., 2013, 62, 889 CrossRef CAS PubMed; (b) M. Jo, M. H. Park, P. S. Kollipara, B. J. An, H. S. Song, S. B. Han, J. H. Kim, M. J. Song and J. T. Hong, Toxicol. Appl. Pharmacol., 2012, 258, 72 CrossRef CAS PubMed.
- (a) S. J. Irudayam and M. L. Berkowitz, Biochim. Biophys. Acta, 2011, 1808, 2258 CrossRef CAS PubMed; (b) G. Wiedman, S. Y. Kim, E. Zapata-Mercado, W. C. Wimley and K. Hristova, J. Am. Chem. Soc., 2017, 139, 937 CrossRef CAS PubMed.
- A. J. Krauson, J. He and W. C. Wimley, J. Am. Chem. Soc., 2012, 134, 12732 CrossRef CAS PubMed.
- A. J. Krauson, O. M. Hall, T. Fuselier, C. G. Starr, W. B. Kauffman and W. C. Wimley, J. Am. Chem. Soc., 2015, 137, 16144 CrossRef CAS PubMed.
- M. Moreno and E. Giralt, Toxins, 2015, 7, 1126 CrossRef CAS PubMed.
- G. Wiedman, T. Fuselier, J. He, P. C. Searson, K. Hristova and W. C. Wimley, J. Am. Chem. Soc., 2014, 136, 4724 CrossRef CAS PubMed.
- (a) W. H. Fishman, Clin. Biochem., 1990, 23, 99 CrossRef CAS PubMed; (b) J. L. Millan, Mammalian Alkaline Phosphatases: From Biology to Applications in Medicine and Biotechnology, Wiley-VCH, Weinheim, 2006 Search PubMed.
- (a) S. M. Lim, Y. N. Kim, K. H. Park, B. Kang, H. J. Chon, C. Kim, J. H. Kim and S. Y. Rha, BMC Cancer, 2016, 16, 385 CrossRef PubMed; (b) P. Dua, H. S. Kang, S. M. Hong, M. S. Tsao, S. Kim and D. K. Lee, Cancer Res., 2013, 73, 1934 CrossRef CAS PubMed; (c) A. Burlina, D. Rubin, S. Secchiero, L. Sciacovelli, M. Zaninotto and M. Plebani, Clin. Chim. Acta, 1994, 226, 151 CrossRef CAS PubMed; (d) P. A. Hoisaeter, Acta Oncol., 1991, 30, 171 CrossRef CAS PubMed.
- (a) H. Wang, Z. Feng, D. Wu, K. J. Fritzsching, M. Rigney, J. Zhou, Y. Jiang, K. Schmidt-Rohr and B. Xu, J. Am. Chem. Soc., 2016, 138, 10758 CrossRef CAS PubMed; (b) S. Mizukami, M. Kashibe, K. Matsumoto, Y. Hori and K. Kikuchi, Chem. Sci., 2017, 8, 3047 RSC.
- D. S. Zhao, Y. X. Chen and Y. M. Li, Chem. Commun., 2015, 51, 2095 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Mass spectra and other materials. See DOI: 10.1039/c7sc03217j |
| This journal is © The Royal Society of Chemistry 2017 |