Open Access Article
Open Access ArticleManipulation of cytokine secretion in human dendritic cells using glycopolymers with picomolar affinity for DC-SIGN†
Daniel A.
Mitchell
*ab,
Qiang
Zhang
c,
Lenny
Voorhaar
c,
David M.
Haddleton
 c,
Shan
Herath
d,
Anne S.
Gleinich
a,
Harpal S.
Randeva
ab,
Max
Crispin
e,
Hendrik
Lehnert
f,
Russell
Wallis
g,
Steven
Patterson
d and
C. Remzi
Becer
c,
Shan
Herath
d,
Anne S.
Gleinich
a,
Harpal S.
Randeva
ab,
Max
Crispin
e,
Hendrik
Lehnert
f,
Russell
Wallis
g,
Steven
Patterson
d and
C. Remzi
Becer
 *h
*h
aClinical Sciences Research Laboratories, University of Warwick, Coventry CV2 2DX, United Kingdom. E-mail: d.mitchell@warwick.ac.uk
bUniversity Hospital Coventry, Warwickshire NHS Trust, Coventry CV2 2DX, United Kingdom
cDepartment of Chemistry, University of Warwick, Coventry CV4 7AL, United Kingdom
dChelsea & Westminster Hospital, Imperial College School of Medicine, London SW10 9NH, United Kingdom
eGlycobiology Institute, University of Oxford, Oxford OX1 3QU, United Kingdom
fUniversität zu Lübeck, 23562 Lübeck, Germany
gDepartment of Biochemistry, University of Leicester, Leicester LE1 9HN, United Kingdom
hSchool of Engineering and Materials Science, Queen Mary University, London E1 4NS, United Kingdom. E-mail: r.becer@qmul.ac.uk
First published on 16th August 2017
Abstract
The human C-type lectin DC-SIGN (CD209) is a significant receptor on the surface of dendritic cells (DCs) – crucial components of host defense that bridge the innate and adaptive immune systems. A range of linear glycopolymers, constructed via controlled radical polymerization techniques have been shown to interact with DC-SIGN with affinities in the physiologically active range. However, these first generation glycopolymers possess limited structural definition and their effects on DCs were not known. Here we report the development of star-shaped mannose glycopolymers with the aim of targeting the clustered domain arrangement of DC-SIGN and these were shown to bind with picomolar affinity. Increased secretion of IL-10 with simultaneous decrease in secreted IL-12p70 occurred in activated DCs incubated with star-shaped glycopolymers – a cytokine secretion pattern characteristic of wound-healing tissue environments. Incorporating stellar architecture into glycopolymer design could be key to developing selective and very high-affinity therapeutic materials with distinct immunomodulatory and tissue repair potential.
Introduction
The recognition of repeating molecular patterns is a major function of the human immune system, enabling it to interact with and interpret various biological structures. These include the surfaces of pathogens such as viruses, fungi and bacteria, in addition to host structures such as glycoproteins and apoptotic cells.1,2 C-type lectins are a major class of pattern recognition molecules in humans that interact with complex carbohydrates such as microbial polysaccharides and oligosaccharides present on human and viral glycoproteins and glycolipids.3 In particular, the C-type lectin DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin; CD209; CLEC4L) is significantly implicated in human disease through its interactions with viral carbohydrates on HIV and mycobacterial lipoarabinomannans (e.g. tuberculosis), in addition to its ability to transduce intracellular signaling events and influence dendritic cell responses.4–7 DC-SIGN is also important in the proper responses to specific apoptotic cell uptake by dendritic cells (DCs).2 DCs are especially important components of the human immune system owing to their roles as highly efficient surveillance entities and professional antigen presenting cells. DCs are the only cell type with the ability to activate naïve T lymphocytes, making them very powerful bridges between innate and adaptive immunity.8 NMR analyses of DC-SIGN-oligosaccharide interactions in solution suggest discrete binding modes of the carbohydrate-recognition domain for distinct glycan types that in turn are thought to drive different cell signaling pathways.9 Furthermore, the distribution of DCs (and DC-SIGN-positive cells such as selected macrophage subpopulations) within discrete regions of tissues of the human body makes them attractive targets for site-specific therapeutic intervention in a broad range of diseases with immune system involvement.10,11Designed glycopolymers have been successfully generated for high affinity interactions with human DC-SIGN.12–15 This has included the development of size- and sequence-controlled polymers, in addition to conformation-controlled polymers and glycoconjugates, collectively offering a diverse range of strategies for DC-SIGN targeting and exploitation. However, the concept of polymer architecture as a rational design strategy for DC-SIGN targeting with these classes of glycopolymer has yet to be employed and furthermore, the impact of advanced glycopolymers on dendritic cell function has yet to be investigated (Fig. 1). Knowledge of the tetrameric structure of DC-SIGN, wherein four mannoside-selective carbohydrate recognition domains (CRDs) are clustered at the top of a coiled-coil stalk projecting from the cell surface,16,17 allows for the rational construction of novel glycopolymers for further affinity enhancement and potential biological impact. Understanding of the role of DC-SIGN stimulation in dendritic cell responses holds potential for efficient, high affinity ligands to serve as immunomodulators as well as antiviral agents. The main text of the article should appear here with headings as appropriate.
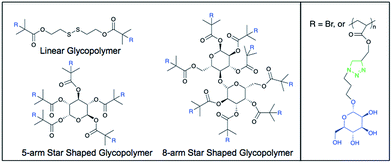 | ||
| Fig. 1 Representative chemical structures of the mannose glycopolymers. R = Br in the initiator structure. | ||
Materials and methods
Instrumentation
NMR spectra were recorded on a Bruker DPX-300 and DPX-400 using deuterated solvents. FTIR spectra were recorded on a Bruker Vector 22 FTIR spectrometer. For SEC a Varian PL 390-LC system was used, equipped with refractive index and viscosimetry detectors, using either CHCl3 or DMF (LiBr 1 g l−1) as the eluent (1 ml min−1). Data analysis was performed using Cirrus software and molecular weights were determined relative to Polymer Labs pMMA standards. The LCST of the glycopolymers was determined by measuring the cloud point of a 5 mg ml−1 solution of the polymer in water with a Stanford Research Systems OptiMelt MPA100.Synthesis of 2-methacrylic acid 3-trimethylsilanyl-prop-2-ynyl ester (TMSMA)
A flask was charged with 3-trimethylsilanyl-prop-2-yn-1-ol (25.0 g, 195 mmol) and triethylamine (41.0 ml, 294 mmol) in THF (250 ml) and cooled with dry ice. A solution of 2-methylacryloyl chloride (23.0 ml, 235 mmol) in THF (50 ml) was added drop wise. The mixture was stirred for 1 hour while cooling with dry ice and stirred overnight at room temperature, during which yellow coloured salts were formed. The mixture was purified on a basic Al2O3 column and the solvent was removed under reduced pressure, leaving behind a clear yellow liquid. The yield was 35.0 g (178 mmol, 92%) 1H NMR (CDCl3, 298 K, 300 MHz) δ (ppm) = 0.18 (s, 9H, Si(CH3)3); 1.96 (m, 3H, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2); 4.76 (s, 2H, OCH2); 5.62 (m, 1H, C
CH2); 4.76 (s, 2H, OCH2); 5.62 (m, 1H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHH); 6.17 (m, 1H, C
CHH); 6.17 (m, 1H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHH).
CHH).
Typical atom transfer radical polymerization procedure
A Schlenk tube was charged with initiator (1.0 mmol), monomer (50 mmol), N-(ethyl)-2-pyridylmethanimine ligand (2.0 mmol) and toluene (same volume as monomer). Two different methods have been used to remove the oxygen from the solution, which have been found to be equally effective. The first method was a freeze–pump–thaw cycle, which was executed three times. The second method was bubbling the solution with nitrogen for at least 20 minutes. A second Schlenk tube was charged with Cu(I)Br (1.0 mmol) and a stirring bar and the oxygen was removed by evacuating and subsequently filling the tube with nitrogen three times. The solution was transferred to the second tube using a cannula and a vacuum to start the transfer. The solution was heated to 90 °C while being kept under nitrogen atmosphere for the entire duration of the reaction. Samples were taken periodically using a degassed syringe in case of the kinetic experiments. The polymerization was stopped by cooling the mixture to room temperature and exposing it to oxygen. The mixture was diluted with CH2Cl2 (approximately same volume as mixture) and passed through a basic Al2O3 column. The polymer was precipitated into a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 vol/vol methanol/water mixture (pTMSMA). The solid was isolated by filtration and dried in a vacuum oven overnight. In most experiments with the star-shaped initiators the amount of toluene used was higher to prevent star–star coupling. This was either twice (for 5-arm star) or three times (8-arm star) the volume of the monomer.
1 vol/vol methanol/water mixture (pTMSMA). The solid was isolated by filtration and dried in a vacuum oven overnight. In most experiments with the star-shaped initiators the amount of toluene used was higher to prevent star–star coupling. This was either twice (for 5-arm star) or three times (8-arm star) the volume of the monomer.
Deprotection of pTMSMA
The pTMSMA (0.5 g) was dissolved in THF (35 ml) and glacial acetic acid (0.22 ml, 1.5 equiv. with respect to the TMS groups) was added. The solution was bubbled with nitrogen and cooled with dry ice for at least 15 minutes. A 0.2 M solution of TBAF in THF (50 ml, 4.0 equiv.) was added dropwise with a syringe while stirring the solution and cooling with dry ice. The dry ice was removed after 30 minutes and the mixture was stirred overnight at room temperature. The mixture was concentrated under reduced pressure, purified on a silica column and concentrated again. The polymer was precipitated in petroleum ether cooled with dry ice and acetone. The white powder was isolated by filtration and dried in a vacuum oven overnight. The deprotection was confirmed by the appearance of a peak at 2.5 ppm in the 1H NMR and the disappearance of the 0.2 ppm peak.Synthesis of α-mannose azide
Sodium azide (7.26 g, 111 mmol), D-(+)-mannose (2.00 g, 11.1 mmol) and triethylamine (15.5 ml, 111 mmol) were dissolved in water (40 ml) and cooled to 0 °C. 2-Chloro-1,3-dimethylimidazolinium chloride (5.61 g, 33.3 mmol) was added and the mixture was stirred for 1 hour at 0 °C. The solvent was removed under reduced pressure and EtOH (40 ml) was added. The solids were removed by filtration and the solution was purified on a long Amberlite IR-120 column, using EtOH as the eluent. The mixture was checked with FTIR to confirm the removal of all sodium azide (ν = 2030 cm−1). The solvent was removed under reduced pressure, water (30 ml) was added and the mixture was washed with dichloromethane (5 × 15 ml). The solvent was removed under reduced pressure, water (10 ml) was added and the solution was freeze-dried overnight to give α-mannose azide (1.59 g, 78 mmol, 70%) as an off-white solid. 1H NMR (D2O, 298 K, 400 MHz) δ (ppm) = 5.45 (1H), 3.95–3.60 (6H). 13C NMR (D2O, 298 K, 400 MHz) δ (ppm) = 89.72 (C1), 74.65, 69.86, 69.77, 66.40, 60.83 (C6). FT-IR (H2O) ν (cm−1) = 2112 (N3).Mannose azide clicking to propargyl polymers
A solution containing propargyl polymer (100 mg, 0.81 mmol propargyl groups), mannose azide (213 mg, 1.05 mmol) and CuBr (116 mg, 0.81 mmol) in DMSO (10 ml) was bubbled with nitrogen for 20 minutes. Ethyl ligand (216 mg, 1.61 mmol) was added and the solution was bubbled with nitrogen for several more minutes. The solution was subsequently stirred at ambient temperature for two days. The mixture was purified by dialysis (MWCO: 1000) against distilled water for two days, while changing the water at least four times. It was then concentrated under reduced pressure and freeze-dried overnight.Glycopolymer synthesis and characterization
Before the click reactions could take place, the propargyl groups of the pTMSMA polymers need to be deprotected by removal of the TMS group. This was performed by reacting the polymer with CH3COOH and TBAF in THF. The deprotection can be followed by 1H NMR (Fig. S1†). The peak of the TMS group at 0.2 ppm disappears and a peak appears at 2.5 ppm for the propargylic proton. After removing the water from the reaction mixture, EtOH was added to filter the unreacted sodium azide from the mixture. However, some of the sodium azide was still soluble in EtOH, as is shown in the FTIR spectrum in Fig. S2.† As sodium azide can form explosive compounds when it is brought into contact with halogenated solvents, it was necessary to remove this from the mixture before continuing with subsequent purification steps. It was found that this last trace of sodium azide could be removed on an Amberlite IR-120 column using EtOH as the eluent. TEA was also removed from the mixture with this column. The remaining DMC was removed by washing the mixture in water with CH2Cl2. Mannose azide was attached to each of the different synthesized propargyl polymers using click chemistry. The reaction can be observed using 1H NMR through the appearance of a triazole peak at 8.30 ppm (Fig. S3†). The peaks at 6.0 ppm and 5.4–4.3 ppm are from the mannose. The kinetic plot for the polymerization of TMSMA shows that it takes roughly 15 minutes for the polymerization to initiate (Fig. S5†). After that the conversion is linear up to at least 95%. The increase of Mn with conversion is almost linear within the margin of error. PDI values are all very close together around 1.3. The homopolymerization of TMSMA using the 8-arm initiator was performed in 75% solvent to avoid coupling of the star polymers. The first-order kinetic plot is linear up to 50% conversion. A short initiation period was observed that was also seen in the homopolymerization of TMSMA with the disulfide initiator. The increase in molecular weight is mostly linear with relation to the conversion (PDI ∼ 1.25). The theoretical molecular weight of the synthesized linear, 5 arm and 8 arm glycopolymers have been calculated as 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 g mol−1, 37
700 g mol−1, 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 g mol−1, and 39
100 g mol−1, and 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 g mol−1, respectively. The measured number average molecular weight of the linear, 5-arm, and 8-arm glycopolymers were found as 41
600 g mol−1, respectively. The measured number average molecular weight of the linear, 5-arm, and 8-arm glycopolymers were found as 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, 42
000 g mol−1, 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 g mol−1, and 45
100 g mol−1, and 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 g mol−1, respectively.
600 g mol−1, respectively.
Protein expression and surface plasmon resonance
Soluble recombinant DC-SIGN was generated in E. coli and purified via affinity chromatography using mannose–sepharose, as described previously and immobilized16 on GLM sensor chips via amine coupling for use in the Bio-Rad ProteOn XPR36 instrument (Bio-Rad Laboratories, Hercules, USA).12 Glycopolymer samples were dissolved in running buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 5 mM CaCl2, 0.01% (v/v) Tween-20, 0.01% (w/v) NaN3) and flowed over sensor chips at a flow rate of 25 μl ml−1 at 25 °C. Sensorgram data were collected and reference subtraction performed against a blank channel treated with amine coupling activators and subsequently blocked with 1 M ethanolamine. Kinetic parameters were determined using the Bia-evaluation software (GE Healthcare) utilizing heterogeneous ligand fitting models.Blood samples
Peripheral blood mononuclear cells (PBMCs) were isolated from blood leukocyte cones (National Blood Transfusion Service, UK) by density gradient centrifugation with Lymphoprep™ 1077 (PAA Laboratories, GmbH, UK) followed by a 50% Percoll (Sigma, UK) gradient to obtain a high-density fraction enriched in lymphocytes and a low-density fraction enriched in monocytes. Fractions were aliquoted and stored in liquid N2 until required.Generation of dendritic cells
Monocyte derived dendritic cells (moDCs) were isolated from the monocyte rich fraction by magnetic bead separation using anti-CD14 coated magnetic beads (Miltenyi Biotec). Purified monocytes were cultured in complete medium (RPMI containing 10% FBS, 100 IU ml−1 penicillin, 0.1 mg ml−1 streptomycin, and 2 mM L-glutamine; Sigma-Aldrich) supplemented with GM-CSF (100 ng ml−1) and IL-4 (50 ng mLl−1; R&D Systems), this being replenished every other day. On day 7, DCs were harvested, washed twice and resuspended in serum-free RPMI-1640 prior to staining for flow cytometry.Flow cytometry
Dendritic cells were harvested, washed and stained with either 5-arm polymer-FITC, 8-arm polymer-FITC, a linear polymer-FITC or gp120-FITC (generated from soluble recombinant gp120 produced in HEK cells as previously described) at the concentrations indicated in the results. To assess the ability of the polymers to bind to the gp120 site, DCs were incubated with unlabeled polymer (at the concentrations indicated in the results) for 45 minutes at room temperature and then treated with gp120-FITC (1 μg ml−1) for a further 30 minutes at room temperature. To determine binding of gp120 to CD4+ cells, DCs and lymphocytes were firstly incubated with unlabeled gp120 at 1 μg ml−1 for 45 min at room temperature and then stained with anti-CD4-APC, anti-CD3-Percp, and gp120-FITC for a further 30 minutes. Following all the incubations, cells were washed and fixed with BD stabilizing fixative (BD Biosciences). Unstained DCs were used as a control. One hundred thousand cells were acquired within a live gate using a 3-laser configuration LSRII flow cytometer (BD Biosciences, USA), and analysed by FlowJo (Tree Star Inc, USA).Cytokine assays
Sets of dendritic cell cultures, derived from four separate donors, were incubated with 1 μg ml−1 gp120 and also defined quantities of the three species of glycopolymer (8-arm, 5-arm and linear; at 1, 3 and 10 μg ml−1). After 24 hours, one group of DC cultures was activated with 1 μg ml−1 lipopolysaccharide (LPS; Sigma, Poole, UK) and 20 ng ml−1 interferon gamma (IFN-γ), the other left unstimulated. Supernatants (100 μl) from the assorted cultures were recovered after a further 24 hours via centrifugation at 2000 rpm. Samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in DMEM cell culture medium without supplements and duplicate measurements were made using Luminex bead immunoassay. A panel of nine cytokines was analyzed simultaneously using the commercial Bio-Rad Th1 and Th2 kit within the Bio-plex 200 Multiplex System in accordance with manufacturer instructions (Bio-Rad Laboratories, Hercules, USA). Quantitative readouts were obtained for the following cytokines: tumor necrosis factor alpha (TNF-α), IFN-γ, interleukins (IL)-2, IL-4, IL-5, IL-10, IL-12p70, IL-13, and granulocyte/monocyte-colony stimulating factor (GM-CSF).
1 in DMEM cell culture medium without supplements and duplicate measurements were made using Luminex bead immunoassay. A panel of nine cytokines was analyzed simultaneously using the commercial Bio-Rad Th1 and Th2 kit within the Bio-plex 200 Multiplex System in accordance with manufacturer instructions (Bio-Rad Laboratories, Hercules, USA). Quantitative readouts were obtained for the following cytokines: tumor necrosis factor alpha (TNF-α), IFN-γ, interleukins (IL)-2, IL-4, IL-5, IL-10, IL-12p70, IL-13, and granulocyte/monocyte-colony stimulating factor (GM-CSF).
Research using human material was carried out in accordance with institutional guidelines issued by the University of Warwick and Imperial College London, and with the principles expressed in the Declaration of Helsinki. Ethical approval was obtained from the Riverside Research Ethics Committee and informed written consent was obtained prior to patient blood collection. Data from human-derived material were processed anonymously.
Results and discussion
Carbohydrates in the form of oligosaccharides and polysaccharides represent a large, crucial and exciting family of molecules in the development of novel therapeutics. Uncovering the roles of complex carbohydrates in human physiology and disease coincides with the advancement of our understanding of carbohydrate-binding receptors such as DC-SIGN. Initially identified as a molecule exploited by HIV-1 in order to promote viral trafficking and within-host survival, it has since emerged recently that DC-SIGN is central to important physiological processes such as healthy responses during apoptotic cell clearance and foeto-maternal tolerance.2,18 Studies into the impact of glycan ligands for DC-SIGN on immune cells indicate that this receptor is capable of driving anti-inflammatory and immunosuppressive cell signaling with major effects on gene expression and cellular responses.6,7 Whilst undesirable in the context of certain infections, these responses are beneficial in the active processes of wound healing and the resolution of inflammatory episodes. It is becoming increasingly clear that these tissue repair and protection mechanisms are highly active and require advanced immune system involvement. Of significance is the association of molecules such as DC-SIGN with tissue environments such as placenta, uterus and gut, where tight control of tissue growth & stability and blood supply in the presence of abundant non-self material is essential.11Understanding the structure and function of DC-SIGN allows us to design and prepare advanced synthetic ligands for this molecule and investigate opportunities for exploiting its anti-inflammatory properties. The strategy of generating defined star-shaped glycopolymers (GPs) to achieve enhanced binding to the tetrameric configuration of DC-SIGN has been very successful, showing increases in affinity exceeding an order of magnitude, in addition to demonstrating strong interactions with DCs. We speculate that the star-shaped architecture spans the four CRDs within the DC-SIGN tetramer, increasing the strength of the interaction and potentially influencing the consequences of DC-SIGN engagement. It is assumed that DC-SIGN can only transduce signals when clustered in multimeric complexes with multiple glycan ligands expressed on a large surface area, although small ligands such as Lewis-x oligosaccharides have been shown to drive signaling and gene expression responses in DCs via DC-SIGN engagement. Furthermore, NMR analyses of DC-SIGN–oligosaccharide interactions in solution suggest discrete binding modes of the carbohydrate-recognition domain for distinct glycan types that in turn are thought to drive different cell signaling pathways. Therefore, the consequences of DC-SIGN engagement by glycopolymers could be influenced and even tuned by variations in sugar composition, architecture and size.
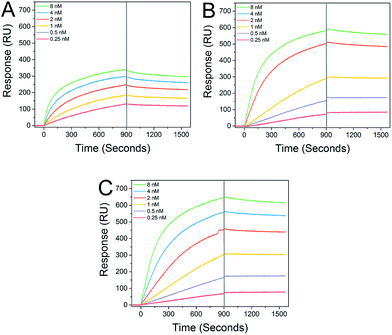
Star-shaped GPs bind to recombinant DC-SIGN with sub-nanomolar affinity. Interaction analysis in real time via surface plasmon resonance (SPR) demonstrated distinct and strong binding between immobilized DC-SIGN and all of the GPs examined (Fig. 2), consistent with previous studies.12 As expected, the star-shaped GPs showed greater affinity compared with the linear GP of equal molecular weight and dissociation rates (koff) were markedly slow, in keeping with strong multivalent interactions with the oligomeric DC-SIGN protein. Apparent KD values of 65.1 pM and 72.5 pM were determined for the star-shaped 5-arm and 8-arm glycopolymers, respectively (Table 1).
| Run | Type | k on (M−1 s−1) | k off (s−1) | K D (M) |
|---|---|---|---|---|
| P1 | Linear | 1.24 × 106 | 1.34 × 10−4 | 1.08 × 10−10 |
| P2 | 5-Arm star | 1.01 × 106 | 7.33 × 10−5 | 7.25 × 10−11 |
| P3 | 8-Arm star | 0.93 × 106 | 6.03 × 10−5 | 6.51 × 10−11 |
Star-shaped glycopolymers show improved and dose-dependent binding to human dendritic cells
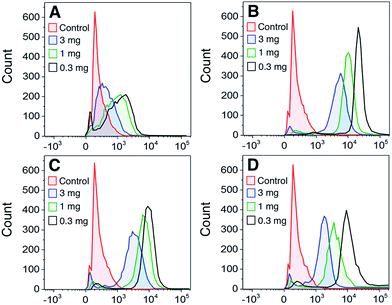
To determine whether the polymers bound directly to dendritic cells (DCs), DCs were stained with FITC-labeled polymers at a range of polymer concentrations. Fig. 3A–C shows that all GPs bound to DCs to some degree with the 5-arm and 8-arm glycopolymers binding better than the linear polymer. We also assessed the binding of gp120 to DCs and found that binding followed a similar profile to the star-shaped glycopolymers (Fig. 3D). In a direct comparison between the polymers and gp120, we found that the 5-arm and 8-arm GPs bound with a similar intensity as compared to the gp120 at the highest dose of 10 μg ml−1 (see ESI†).In addition to assessing the intrinsic binding of GPs to DCs we also determined whether they exerted any measurable influence on the activation status of the treated DCs by analyzing their cell-surface markers. We found that following 24 hours of exposure to the polymers generated here, there was no significant change in DC expression of the canonical activation markers CD80, CD83, CD86 or HLA-DR when examining cells from three separate donors.
There is a likelihood that these mannose glycopolymers also interact with dendritic cell and macrophage subpopulations that express the mannose receptor (CD206), in addition to specialised endothelial cells that express receptors such as DC-SIGNR (CD299). Where the very high affinity interactions between the star-shaped glycopolymers and DC-SIGN could possess significant selectivity would be in discrete tissue spaces such as inflamed synovium associated with rheumatoid arthritis, where salient subpopulations of DC-SIGN-positive macrophages are upregulated.19
Competition of glycopolymers with gp120 for binding sites on dendritic cells due to cellular CD4 expression
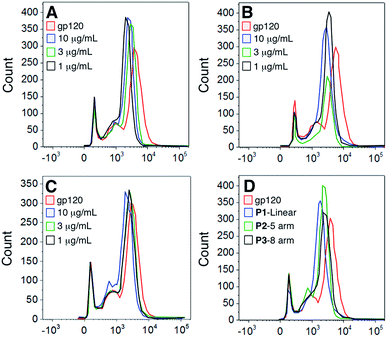
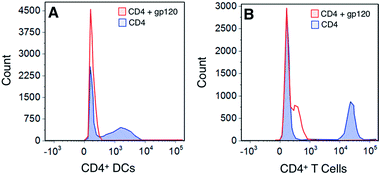
To determine whether the polymers competed with the gp120 for the same cellular binding site, we incubated DCs with unlabeled polymers and then stained the cells with FITC-labeled gp120 (Fig. 4). Our results show that although there was a slight shift in binding when polymer was present, none of the polymers significantly abrogated gp120 binding.To assess whether this was due to the gp120 using more than one binding partner on cell surfaces, we looked at determining the expression of the major gp120 ligand – CD4 – on both DCs and T cells. Our results show that gp120 is able to bind to CD4 molecules on both DCs and T cells – a site to which gp120 binding is not carbohydrate-dependent (Fig. 5). Consequently our results demonstrate that the polymers bind to separate sites on cells from those utilized and possibly preferred by gp120.
The flow cytometry studies involving HIV gp120 interactions with DCs raise a very important issue in the use of glycopolymers as agents for viral blockade. The presence of CD4 on moDCs (and also native peripheral DC populations) serves as a very effective adhesion route for HIV, such that polymer-based barrier preparations could be compromised in vivo. This would be consistent with in vivo findings that DC-SIGN is associated with parenteral but not mucosal trafficking of HIV.20 However, it is yet to be known whether the presence of glycopolymers could affect HIV infection of CD4+ T cells in trans and whether glycopolymers would be effective in blocking DC interactions with other dangerous glycosylated viruses that are known to interact with DC-SIGN such as hepatitis C virus (HCV), Ebola and Dengue.
Glycopolymers modulate cytokine production by activated DCs
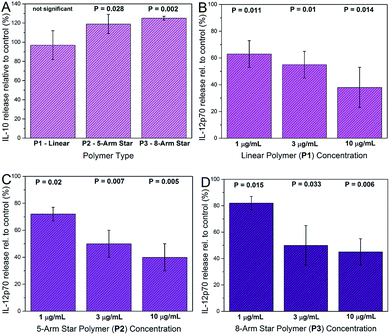
Cytokine analysis was carried out on supernatants recovered from DCs exposed to glycopolymers and consensus activation stimuli (LPS plus IFN-γ). Prior to supernatant harvesting, the DCs appeared healthy and normal in the presence of glycopolymers, suggesting that these materials were tolerated and not markedly toxic at the quantities used (1–10 μg ml−1). Measured cytokine secretion levels were negligible in all of the DC cultures that were not stimulated with LPS and IFN-γ, suggesting that the GPs themselves do not invoke major secreted DC activation responses. In DC cultures activated with LPS and IFN-γ, key cytokines such as TNF-α were up-regulated in all cultures, as expected, and the exposure of DCs to star-shaped GPs led to significant changes in the secretion of two cytokines – IL-10 and IL-12p70. Levels of secreted IL-10 were significantly increased only in supernatants from activated DCs exposed to the 8-arm and 5-arm GPs, and only at the highest concentration used (10 μg ml−1; Fig. 6A). Equivalent quantities of the linear GP did not invoke significant changes in IL-10 secretion. Levels of IL-12p70 were significantly reduced by all three species of GP in a dose dependent manner (Fig. 6B–D). Interestingly, the linear GP showed an effect in reducing IL-12p70 secretion (Fig. 6B). This suggests that whilst showing lower levels of binding to DC-SIGN and to DCs, linear GPs may still influence certain DC responses.The changes in cytokine secretion by DCs invoked by the star-shaped polymers are consistent with those observed in mammalian wound healing. IL-10 is a potent immunosuppressive cytokine with broad effects on inflammation and its mediators.21 IL-12p70 is a pro-inflammatory cytokine and a potent inhibitor of angiogenesis (blood vessel growth).22 The combination of IL-10 increase and IL-12 decrease represents a rapid and effective means of reducing inflammation and restoring organized blood flow to a site of tissue injury. In general, qualitative changes in secretion of these two cytokines are in keeping with the consequences of DC-SIGN engagement by biologically-sourced mannosylated ligands. However, a key difference is that in the case of the glycopolymers, the chemical definition is very precise. In contrast, biological high mannose ligands such as gp120, yeast mannan and mycobacterial lipoarabinomannan are notoriously heterogeneous.23–25 Furthermore, the low cost of polymer precursors and the simple synthesis workflow make the glycopolymers described here an attractive and affordable means of generating abundant, water-soluble material with advanced immunological activity.
Conclusions
To summarize, we report the synthesis and characterization of highly water-soluble advanced glycopolymer sets of defined size and shape designed to interact profoundly with the key human lectin DC-SIGN. Biophysical studies demonstrate remarkable glycopolymer-DC-SIGN interactions with affinities in the picomolar range and positive binding to human monocyte-derived dendritic cells. Star-shaped glycopolymers invoke significant changes in the production of two key cytokines, IL-10 and IL-12p70, when incubated with dendritic cells at pharmacologically realistic concentrations. Importantly, IL-10 levels rise and IL-12 levels fall in response to the glycopolymers and this cytokine release pattern is reminiscent of tissue environments associated with active wound healing, anti-inflammation, and also healthy pregnancy at the feto-maternal interface. In our experiments, however, the glycopolymers do not affect the ability of HIV gp120 to interact with monocyte-derived dendritic cells – previously a major direction for both DC-SIGN and glycopolymer research. We propose novel roles for glycopolymers of this kind, shifting the focus away from HIV prophylaxis towards the treatment of conditions that require immune modulation that promotes wound healing and inflammatory resolution. Examples of such conditions could include burns & chronic skin lesions, in addition to chronic inflammatory diseases with immunological involvement such as rheumatoid arthritis, multiple sclerosis and Crohn's disease.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the European Commission (EU-ITN EuroSequences Proposal No: 642083 to C. R. B.) and EPSRC (EP/P009018/1). ASG is supported by a Studentship Award from Coventry General Charities, UK (Registered Charity Number 216235).Notes and references
- Y. van Kooyk, A. Engering, A. N. Lekkerkerker, I. S. Ludwig and T. B. Geijtenbeek, Curr. Opin. Immunol., 2004, 16, 488–493 CrossRef CAS PubMed.
- S. K. Pathak, A. E. Skold, V. Mohanram, C. Persson, U. Johansson and A. L. Spetz, J. Biol. Chem., 2012, 287, 13731–13742 CrossRef CAS PubMed.
- M. E. Taylor and K. Drickamer, Methods Enzymol., 2003, 363, 3–16 CAS.
- T. B. Geijtenbeek, D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor and Y. van Kooyk, Cell, 2000, 100, 587–597 CrossRef CAS PubMed.
- T. B. Geijtenbeek, S. J. Van Vliet, E. A. Koppel, M. Sanchez-Hernandez, C. M. Vandenbroucke-Grauls, B. Appelmelk and Y. Van Kooyk, J. Exp. Med., 2003, 197, 7–17 CrossRef CAS PubMed.
- S. I. Gringhuis, J. den Dunnen, M. Litjens, M. van der Vlist and T. B. Geijtenbeek, Nat. Immunol., 2009, 10, 1081–1088 CrossRef CAS PubMed.
- S. I. Gringhuis, J. den Dunnen, M. Litjens, B. van Het Hof, Y. van Kooyk and T. B. Geijtenbeek, Immunity, 2007, 26, 605–616 CrossRef CAS PubMed.
- R. M. Steinman, Cell, 2000, 100, 491–494 CrossRef CAS PubMed.
- K. Pederson, D. A. Mitchell and J. H. Prestegard, Biochemistry, 2014, 53, 5700–5709 CrossRef CAS PubMed.
- E. J. Soilleux, L. S. Morris, B. Lee, S. Pohlmann, J. Trowsdale, R. W. Doms and N. Coleman, J. Pathol., 2001, 195, 586–592 CrossRef CAS PubMed.
- E. J. Soilleux, L. S. Morris, G. Leslie, J. Chehimi, Q. Luo, E. Levroney, J. Trowsdale, L. J. Montaner, R. W. Doms, D. Weissman, N. Coleman and B. Lee, J. Leukocyte Biol., 2002, 71, 445–457 CAS.
- C. R. Becer, M. I. Gibson, J. Geng, R. Ilyas, R. Wallis, D. A. Mitchell and D. M. Haddleton, J. Am. Chem. Soc., 2010, 132, 15130–15132 CrossRef CAS PubMed.
- J. Huang, Q. Zhang, G. Z. Li, D. M. Haddleton, R. Wallis, D. Mitchell, A. Heise and C. R. Becer, Macromol. Rapid Commun., 2013, 34, 1542–1546 CrossRef CAS PubMed.
- Q. Zhang, J. Collins, A. Anastasaki, R. Wallis, D. A. Mitchell, C. R. Becer and D. M. Haddleton, Angew. Chem., 2013, 52, 4435–4439 CrossRef CAS PubMed.
- Q. Zhang, L. Su, J. Collins, G. Chen, R. Wallis, D. A. Mitchell, D. M. Haddleton and C. R. Becer, J. Am. Chem. Soc., 2014, 136, 4325–4332 CrossRef CAS PubMed.
- D. A. Mitchell, A. J. Fadden and K. Drickamer, J. Biol. Chem., 2001, 276, 28939–28945 CrossRef CAS PubMed.
- H. Feinberg, Y. Guo, D. A. Mitchell, K. Drickamer and W. I. Weis, J. Biol. Chem., 2005, 280, 1327–1335 CrossRef CAS PubMed.
- P. Hsu, B. Santner-Nanan, J. E. Dahlstrom, M. Fadia, A. Chandra, M. Peek and R. Nanan, Am. J. Pathol., 2012, 181, 2149–2160 CrossRef CAS PubMed.
- T. Sokka, E. Krishnan, A. Hakkinen and P. Hannonen, Arthritis Rheum., 2003, 48, 59–63 CrossRef PubMed.
- M. P. Martin, M. M. Lederman, H. B. Hutcheson, J. J. Goedert, G. W. Nelson, Y. van Kooyk, R. Detels, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien and M. Carrington, J. Virol., 2004, 78, 14053–14056 CrossRef CAS PubMed.
- A. King, S. Balaji, L. D. Le, T. M. Crombleholme and S. G. Keswani, Adv. Wound Care, 2014, 3, 315–323 CrossRef PubMed.
- A. Naldini and F. Carraro, Curr. Drug Targets: Inflammation Allergy, 2005, 4, 3–8 CrossRef CAS PubMed.
- T. Mizuochi, M. W. Spellman, M. Larkin, J. Solomon, L. J. Basa and T. Feizi, Biochem. J., 1988, 254, 599–603 CrossRef CAS PubMed.
- C. E. Ballou, Adv. Enzymol. Relat. Areas Mol. Biol., 1974, 40, 239–270 CAS.
- D. Chatterjee and K. H. Khoo, Glycobiology, 1998, 8, 113–120 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc01515a |
| This journal is © The Royal Society of Chemistry 2017 |