Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multi-target-directed phenol–triazole ligands as therapeutic agents for Alzheimer's disease†
Michael R.
Jones
ab,
Emilie
Mathieu
a,
Christine
Dyrager
 a,
Simon
Faissner
bc,
Zavier
Vaillancourt
a,
Kyle J.
Korshavn
d,
Mi Hee
Lim
a,
Simon
Faissner
bc,
Zavier
Vaillancourt
a,
Kyle J.
Korshavn
d,
Mi Hee
Lim
 e,
Ayyalusamy
Ramamoorthy
e,
Ayyalusamy
Ramamoorthy
 df,
V.
Wee Yong
b,
Shigeki
Tsutsui
b,
Peter K.
Stys
b and
Tim
Storr
df,
V.
Wee Yong
b,
Shigeki
Tsutsui
b,
Peter K.
Stys
b and
Tim
Storr
 *a
*a
aDepartment of Chemistry, Simon Fraser University, V5A1S6, Burnaby, BC, Canada. E-mail: tim_storr@sfu.ca
bDepartment of Clinical Neurosciences, Hotchkiss Brain Institute, Cumming School of Medicine, University of Calgary, Calgary, Canada
cDepartment of Neurology, St. Josef-Hospital, Ruhr-University, Bochum, Germany
dDepartment of Chemistry, University of Michigan, Ann Arbor, USA
eDepartment of Chemistry, Ulsan National Institute of Science and Technology (UNIST), Ulsan, Korea
fDepartment of Biophysics, University of Michigan, Ann Arbor, USA
First published on 5th June 2017
Abstract
Alzheimer's disease (AD) is a multifactorial disease that is characterized by the formation of intracellular neurofibrillary tangles and extracellular amyloid-β (Aβ) plaque deposits. Increased oxidative stress, metal ion dysregulation, and the formation of toxic Aβ peptide oligomers are all considered to contribute to the etiology of AD. In this work we have developed a series of ligands that are multi-target-directed in order to address several disease properties. 2-(1-(3-Hydroxypropyl)-1H-1,2,3-triazol-4-yl)phenol (POH), 2-(1-(2-morpholinoethyl)-1H-1,2,3-triazol-4-yl)phenol (PMorph), and 2-(1-(2-thiomorpholinoethyl)-1H-1,2,3-triazol-4-yl)phenol (PTMorph) have been synthesized and screened for their antioxidant capacity, Cu-binding affinity, interaction with the Aβ peptide and modulation of Aβ peptide aggregation, and the ability to limit Aβ1–42-induced neurotoxicity in human neuronal culture. The synthetic protocol and structural variance incorporated via click chemistry, highlights the influence of R-group modification on ligand-Aβ interactions and neuroprotective effects. Overall, this study demonstrates that the phenol–triazole ligand scaffold can target multiple factors associated with AD, thus warranting further therapeutic development.
Introduction
Healthcare advances across the globe have increased life expectancy, facilitating increases in the prevalence of neurodegenerative diseases such as Alzheimer's disease (AD).1,2 In Canada for example, ca. 5% of the population over 65 years of age have AD, increasing to 25% over the age of 85 years old.3 Currently, there are no approved disease-modifying therapeutic interventions other than symptomatic treatments such as cholinesterase inhibitors.4AD is formally characterized by two distinct neuropathological features: extracellular amyloid-β (Aβ) plaques and intracellular neurofibrillary tangles (NFTs).5,6 The major constituent of Aβ plaques is the Aβ peptide of ca. 38–43 amino acid residues, with Aβ1–40 and Aβ1–42 being the most abundant forms. NFTs result from the hyperphosphorylation of tau proteins. Oxidative stress is linked to the formation of both of these pathological features,7 and recent reports suggest an inter-relationship between Aβ and NFTs.8
AD is a multifactorial disease where oxidative stress, altered Aβ clearance mechanisms, Aβ and tau aggregation, and dysregulated metal ions play a role in disease etiology.9 The N-terminus of the Aβ peptide exhibits a relatively high affinity for Cu (Kd = 10−11 to 10−7) and Zn (Kd = 10−9 to 10−6) where residues His6, His13, and His14 are located.10–13 The concentration of metal ions in Aβ plaque deposits is 3–5 fold higher in comparison to age-matched healthy parenchyma, suggesting that Aβ plaques act as metal reservoirs, as overall metal concentrations in the brain are not altered.14 The interaction between metal ions and the Aβ peptide drastically alters the Aβ aggregation profile; interaction with Zn affords amorphous high molecular weight species, while interaction with Cu affords neurotoxic oligomers.15 In the case of CuAβ, peptide binding promotes CuII/CuI redox cycling, and generation of reactive oxygen species (ROS), potentially implicating these metalated species in oxidative stress, and neuronal toxicity.16–18
Small molecule chemical agents that can address multiple factors associated with AD etiology may play a key role in the development of new effective therapeutic strategies. Specifically, developing multifunctional agents that can act on multiple disease pathways could provide key information on the interrelationship between these pathways, enhancing our overall understanding of AD etiology. The rational design of multifunctional metal binding agents has become a promising therapeutic strategy.19–27 Previously, we developed several multifunctional frameworks based on pyridine–triazole and quinoline–triazole scaffolds (Fig. 1). The pyridine–triazole frameworks were shown to alter metal–Aβ interactions and associated Aβ aggregation.28 The quinoline–triazole analogues were developed to provide an enhanced interaction with the hydrophobic region of the Aβ peptide.29 In a significant advance, we report herein a series of multi-target directed ligands possessing a phenol–triazole framework (Fig. 1). These ligands exhibit an enhanced affinity for Cu in comparison to the pyridine/quinoline analogues, and in addition, contain an antioxidant phenolic group. The phenol–triazoles were shown to modulate the Aβ aggregation profile in the presence and absence of added Cu based on gel electrophoresis and transmission electron microscopy. 2-D NMR and molecular modelling studies were employed to provide further insight into the ligand–peptide interaction. Finally, one compound in the series (POH) was shown to reverse Aβ1–42 neurotoxicity in primary neurons.
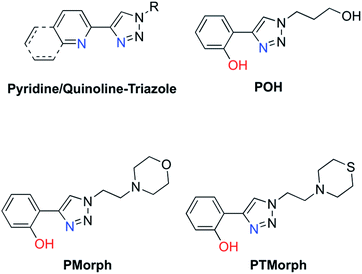 | ||
| Fig. 1 Chemical structures of relevant triazole-containing ligands including POH, PMorph, and PTMorph. | ||
Results and discussion
Ligand properties
AD drug development has targeted several different pathophysiological pathways, including Aβ peptide aggregation, tau protein hyperphosporylation, oxidative stress, cholinesterase inhibition, and metal ion dyshomeostasis.30 The lack of a single defined target suggests that an effective therapeutic intervention may require the ability to address several different factors associated with the disease. Herein, a series of phenol–triazoles (POH, PMorph and PTMorph, Fig. 1) were designed to bind Cu, modulate metal–peptide interactions, and limit the generation of reactive oxygen species (ROS) via the phenol moiety. The phenol–triazole series were synthesized in a modular fashion using Huisgen's 1,3-dipolar cycloaddition, also known as click chemistry.31,32 Reaction of an alkyne substituted phenol and azides with different peripheral R-groups affords a bidentate metal-binding site (Scheme S1†).We next determined the ligand acidity constants via a combination of variable pH UV-Vis and 1H NMR spectroscopies (Fig. S1–S3†). The phenol pKa values were thus determined to be within the range of 9.54–9.55 (Table S1†), comparable to the deprotonation of free phenol (pKa = 9.98 (ref. 33)). The additional pKa values for the peripheral morpholine (PMorph) and thiomorpholine (PTMorph) functions were determined to be 5.5 and 5.6, respectively. These values are lower than free morpholine and thiomorpholine (pKa morpholine = 8.36, thiomorpholine = 9.0) as has been previously reported in similar systems.29,34–36 Overall, each ligand was found to be neutral at physiological pH 7.4, which is optimal for passive diffusion across the BBB. To further investigate the drug-like properties of the phenol–triazoles, several physicochemical parameters were calculated. Each phenol–triazole ligand was determined to comply with Lipinski's rules and log BB for drug-likeness and BBB penetration (Table S2†).37,38 Overall, the phenol–triazole series exhibit promising physicochemical properties warranting further development.
Metal binding affinity
We next investigated the Cu-binding affinity of the bidentate phenol–triazole ligands in solution using variable pH UV-vis titrations. The Aβ peptide exhibits a high affinity for Cu(II) (Kdca. 10−11 to 10−7)12,13 and thus in order to compete with Aβ for Cu, either via de-metallation or ternary complex formation, ligands should exhibit a Cu Kd value in the 10−12 to 10−8 range.12 This range is appropriate to disrupt Cu–Aβ interactions, while limiting sequestration of essential metal ions from metalloproteins.39The stoichiometry of the Cu(II) – phenol–triazole complexes in PBS (pH 7.4) was initially determined using Jobs plot analysis (Fig. S4†).40 For the example ligand PMorph, a broad maxima is observed between ca. 0.3–0.6 mole fraction of Cu(II), suggesting the formation of both 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 PMorph–Cu(II) complexes in solution (vide infra).41,42 Further, both 1
2 PMorph–Cu(II) complexes in solution (vide infra).41,42 Further, both 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 binding stoichiometries were observed for the three phenol–triazoles by both Jobs plot and ESI-MS analysis (data not shown). Measurement of the binding affinity of the phenol–triazole ligands for Cu(II) was completed using variable pH spectrophotometric titrations and speciation modelling using HypSpec (Fig. 2).43 The ligand pKa values, as well as the hydrolysis reactions of free Cu(II), were included as constants in the calculations.44 Modelling of the data for POH shows significant log
2 binding stoichiometries were observed for the three phenol–triazoles by both Jobs plot and ESI-MS analysis (data not shown). Measurement of the binding affinity of the phenol–triazole ligands for Cu(II) was completed using variable pH spectrophotometric titrations and speciation modelling using HypSpec (Fig. 2).43 The ligand pKa values, as well as the hydrolysis reactions of free Cu(II), were included as constants in the calculations.44 Modelling of the data for POH shows significant log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K values for both 1
K values for both 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 Cu
2 Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L ratios (where L is deprotonated) as shown in Table 1. As expected, the calculated log
L ratios (where L is deprotonated) as shown in Table 1. As expected, the calculated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K values are similar across the phenol–triazole series (Table 1), reflecting a common metal-binding motif. A representative speciation diagram for POH (Fig. 2) shows that at physiological pH 1
K values are similar across the phenol–triazole series (Table 1), reflecting a common metal-binding motif. A representative speciation diagram for POH (Fig. 2) shows that at physiological pH 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 Cu
2 Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L species predominate (in agreement with Jobs plot analysis) with negligible free Cu. At higher pH, the [CuL2(OH)]− species becomes relevant. Similar speciation diagrams for PMorph and PTMorph are shown in the ESI (Fig. S5 and S6†). Using the solution speciation diagrams, the concentration of unchelated Cu(II) (pCu = −log([Cuunchelated])) at pH 7.4 and total Cu concentration can be calculated (Table 1). The pCu value is a direct estimate of the ligand–Cu affinity by taking into account all relevant equilibria, and can therefore be used to compare the metal-binding affinity among the various ligands, including the Aβ peptide. The pCu values are similar across the phenol–triazole series (6.6–6.9), and represent approximate dissociation constants (nM to μM range) that compare favourably with the Kd values reported for Cu–Aβ species.12,13 The Cu-binding affinity for the phenol–triazole ligands are thus significantly stronger than those reported for the quinoline–triazole analogues.29 A Cu-competition assay was then performed in which 2 eq. of each phenol–triazole ligand was added to pre-formed Cu–Aβ1–16 and Cu–Aβ1–42 species. The Cu complex of the phenol–triazole ligands displays an absorption peak at ca. 320 nm (Fig. S7–S9†), and upon addition of ligand to solutions containing Cu–Aβ1–42/1–16, an increase at 320 nm is observed. This data suggests an interaction of the phenol–triazole ligands with Cu. A baseline increase in the Aβ1–42 experiments is likely due to aggregate formation and associated light scattering. On the basis of this data we expect the phenol–triazole ligands to have the appropriate Cu-binding affinity to interact with Cu in the presence of the Aβ peptide.
L species predominate (in agreement with Jobs plot analysis) with negligible free Cu. At higher pH, the [CuL2(OH)]− species becomes relevant. Similar speciation diagrams for PMorph and PTMorph are shown in the ESI (Fig. S5 and S6†). Using the solution speciation diagrams, the concentration of unchelated Cu(II) (pCu = −log([Cuunchelated])) at pH 7.4 and total Cu concentration can be calculated (Table 1). The pCu value is a direct estimate of the ligand–Cu affinity by taking into account all relevant equilibria, and can therefore be used to compare the metal-binding affinity among the various ligands, including the Aβ peptide. The pCu values are similar across the phenol–triazole series (6.6–6.9), and represent approximate dissociation constants (nM to μM range) that compare favourably with the Kd values reported for Cu–Aβ species.12,13 The Cu-binding affinity for the phenol–triazole ligands are thus significantly stronger than those reported for the quinoline–triazole analogues.29 A Cu-competition assay was then performed in which 2 eq. of each phenol–triazole ligand was added to pre-formed Cu–Aβ1–16 and Cu–Aβ1–42 species. The Cu complex of the phenol–triazole ligands displays an absorption peak at ca. 320 nm (Fig. S7–S9†), and upon addition of ligand to solutions containing Cu–Aβ1–42/1–16, an increase at 320 nm is observed. This data suggests an interaction of the phenol–triazole ligands with Cu. A baseline increase in the Aβ1–42 experiments is likely due to aggregate formation and associated light scattering. On the basis of this data we expect the phenol–triazole ligands to have the appropriate Cu-binding affinity to interact with Cu in the presence of the Aβ peptide.
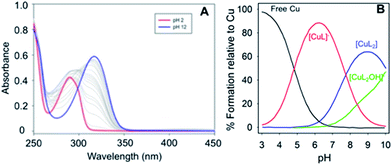 | ||
Fig. 2 (Left) Variable pH UV-vis titration of POH (75 μM) and CuCl2 (37.5 μM) where the red spectrum represents pH 2 and the blue spectrum at pH 12. (Right) Using HypSpec and HySS,43,45 the variable pH data were fit to a model including a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2 1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ligand 1 ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu species along with free Cu and a Cu(PMorph)2OH component at high pH. At physiological pH 7.4, very little free Cu is present and a combination of 1 Cu species along with free Cu and a Cu(PMorph)2OH component at high pH. At physiological pH 7.4, very little free Cu is present and a combination of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2 1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ligand 1 ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu species are present. Cu species are present. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K's) of each ligand with Cu2+
K's) of each ligand with Cu2+
| Reaction | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K |
||
|---|---|---|---|
| POH | PMorph | PTMorph | |
| a pCu was calculated using pCu = (−log[Cu2+]free), where [Cu2+]free is determined from the HySS model.45 | |||
| Cu2+ + L− = [CuL]+ | 8.97(2) | 8.90(3) | 8.77(7) |
| Cu2+ + 2L− = [CuL2] | 15.46(3) | 15.65(2) | 15.16(9) |
| [CuL2(H2O)] = [CuL2(OH)]− + H+ | −14.36(3) | −13.98(9) | −12.96(2) |
| pCua | 6.9 | 6.8 | 6.6 |
Anti-oxidant assays
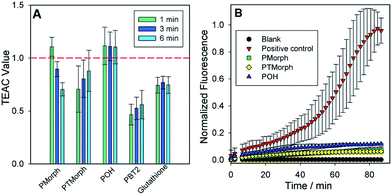
To further evaluate the multifunctional nature of the phenol–triazoles, we investigated their antioxidant capability using several different assays.46–48 In the first assay, the antioxidant activity of the phenolic functions were evaluated using a Trolox-Equivalent Antioxidant Capacity (TEAC) assay. The TEAC assay has been used to quantify the antioxidant activity of biological fluids, extracts, and pure compounds by measuring the disappearance of the ABTS+˙ radical cation via UV-vis spectroscopy.49 Trolox, a water-soluble vitamin-E analogue, is used as a standard against which each ligand is compared. In addition, each phenol–triazole ligand was compared with glutathione and PBT2, an 8-hydroxyquinoline derivative, that has shown promise as an Alzheimer's disease therapeutic.50,51 Each ligand exhibited TEAC values comparable to Trolox and slightly enhanced in comparison to PBT2 (Fig. 3).In two other complimentary antioxidant assays, we measured the ability of the phenol–triazole ligands to bind Cu and limit hydroxyl radical formation from aqueous Cu in the presence of dioxygen. Firstly, using a fluorescent coumarin carboxylic acid (CCA) assay, in which the highly reactive hydroxyl radical specifically hydroxylates CCA at the 7-position to form a fluorescent product,52,53 aerobic aqueous Cu solutions in the presence of a physiologically-relevant concentration of ascorbate54 exhibit significant fluorescence over a short period (Fig. 3). The addition of 2 eq. POH, PMorph, or PTMorph to the Cu/ascorbate/CCA solutions significantly reduces the observed fluorescence (Fig. 3), consistent with either the high affinity of these ligands for Cu (vide supra), or direct reaction of the ligands with the generated ˙OH. A complimentary ascorbate (Asc) reduction assay55,56 was employed to discern if Cu chelation was responsible for the observed response in the CCA experiments. In the absence of ligands, Asc consumption in the presence of Cu is rapid (Fig. S10†), however, upon addition of 3 eq. of the high affinity chelator DTPA, Asc consumption is arrested. We observed only a small decrease in Asc consumption upon addition of 10 eq. of phenol–triazole ligands to a Cu/Asc system (Fig. S10†), signifying that the compounds act primarily as antioxidants, and not via Cu redox silencing, in the CCA assay. Ascorbate consumption was also evaluated in the presence of Aβ1–16, which contains the metal binding region for Cu (Fig. S11†). When comparing Cu–Aβ1–16vs. Cu–Aβ1–16 + 10 eq. of phenol–triazole ligand, no change in the rate of ascorbate consumption was observed. This suggests that the ligands do not limit Cu2+/+ redox cycling and therefore, act primarily as antioxidants, reacting with Cu-generated ROS.
Ligand–Aβ peptide interactions
The direct interaction of the phenol–triazole ligands with monomeric Aβ1–40 was investigated using 2-D 1H–15N SOFAST-HMQC NMR experiments.29,57 The less aggregation prone Aβ1–40 peptide length was used here to limit aggregation during data collection and ensure all shifts are solely the result of peptide–ligand interactions and not peptide aggregation. Incubation with one of POH, PMorph, or PTMorph resulted in small but detectable chemical shift changes for specific peptide residues (see ESI for experimental details†).Interestingly, POH was determined to exhibit significant chemical shift changes distributed across the entire peptide length, including Aβ residues E3, D7, Y10, V18, F20, G33, V36, and G38 (Fig. 4). In the case of PMorph, significant chemical shift changes were observed for Aβ residues D7, F19, D23, and N27 (Fig. S12†). The hydrophobic region of the Aβ peptide, encompassing residues 17–21, and in particular F19, is considered to play a critical role in the initial stages of peptide aggregation, and thus the interaction of PMorph with F19 may play a significant role in mediating the Aβ aggregation process (vide infra).58–63PTMorph demonstrates similar interactions to those observed with PMorph, including significant chemical shift changes of Aβ residues D7, V18, F19, N27, and G33 (Fig. S13†). The similar interaction of PMorph and PTMorph with Aβ is not surprising as the only structural difference is the O for S heteroatom substitution.
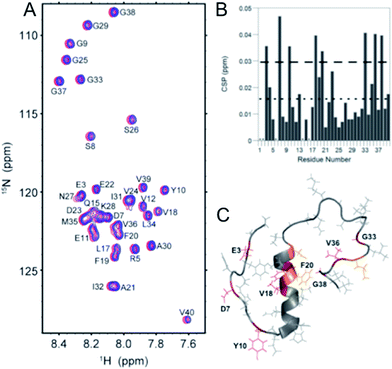 | ||
| Fig. 4 2-D 1H–15N SOFAST NMR experiments using 15N-labeled Aβ1–40 and 0–10 eq. POH. (A) 2-D 1H–15N SOFAST NMR spectra showing the assignment of specific amino acid residues in the Aβ1–40 peptide. (B) Summary of the specific amino acid residues that have shifted at 10 eq. POH. The dotted line represents the average CSP while the dashed line is the average + one standard deviation, which was used to identify statistically relevant Chemical Shift Perturbations (CSP). (C) Aβ1–40 solution NMR structure (PDB: 2LFM) highlighting specific amino acid residues that have significant CSP shifts (Red, >0.03 ppm shift) and moderate CSP shifts (orange, between 0.02 and 0.03 ppm). | ||
Although limited to three ligands, the 2-D NMR results suggest that the triazole R-group plays a significant role in dictating Aβ peptide interactions (Fig. S14†), with the propanol group of POH affording non-specific interactions, while the morpholine and thiomorpholine heterocycles confer a higher degree of selectivity for Aβ peptide residues. The direct interaction of the phenol–triazoles with the Aβ peptide, as supported by the 2-D NMR studies, suggest that these ligands may modulate the Aβ aggregation profile in solution, even in the absence of Cu (vide infra).
Aβ peptide aggregation experiments
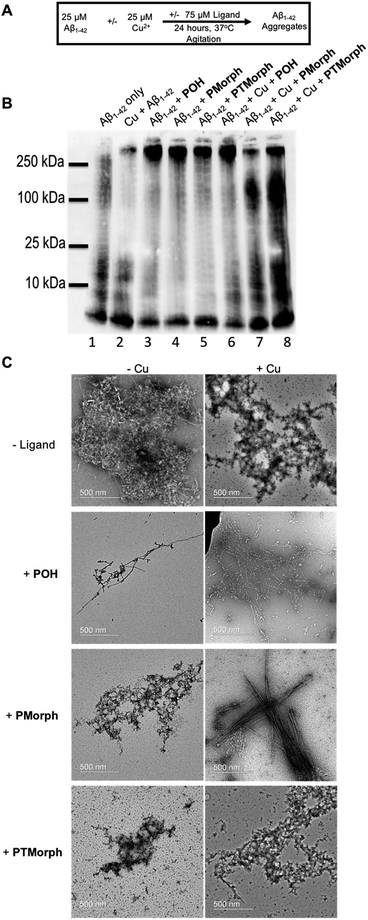
Modulation of the Aβ peptide aggregation pathway offers a significant opportunity for drug development. Shifting the Aβ aggregation pathway away from toxic intermediates, such as soluble oligomeric species,64–68 may lead to a decrease in neuronal cell death.To further explore the multifunctional nature of the phenol–triazole ligands we investigated the effects of these ligands on Aβ peptide aggregation in the presence and absence of Cu ions (Fig. 5A). Using native gel electrophoresis/western blotting to visualize the size distribution of Aβ species, in conjunction with transmission electron microscopy (TEM) to examine Aβ aggregate morphology, we assessed the ability of the phenol–triazole ligands to modulate Aβ peptide aggregation.
Incubation of the phenol–triazole ligands with the Aβ peptide results in significant changes to the size distribution of peptide aggregates (Fig. 5B), highlighting the importance of the interactions observed in the 2-D NMR experiments. Incubation of Aβ1–42 alone over 24 h (lane 1) affords both high and low molecular weight species, and significant fibril formation as observed by TEM (Fig. 5C), in line with previous reports.42,69,70 Addition of the phenol–triazole ligands significantly alters the aggregation profile towards high molecular weight species (Fig. 5B, lanes, 3, 4 and 5), and in addition the aggregate morphology is now amorphous as ascertained by TEM (Fig. 5C). These results show that the presence of the phenol–triazole ligands limits the formation of oligomeric species (10–100 kDa range), which have been implicated to play a major role in Aβ-associated toxicity.71,72
We next investigated the effect of the phenol–triazole ligands on Aβ peptide aggregation in the presence of Cu ions. The metal competition assays (Fig. S7–S9†) suggest that the phenol–triazole ligands only partially demetallate Cu–Aβ and thus we investigated the effect of the ligands on aggregation of metalated peptide species. Incubation of Aβ1–42 with 1 eq. of Cu affords primarily oligomeric species in comparison to Aβ1–42 alone at 24 h (Fig. 5B, lane 1 vs. 2).15 In addition, aggregate morphology has changed from fibrillar to amorphous (Fig. 5C). Incubation of Cu–Aβ peptide solutions with the phenol–triazole ligands leads to significant changes in the aggregation profile (Fig. 5B, lane 2, vs. lanes 6, 7, and 8). For POH, a significant shift towards high molecular weight species is observed, with a similar soluble aggregate profile to that observed in the peptide only experiment (Fig. 5B, lane 3 vs. lane 6). Interestingly, TEM analysis shows primarily fibrillar aggregates for Cu–Aβ in the presence of POH (Fig. 5C). While this data is consistent with Cu sequestration by the ligand and aggregation of the peptide,73 it is likely that ligand–peptide interactions are the dominant factor that controls the aggregation process of both Aβ1–42 and Cu–Aβ1–42.
For PMorph and PTMorph, similar Aβ-aggregation profiles are observed in the presence of Cu (Fig. 5B, lanes 7 and 8), which differ significantly from the Cu–Aβ experiment (Fig. 5B, lane 2), and also from the POH experiment (Fig. 5, lane 6). Interestingly, PMorph and PTMorph shift Aβ aggregation towards distinctly different high molecular weight species in comparison to POH (ca. 100 kDa vs. 250 kDa). TEM analysis of the Cu–Aβ aggregation experiment in the presence of PMorph shows the formation of fibrils (Fig. 5C), while for PTMorph both fibrils and amorphous aggregates are observed (Fig. 5C). The three phenol–triazole ligands exhibit similar Cu affinity constants and thus the different aggregation profiles observed for the ligands in the presence of Cu–Aβ may result from distinct interactions of the triazole R-groups with the Aβ peptide, leading to ternary complex formation,19,74,75 and/or interactions with specific aggregates, such as oligomeric Aβ.
Attenuation of Aβ-induced neurotoxicity in human neuronal culture
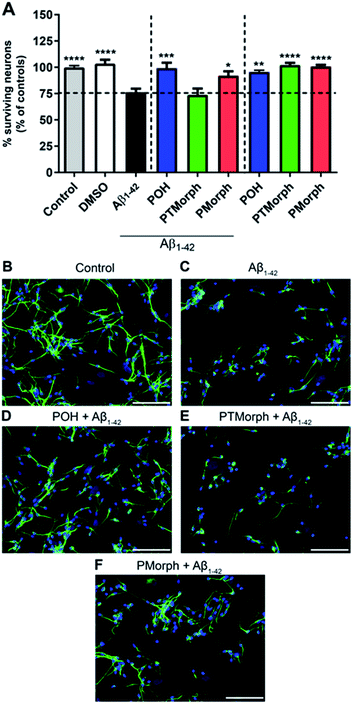
To investigate whether the phenol–triazole ligands protect against Aβ1–42-mediated neurotoxicity, human fetal neurons (HFN) were treated with POH, PTMorph and PMorph for 1 hour prior to Aβ1–42 application. Treatment with monomeric Aβ1–42 induced significant cell death (Fig. 6, 24% fewer MAP-2 positive neurons compared to the control condition, p < 0.0001). Upon treatment with 75 μM POH, complete prevention of Aβ1–42-induced neurotoxicity was achieved (Fig. 6, p < 0.001). PMorph was also able to protect against Aβ1–42-induced neurotoxicity (p < 0.05), while PTMorph demonstrated no protective effect in comparison to Aβ1–42. When 75 μM of ligand was incubated with HFN for 24 hours in the absence of any Aβ1–42, similar cell viability was observed in comparison to the control, indicating that these ligands are not toxic at the experimental concentration used.We next assessed the neurotoxicity of Aβ1–42 premixed with Cu (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 5, 10 and 25 μM (Fig. S15†). While Cu alone was not toxic at these concentrations, Cu–Aβ1–42 exhibited significant toxicity at all concentrations studied. Pre-incubation of the HFN with the phenol–triazole ligands before addition of Cu–Aβ1–42 did not have a neuroprotective effect at the 24 h timepoint (Fig. S15†). Overall, these results are consistent with the previously reported toxicity of Cu–Aβ1–42 species,15,76,77 and show that under the HFN assay conditions the phenol–triazole ligands cannot prevent Cu–Aβ1–42 toxicity. While not effective in limiting the toxicity of Cu–Aβ1–42 in neurons, the phenol–triazole ligands are effective under metal-free conditions. Overall, POH demonstrates the best neuroprotective properties when in the presence of Aβ1–42, conferring complete protection, followed by PMorph, with PTMorph being ineffective in this assay.
1) at 5, 10 and 25 μM (Fig. S15†). While Cu alone was not toxic at these concentrations, Cu–Aβ1–42 exhibited significant toxicity at all concentrations studied. Pre-incubation of the HFN with the phenol–triazole ligands before addition of Cu–Aβ1–42 did not have a neuroprotective effect at the 24 h timepoint (Fig. S15†). Overall, these results are consistent with the previously reported toxicity of Cu–Aβ1–42 species,15,76,77 and show that under the HFN assay conditions the phenol–triazole ligands cannot prevent Cu–Aβ1–42 toxicity. While not effective in limiting the toxicity of Cu–Aβ1–42 in neurons, the phenol–triazole ligands are effective under metal-free conditions. Overall, POH demonstrates the best neuroprotective properties when in the presence of Aβ1–42, conferring complete protection, followed by PMorph, with PTMorph being ineffective in this assay.
Summary
In this report, we describe the development of a series of phenol–triazole ligands that target multiple factors associated with AD etiology. The modular synthetic strategy afforded ligands that exhibit favourable physicochemical properties via both experiments (pKa values) and calculations (drug-likeness), while also displaying antioxidant activity. The three phenol–triazole ligands were shown to interact with the Aβ peptide via 2-D NMR studies, with the PMorph and PTMorph derivatives displaying similar interactions with specific peptide residues, including F19. Interestingly, POH displays a larger number of interactions distributed across the length of the Aβ peptide. The three ligands alter the Aβ peptide aggregation profile in a similar manner via limiting oligomer formation in favour of amorphous high molecular weight species. The Cu-binding affinity of the phenol–triazoles was determined to be of appropriate strength to compete with the Aβ peptide, and as expected the ligands altered the Cu–Aβ aggregation profile. In the presence of Cu, only POH significantly reduced Aβ oligomer formation promoting the formation of large molecular weight aggregates. These results highlight that the triazole R-group offers a significant opportunity to tune the biological properties of the ligand scaffold. Encouragingly, POH, and to a lesser extent PMorph, exhibited a protective effect against Aβ1–42-induced neurotoxicity in human neuronal culture, while these ligands were not able to rescue the neurotoxicity associated with Cu–Aβ1–42 under our conditions. Altogether, these promising results strongly suggest that the multifunctional phenol–triazole ligand scaffold, and in particular POH, warrants further investigation in an animal model to interrogate mechanisms of action and efficacy.Acknowledgements
This work was supported by a Natural Sciences and Engineering Research Council (NSERC) Discovery Grant, a Michael Smith Career Investigator Award, and a New Investigator Grant from the Alzheimer's Association (NIRG-15-362537) and Brain Canada (to T. S.); the Alzheimer's Society of Canada for a Biomedical Doctoral Scholarship and the Alberta Prion Research Institute (APRI) and Alzheimer Society of Alberta and Northwest Territories (ASANT) for the Ed and Joyce Lyons postdoctoral fellowship (to M. R. J.); an international postdoctoral grant from the Swedish Research Council (Dnr: 350-2012-239 (to C. D.); ENS Cachan for a doctoral fellowship (E. M.); an operating grant from the Canadian Institutes of Health Research (CIHR) (to V. W. Y.); APRI and ASANT research grants (to S. T. and P. K. S.); and Tier I CRC (to P. K. S.). This work was also supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2014S1A2A2028270) (to M. H. L. and A. R.) and partly by the National Institutes of Health (AG048934 to A. R.).Notes and references
- Alzheimer's Association, Alzheimer's & Dementia, 2014, vol. 10, pp. e47–e92 Search PubMed.
- Alzheimer's Association, Alzheimer's & Dementia, 2015, vol. 11, pp. 332–384 Search PubMed.
- http://www.alzheimer.ca/ .
- A. Kumar and A. S. Ekavali, Pharmacol. Rep., 2015, 67, 195–203 CrossRef CAS PubMed.
- J. Hardy and D. J. Selkoe, Science, 2002, 297, 353–356 CrossRef CAS PubMed.
- D. J. Selkoe, Neuron, 1991, 6, 487–498 CrossRef CAS PubMed.
- H. W. Querfurth and F. M. LaFerla, N. Engl. J. Med., 2010, 362, 329–344 CrossRef CAS PubMed.
- A. Ittner, S. W. Chua, J. Bertz, A. Volkerling, J. van der Hoven, A. Gladbach, M. Przybyla, M. Bi, A. van Hummel, C. H. Stevens, S. Ippati, L. S. Suh, A. Macmillan, G. Sutherland, J. J. Kril, A. P. G. Silva, J. Mackay, A. Poljak, F. Delerue, Y. D. Ke and L. M. Ittner, Science, 2016, 354, 904–908 CrossRef CAS PubMed.
- K. Iqbal and I. Grundke-Iqbal, Alzheimer's Dementia, 2010, 6, 420–424 CrossRef CAS PubMed.
- K. J. Barnham and A. I. Bush, Chem. Soc. Rev., 2014, 43, 6727–6749 RSC.
- K. P. Kepp, Chem. Rev., 2012, 112, 5193–5239 CrossRef CAS PubMed.
- M. G. Savelieff, A. S. DeToma, J. S. Derrick and M. H. Lim, Acc. Chem. Res., 2014, 47, 2475–2482 CrossRef CAS PubMed.
- M. G. Savelieff, S. Lee, Y. Liu and M. H. Lim, ACS Chem. Biol., 2013, 8, 856–865 CrossRef CAS PubMed.
- M. A. Lovell, J. D. Robertson, W. J. Teesdale, J. L. Campbell and W. R. Markesbery, J. Neurol. Sci., 1998, 158, 47–52 CrossRef CAS PubMed.
- A. K. Sharma, S. T. Pavlova, J. Kim, J. Kim and L. M. Mirica, Metallomics, 2013, 5, 1529–1536 RSC.
- C. Cheignon, P. Faller, D. Testemale, C. Hureau and F. Collin, Metallomics, 2016, 8, 1081–1089 RSC.
- J. T. Pedersen, S. W. Chen, C. B. Borg, S. Ness, J. M. Bahl, N. H. H. Heegaard, C. M. Dobson, L. Hemmingsen, N. Cremades and K. Teilum, J. Am. Chem. Soc., 2016, 138, 3966–3969 CrossRef CAS PubMed.
- K. Reybier, S. Ayala, B. Alies, J. V. Rodrigues, S. Bustos Rodriguez, G. La Penna, F. Collin, C. M. Gomes, C. Hureau and P. Faller, Angew. Chem., Int. Ed., 2016, 55, 1085–1089 CrossRef CAS PubMed.
- M. W. Beck, J. S. Derrick, R. A. Kerr, S. B. Oh, W. J. Cho, S. J. C. Lee, Y. Ji, J. Han, Z. A. Tehrani, N. Suh, S. Kim, S. D. Larsen, K. S. Kim, J.-Y. Lee, B. T. Ruotolo and M. H. Lim, Nat. Commun., 2016, 7, 13115 CrossRef CAS PubMed.
- A. K. Sharma, S. T. Pavlova, J. Kim, D. Finkelstein, N. J. Hawco, N. P. Rath, J. Kim and L. M. Mirica, J. Am. Chem. Soc., 2012, 134, 6625–6636 CrossRef CAS PubMed.
- C. Rodríguez-Rodríguez, N. Sánchez de Groot, A. Rimola, Á. Álvarez-Larena, V. Lloveras, J. Vidal-Gancedo, S. Ventura, J. Vendrell, M. Sodupe and P. González-Duarte, J. Am. Chem. Soc., 2009, 131, 1436–1451 CrossRef PubMed.
- K. M. Lincoln, T. E. Richardson, L. Rutter, P. Gonzalez, J. W. Simpkins and K. N. Green, ACS Chem. Neurosci., 2012, 3, 919–927 CrossRef CAS PubMed.
- J. S. Derrick and M. H. Lim, ChemBioChem, 2015, 16, 887–898 CrossRef CAS PubMed.
- M. A. Telpoukhovskaia and C. Orvig, Chem. Soc. Rev., 2013, 42, 1836–1846 RSC.
- C. Rodríguez-Rodríguez, M. Telpoukhovskaia and C. Orvig, Coord. Chem. Rev., 2012, 256, 2308–2332 CrossRef.
- L. R. Perez and K. J. Franz, Dalton Trans., 2010, 39, 2177–2187 RSC.
- K. J. Franz, Curr. Opin. Chem. Biol., 2013, 17, 143–149 CrossRef CAS PubMed.
- M. R. Jones, E. L. Service, J. R. Thompson, M. C. P. Wang, I. J. Kimsey, A. S. DeToma, A. Ramamoorthy, M. H. Lim and T. Storr, Metallomics, 2012, 4, 910–920 RSC.
- M. R. Jones, C. Dyrager, M. Hoarau, K. J. Korshavn, M. H. Lim, A. Ramamoorthy and T. Storr, J. Inorg. Biochem., 2016, 158, 131–138 CrossRef CAS PubMed.
- A. Agis-Torres, M. Sölhuber, M. Fernandez and J. M. Sanchez-Montero, Curr. Neuropharmacol., 2014, 12, 2–36 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS PubMed.
- C. W. Tornoe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057–3064 CrossRef CAS PubMed.
- M. D. Liptak, K. C. Gross, P. G. Seybold, S. Feldgus and G. C. Shields, J. Am. Chem. Soc., 2002, 124, 6421–6427 CrossRef CAS PubMed.
- A. G. Cook, L. R. Wesner and S. L. Folk, J. Org. Chem., 1997, 62, 7205–7209 CrossRef CAS PubMed.
- H. K. Hall, J. Am. Chem. Soc., 1957, 79, 5441–5444 CrossRef CAS.
- R. E. Martin, B. Plancq, O. Gavelle, B. Wagner, H. Fischer, S. Bendels and K. Müller, ChemMedChem, 2007, 2, 285–287 CrossRef CAS PubMed.
- P. Leeson, Nature, 2012, 481, 455–456 CrossRef CAS PubMed.
- D. E. Clark and S. D. Pickett, Drug Discovery Today, 2000, 5, 49–58 CrossRef CAS PubMed.
- T. Storr, M. Merkel, G. X. Song-Zhao, L. E. Scott, D. E. Green, M. L. Bowen, K. H. Thompson, B. O. Patrick, H. J. Schugar and C. Orvig, J. Am. Chem. Soc., 2007, 129, 7453–7463 CrossRef CAS PubMed.
- C. Y. Huang, Methods Enzymol., 1982, 87, 509–525 CAS.
- Z. D. Hill and P. MacCarthy, J. Chem. Educ., 1986, 63, 162 CrossRef CAS.
- A. K. Sharma, J. Kim, J. T. Prior, N. J. Hawco, N. P. Rath, J. Kim and L. M. Mirica, Inorg. Chem., 2014, 53, 11367–11376 CrossRef CAS PubMed.
- P. S. Gans and A. Vacca, Ann. Chim., 1999, 89, 45–49 CAS.
- C. F. Baes Jr and R. E. Mesmer, The Hydrolysis of Cations, Publishing Co., Malabar, Florida, 1986 Search PubMed.
- L. Alderighi, P. Gans, A. Ienco, D. Peters, A. Sabatini and A. Vacca, Coord. Chem. Rev., 1999, 184, 311–318 CrossRef CAS.
- J. S. Wright, E. R. Johnson and G. A. DiLabio, J. Am. Chem. Soc., 2001, 123, 1173–1183 CrossRef CAS PubMed.
- J. Geng, M. Li, L. Wu, J. Ren and X. Qu, J. Med. Chem., 2012, 55, 9146–9155 CrossRef CAS PubMed.
- C. Cheignon, M. Jones, E. Atrian-Blasco, I. Kieffer, P. Faller, F. Collin and C. Hureau, Chem. Sci., 2017 10.1039/c7sc00809k.
- R. Re, N. Pellegrini, A. Proteggente, A. Pannala, M. Yang and C. Rice-Evans, Free Radical Biol. Med., 1999, 26, 1231–1237 CrossRef CAS PubMed.
- N. G. Faux, C. W. Ritchie, A. Gunn, A. Rembach, A. Tsatsanis, J. Bedo, J. Harrison, L. Lannfelt, K. Blennow, H. Zetterberg, M. Ingelsson, C. L. Masters, R. E. Tanzi, J. L. Cummings, C. M. Herd and A. I. Bush, J. Alzheimer's Dis., 2010, 20, 509–516 CrossRef CAS PubMed.
- L. Lannfelt, K. Blennow, H. Zetterberg, S. Batsman, D. Ames, J. Harrison, C. L. Masters, S. Targum, A. I. Bush, R. Murdoch, J. Wilson and C. W. Ritchie, Lancet Neurol., 2008, 7, 779–786 CrossRef CAS PubMed.
- Y. Manevich, K. D. Held and J. E. Biaglow, Radiat. Res., 1997, 148, 580–591 CrossRef CAS PubMed.
- L. Guilloreau, S. Combalbert, A. Sournia-Saquet, H. Mazarguil and P. Faller, ChemBioChem, 2007, 8, 1317–1325 CrossRef CAS PubMed.
- M. E. Rice, Trends Neurosci., 2000, 23, 209–216 CrossRef CAS PubMed.
- A. Conte-Daban, A. Day, P. Faller and C. Hureau, Dalton Trans., 2016, 45, 15671–15678 RSC.
- B. Alies, I. Sasaki, O. Proux, S. Sayen, E. Guillon, P. Faller and C. Hureau, Chem. Commun., 2013, 49, 1214–1216 RSC.
- A. S. DeToma, J. Krishnamoorthy, Y. Nam, H. J. Lee, J. R. Brender, A. Kochi, D. Lee, V. Onnis, C. Congiu, S. Manfredini, S. Vertuani, G. Balboni, A. Ramamoorthy and M. H. Lim, Chem. Sci., 2014, 5, 4851–4862 RSC.
- A. S. Pithadia and M. H. Lim, Curr. Opin. Chem. Biol., 2012, 16, 67–73 CrossRef CAS PubMed.
- L. O. Tjernberg, C. Lilliehöök, D. J. E. Callaway, J. Näslund, S. Hahne, J. Thyberg, L. Terenius and C. Nordstedt, J. Biol. Chem., 1997, 272, 12601–12605 CrossRef CAS PubMed.
- L. O. Tjernberg, J. Näslund, F. Lindqvist, J. Johansson, A. R. Karlström, J. Thyberg, L. Terenius and C. Nordstedt, J. Biol. Chem., 1996, 271, 8545–8548 CrossRef CAS PubMed.
- C. Haass and D. J. Selkoe, Nat. Rev. Mol. Cell Biol., 2007, 8, 101–112 CrossRef CAS PubMed.
- D. M. Walsh and D. J. Selkoe, J. Neurochem., 2007, 101, 1172–1184 CrossRef CAS PubMed.
- R. Riek and D. S. Eisenberg, Nature, 2016, 539, 227–235 CrossRef PubMed.
- E. K. Pickett, R. M. Koffie, S. Wegmann, C. M. Henstridge, A. G. Herrmann, M. Colom-Cadena, A. Lleo, K. R. Kay, M. Vaught, R. Soberman, D. M. Walsh, B. T. Hyman and T. L. Spires-Jones, J. Alzheimer's Dis., 2016, 53, 787–800 CrossRef CAS PubMed.
- J. C. Stroud, C. Liu, P. K. Teng and D. Eisenberg, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7717–7722 CrossRef CAS PubMed.
- J. Bieschke, M. Herbst, T. Wiglenda, R. P. Friedrich, A. Boeddrich, F. Schiele, D. Kleckers, J. M. Lopez del Amo, B. A. Grüning, Q. Wang, M. R. Schmidt, R. Lurz, R. Anwyl, S. Schnoegl, M. Fändrich, R. F. Frank, B. Reif, S. Günther, D. M. Walsh and E. E. Wanker, Nat. Chem. Biol., 2012, 8, 93–101 CrossRef CAS PubMed.
- I. Benilova, E. Karran and B. De Strooper, Nat. Neurosci., 2012, 15, 349–357 CrossRef CAS PubMed.
- J. Brouillette, R. Caillierez, N. Zommer, C. Alves-Pires, I. Benilova, D. Blum, B. De Strooper and L. Buée, J. Neurosci., 2012, 32, 7852–7861 CrossRef CAS PubMed.
- S. Lee, X. Y. Zheng, J. Krishnamoorthy, M. G. Savelieff, H. M. Park, J. R. Brender, J. H. Kim, J. S. Derrick, A. Kochi, H. J. Lee, C. Kim, A. Ramamoorthy, M. T. Bowers and M. H. Lim, J. Am. Chem. Soc., 2014, 136, 299–310 CrossRef CAS PubMed.
- L. M. F. Gomes, R. P. Vieira, M. R. Jones, M. C. P. Wang, C. Dyrager, E. M. Souza-Fagundes, J. G. Da Silva, T. Storr and H. Beraldo, J. Inorg. Biochem., 2014, 139, 106–116 CrossRef CAS PubMed.
- M. P. Lambert, A. K. Barlow, B. A. Chromy, C. Edwards, R. Freed, M. Liosatos, T. E. Morgan, I. Rozovsky, B. Trommer, K. L. Viola, P. Wals, C. Zhang, C. E. Finch, G. A. Krafft and W. L. Klein, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6448–6453 CrossRef CAS.
- K. L. Viola and W. L. Klein, Acta Neuropathol., 2015, 129, 183–206 CrossRef CAS PubMed.
- M. Mold, L. Ouro-Gnao, B. M. Wieckowski and C. Exley, Sci. Rep., 2013, 3, 1256 CrossRef PubMed.
- J.-S. Choi, J. J. Braymer, R. P. R. Nanga, A. Ramamoorthy and M. H. Lim, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 21990–21995 CrossRef CAS PubMed.
- S.-J. Hyung, A. S. DeToma, J. R. Brender, S. Lee, S. Vivekanandan, A. Kochi, J.-S. Choi, A. Ramamoorthy, B. T. Ruotolo and M. H. Lim, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 3743–3748 CrossRef CAS PubMed.
- G. Meloni, V. Sonois, T. Delaine, L. Guilloreau, A. Gillet, J. Teissie, P. Faller and M. Vasak, Nat. Chem. Biol., 2008, 4, 366–372 CrossRef CAS PubMed.
- J. H. Jhamandas, Z. Li, D. Westaway, J. Yang, S. Jassar and D. MacTavish, Am. J. Pathol., 2011, 178, 140–149 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc01269a |
| This journal is © The Royal Society of Chemistry 2017 |