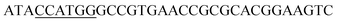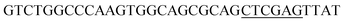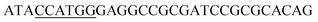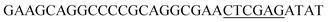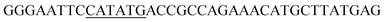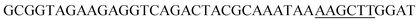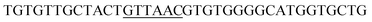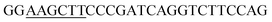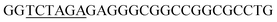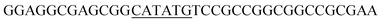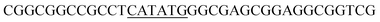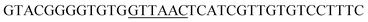Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Halogenation of glycopeptide antibiotics occurs at the amino acid level during non-ribosomal peptide synthesis†
Tiia
Kittilä‡
a,
Claudia
Kittel‡
b,
Julien
Tailhades
 cd,
Diane
Butz
e,
Melanie
Schoppet
acd,
Anita
Büttner
f,
Rob J. A.
Goode
cd,
Diane
Butz
e,
Melanie
Schoppet
acd,
Anita
Büttner
f,
Rob J. A.
Goode
 dg,
Ralf B.
Schittenhelm
dg,
Karl-Heinz
van Pee
e,
Roderich D.
Süssmuth
f,
Wolfgang
Wohlleben
bh,
Max J.
Cryle
dg,
Ralf B.
Schittenhelm
dg,
Karl-Heinz
van Pee
e,
Roderich D.
Süssmuth
f,
Wolfgang
Wohlleben
bh,
Max J.
Cryle
 *acdi and
Evi
Stegmann
*bh
*acdi and
Evi
Stegmann
*bh
aDepartment of Biomolecular Mechanisms, Max Planck Institute for Medical Research, Jahnstrasse 29, 69120 Heidelberg, Germany
bInterfaculty Institute of Microbiology and Infection Medicine Tuebingen, Microbiology/Biotechnology, University of Tuebingen, Auf der Morgenstelle 28, 72076 Tuebingen, Germany. E-mail: evi.stegmann@biotech.uni-tuebingen.de
cEMBL Australia, Monash University, Clayton, Victoria 3800, Australia. E-mail: max.cryle@monash.edu
dThe Monash Biomedicine Discovery Institute, Department of Biochemistry and Molecular Biology, Monash University, Clayton, Victoria 3800, Australia
eInstitut für Chemie, Technische Universität Berlin, 10623 Berlin, Germany
fAllgemeine Biochemie, TU Dresden, 01062 Dresden, Germany
gMonash Biomedical Proteomics Facility, Monash University, Clayton, Victoria 3800, Australia
hGerman Centre for Infection Research (DZIF), Partner Site Tuebingen, Tuebingen, Germany
iARC Centre of Excellence in Advanced Molecular Imaging, Monash University, Clayton, Victoria 3800, Australia
First published on 13th July 2017
Abstract
Halogenation plays a significant role in the activity of the glycopeptide antibiotics (GPAs), although up until now the timing and therefore exact substrate involved was unclear. Here, we present results combined from in vivo and in vitro studies that reveal the substrates for the halogenase enzymes from GPA biosynthesis as amino acid residues bound to peptidyl carrier protein (PCP)-domains from the non-ribosomal peptide synthetase machinery: no activity was detected upon either free amino acids or PCP-bound peptides. Furthermore, we show that the selectivity of GPA halogenase enzymes depends upon both the structure of the bound amino acid and the PCP domain, rather than being driven solely via the PCP domain. These studies provide the first detailed understanding of how halogenation is performed during GPA biosynthesis and highlight the importance and versatility of trans-acting enzymes that operate during peptide assembly by non-ribosomal peptide synthetases.
Introduction
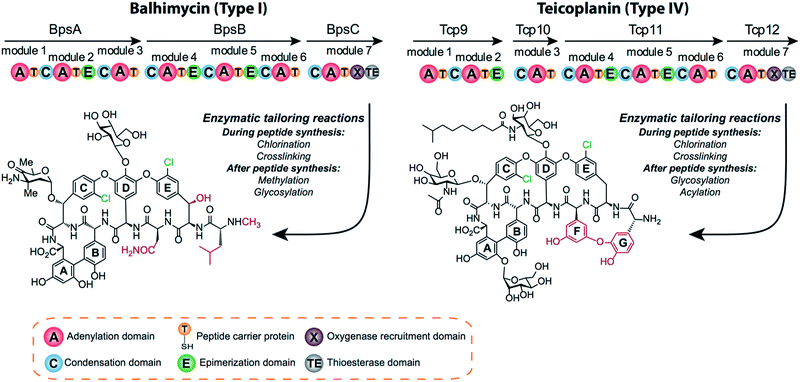
Non-ribosomal peptide synthesis is a highly important route for the production of bioactive peptides, many of which have medicinal value. Some of the most important members of non-ribosomal peptides are known for their antibiotic action: well-known examples include beta-lactam antibiotics such as nocardicin, cyclic peptides such as daptomycin and the glycopeptide antibiotics (GPAs) that include vancomycin and teicoplanin.1 By decoupling peptide synthesis from the ribosome, non-ribosomal peptide synthetase (NRPS) machineries are able to produce peptides from a significantly larger pool of precursors than the standard proteinogenic amino acids.2,3 NRPS-catalysed biosynthesis is based around active domains that are organised into modules. Adenylation (A)-domains catalyse the selection and activation of precursors, which are then loaded onto peptidyl carrier protein (PCP)-domains via a phosphopantetheine linker.4 The resultant thioester linkage remains chemically reactive for peptide bond formation – catalysed by condensation (C)-domains – but retains NRPS biosynthetic intermediates in a protein-bound form.5 In addition to these domains crucial for peptide synthesis, many NRPSs have additional modification domains, such as epimerisation (E)-domains, which alter the stereochemistry of amino acid residues and are a common hallmark of NRPS-produced peptides.6 The final peptide product is detached from the NRPS machinery via hydrolysis or cyclisation catalysed by a thioesterase (TE)-domain, which also offers further options to gain biological activity and compound diversity.7–9The complexity of the products of NRPS biosynthesis not only stems from the direct actions of the NRPS assembly line but also from a great number of further peptide modifications by external enzymes acting in trans. Examples from GPA biosynthesis include the hydroxylation of amino acid side chains, cytochrome P450-catalysed crosslinking of aromatic side chains, glycosylation, sulfation, acylation as well as the incorporation of chlorine atoms into the peptide via the halogenation of aromatic side chains of amino acid residues.10 Many of the modifications appear to be directed towards improving the selectivity of GPAs towards specific bacterial strains but some, for example the installation of crosslinks into the peptide backbone, are essential for their antibiotic activity.10,11 Because of their crucial roles in GPA biosynthesis, the modification enzymes are an appealing target for GPA remodelling.12
GPAs can be divided into several classes based on the heptapeptide backbone structure.10,13 Type I GPAs possess aliphatic residues at positions one and three of the peptide and display three crosslinks between aromatic residues of the peptide, forming the AB, C-O-D and D-O-E rings needed for GPA activity. Type II GPAs share the crosslinking pattern of Type I GPAs but contain aromatic residues at positions one and three of the peptide. Type III and IV GPAs also have aromatic residues at positions one and three of the peptide, but these are crosslinked to form an additional F-O-G ring, with differentiation between these classes due to the presence of an acyl moiety in Type IV GPAs that is absent in Type III GPAs. Balhimycin belongs to the vancomycin-type peptides (Type I GPAs), with balhimycin differing from vancomycin only with regard to the glycosylation pattern, whilst teicoplanin belongs to the Type IV GPAs (Fig. 1).
The chlorine atoms present within the GPA peptides contribute significantly to the binding affinity of the GPAs for their target, the dipeptide terminus of lipid II.14–16 In both Type I and IV GPAs β-hydroxy-tyrosine (OH–Tyr) and/or tyrosine (Tyr) residues are chlorinated in the positions two and six by halogenases that belong to the class of flavin (FADH2)-dependent halogenases. Most GPA gene clusters encode a single halogenase, which was shown in balhimycin biosynthesis to be responsible for chlorination of both positions in the peptide.17 Some GPA gene clusters possess an additional halogenase enzyme that appears to play a role in the chlorination of 4-hydroxyphenylglycine (4-Hpg) residues.18 Whilst insights into the mechanism of FADH2-dependent halogenases have been gained from structural and functional studies,19,20 reconstitution of halogenase activity in vitro has proven to be difficult. Most of the FADH2-dependent halogenases characterised so far are tryptophan halogenases that act on substrates free in solution.21–23 However, halogenases acting on PCP-bound substrates have also been characterised.24,25 In both cases chlorination of the substrate takes place before monomer incorporation to the natural product.
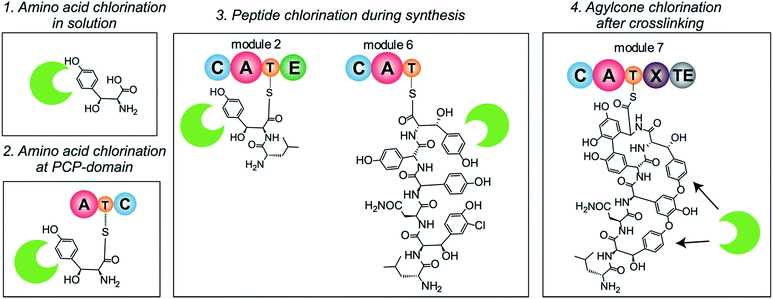
The timing of the chlorination during GPA biosynthesis is still under debate. Four possible scenarios for the incorporation of halide atoms have been postulated and – to some extent – investigated (Fig. 2). In the first scenario, amino acids are chlorinated prior to incorporation by the NRPS machinery; in the second, halogenation of amino acids takes place after loading onto a PCP-domain; in the third, the peptide is chlorinated during peptide synthesis; and in the fourth, halogenation takes place after the peptide aglycone has been formed by P450 (Oxy) enzymes. Previously, the inactivation of the P450 monooxygenases responsible for GPA side chain cyclisation in vivo26–29 strongly suggested that halogenation would take place prior to cyclisation. Furthermore, these scenarios have been studied using two homologous halogenases from Type I GPA biosynthesis: BhaA from balhimycin biosynthesis and VhaA from vancomycin biosynthesis. In vivo gene disruption studies of BhaA have shown that supplementation of the medium by chlorinated amino acids is not able to restore production of the natural, halogenated GPA if the halogenase gene is deleted.30 In contrast, supplementation of the growth medium with non-chlorinated OH–Tyr could overcome the deletion of the genes essential for OH–Tyr production and restore the formation of the halogenated type I GPAs.30 These results strongly suggest that scenario one, chlorination prior to incorporation by module 2 and 6 of the NRPS machinery, is not possible.
Although halogenation of free amino acids has been for most part ruled out, debate concerning the other three scenarios continues. The generation of various halogenated peptide GPA intermediates in a strain where the final condensation domain had been deleted points towards incorporation of the halide atoms during peptide biosynthesis.11 Precedent would suggest a mechanism of action similar to that identified for the halogenase enzymes from pyoluteorin synthesis24 or enediyne synthesis,25 where aminoacyl-PCP domains are the substrates for modification during peptide biosynthesis by the NRPS machinery; other enzymes have also been identified that adopt this approach.31,32 In contrast to this, in vitro studies using a P450 enzyme that crosslinks aromatic side chains from the vancomycin biosynthetic cluster (OxyBvan) showed that halide atoms in PCP-bound hexapeptide substrates made the peptides worse substrates for the first crosslinking step,33 suggesting that chlorination would take place on the aglycone. However, these studies did not take into account the role of the X-domain in recruiting the P450 enzymes to the NRPS-bound peptide34–38 and recent results39 indicate that halogenated peptides are crosslinked equally well when presented by NRPS modules containing the X-domain, making halogenation prior to P450 activity a distinct possibility.
So far, all characterised FADH2-dependent halogenases associated with natural products produced by NRPS machineries act on amino acid/pyrrole substrates rather than on the final product, strongly suggesting the PCP-bound amino acid to be the substrate for halogenases in GPA biosynthesis. However, in vitro experiments using VhaA showed low levels of activity upon PCP-bound hexapeptide substrates40 pointing towards chlorination of the peptide rather than amino acid substrate. Thus, the exact timing of halogenation in GPA biosynthesis has remained unclear until this current study. Here, we are now able to show that halogenation occurs during peptide biosynthesis and that the substrates for the GPA halogenase enzymes are the PCP-bound amino acids, two OH–Tyr residues at amino acid (AA) position 2 and 6 for Type I GPA and two Tyr residues at position 2 and 6 for Type IV GPAs.
Results
Analysis of GPA halogenation in vitro
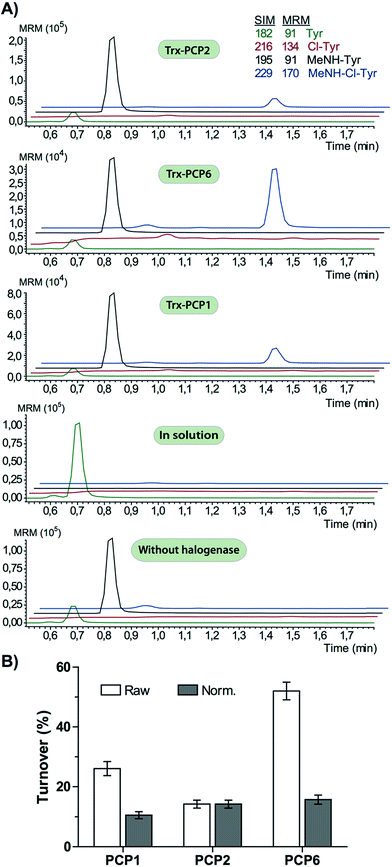
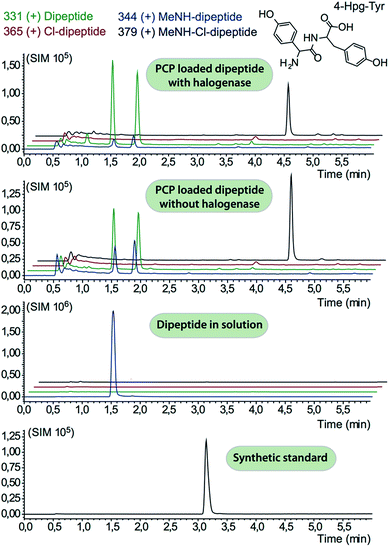
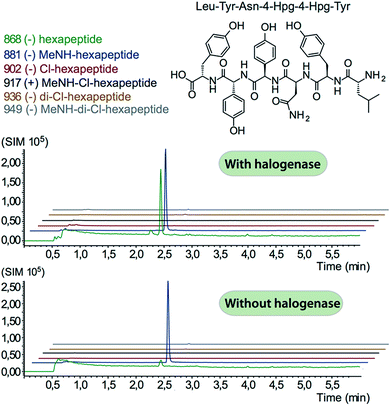
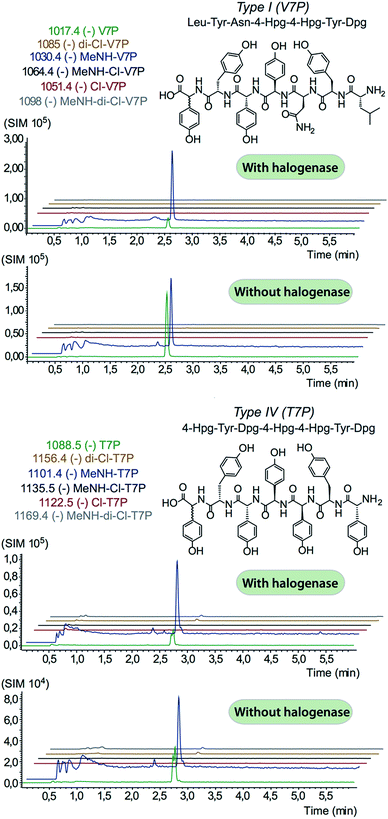
In order to assess the halogenation state of peptide precursors during GPA biosynthesis, we performed in vitro turnover experiments. In this regard, we explored the halogenase enzymes from both the balhimycin producer (Amycolatopsis balhimycina) as well as the teicoplanin producer (A. teichomyceticus). Both enzymes could be overproduced and purified in an FAD-bound form as anticipated (Fig. S1†). The former enzyme was overproduced in Pseudomonas fluorescens BL915 ΔORF1-4 containing the gene bhaA40 in the E. coli-Pseudomonas shuttle vector pCIBhis and the latter in E. coli.We prepared a range of substrates for activity assays: tyrosine, Type IV dipeptide (4-Hpg-Tyr), hexapeptide41 (leucine (Leu)-Tyr-asparagine (Asn)-4-Hpg-4-Hpg-Tyr) and two different heptapeptides42 (Leu-Tyr-Asn-4-Hpg-4-Hpg-Tyr-Dpg and 4-Hpg-Tyr-Dpg-4-Hpg-4-Hpg-Tyr-Dpg) (Fig. 4–6). Substrates were loaded on various PCP domains using the promiscuous phosphopantetheinyl transferase Sfp. Tyrosine was loaded on both PCP-domains that would be expected to interact with the halogenase enzyme (PCP2 and PCP6 from a Type IV NRPS) as well as to a PCP-domain that – based on the GPA chlorination pattern – is not anticipated to interact with the halogenase enzyme (PCP1 from a Type IV NRPS) to test if the PCP-domain is responsible for guiding the halogenase to the correct substrate. Dipeptide was loaded onto a PCP2 domain (Type IV) and hexapeptide on a PCP7 domain (Type I), whilst heptapeptides were loaded on PCP7 domain either from Type I or Type IV NRPSs. Chlorination of tyrosine and dipeptide were also studied using substrates free in solution. We assessed activity using UPLC-MS with single ion monitoring (SIM) and multiple reaction monitoring (MRM), after cleavage of PCP-bound substrates using excess methylamine to afford the methylamide products.
The halogenase from the teicoplanin system (Tcp21) displayed activity upon PCP-bound tyrosine as a substrate. We could observe activity upon tyrosine bound to PCP-domains 1, 2 and 6 from the teicoplanin NRPS machinery, which implies that the halogenase is not solely selective for the PCP domain and that the bound amino acid plays a role in regulating halogenase activity (Fig. 3A). Tyrosine in solution was not chlorinated by Tcp21, which suggests that the interaction with a PCP domain is crucial for catalysis. The loading efficiency differed between the three PCP-domains but the amount of chlorinated tyrosine stayed at the same level in each reaction, which shows that the turnover efficiency appears to be limited by the in vitro efficiency of Tcp21 (Fig. 3B). We also tested whether the halogenase was able to act upon dipeptide substrates. Under the conditions where we observed halogenase activity upon PCP-bound tyrosine, we could show that neither PCP-loaded nor free dipeptide was a substrate for the enzyme (Fig. 4). We also tested longer peptides as potential substrates for halogenation, however – and as with the dipeptide substrates – we did not observe any activity upon these substrates (Fig. 5 and 6). Thus, the results of our in vitro characterisation indicate that PCP-bound amino acids are the actual substrates for halogenation during Type IV GPA biosynthesis. We did not detect activity of the balhimycin halogenase (BhaA) upon any substrates (data not shown), which we attribute to the lack of the hydroxylated side chain of the tyrosine residue (present in Type I GPAs) in the aminoacyl-PCP substrate.10
Construction of a dipeptide producing GPA strain
Since BhaA showed no in vitro activity, we decided to assess the halogenation state in A. balhimycina in vivo. Given that we have elucidated balhimycin biosynthesis in detail,12 have developed tools to construct A. balhimycina mutants and elucidated the resultant intermediates, we generated a balhimycin producer strain that possessed a modified NRPS machinery to investigate halogenation.43–45 As our hypothesis is that the PCP-bound amino acids are substrates for the halogenase during GPA biosynthesis, we modified the balhimycin NRPS in such a way as to address the first peptide intermediate that should contain a halogenated residue – a dipeptide. Three NRPS proteins, BpsA, BpsB and BpsC, are involved in the assembly of the GPA heptapeptide core (Fig. 7). Module 2 is part of BpsA and consists of C-A-PCP (PCP1) and E domains, while module 3 consists of a C-, A- and PCP-domain (PCP3). The C-domain of module 3 is responsible for tripeptide formation from the Leu–OH–Tyr dipeptide and Asn. To terminate the non-ribosomal peptide synthesis after module 2 and hence produce a dipeptide for analysis, significant parts of the C-domain of module 3 were deleted and replaced by TE-domain of BpsC to provoke release of the dipeptide intermediate (Fig. 7). To this end, the fragments mod2left (1289 bp), containing the E-domain and an N-terminal part of the C-domain of module 3, and mod2right (1334 bp) including the C-terminal part of the C-domain and the A-domain of module 3, were amplified by PCR and used for the deletion of the C-domain (see Experimental procedures, Fig. 7).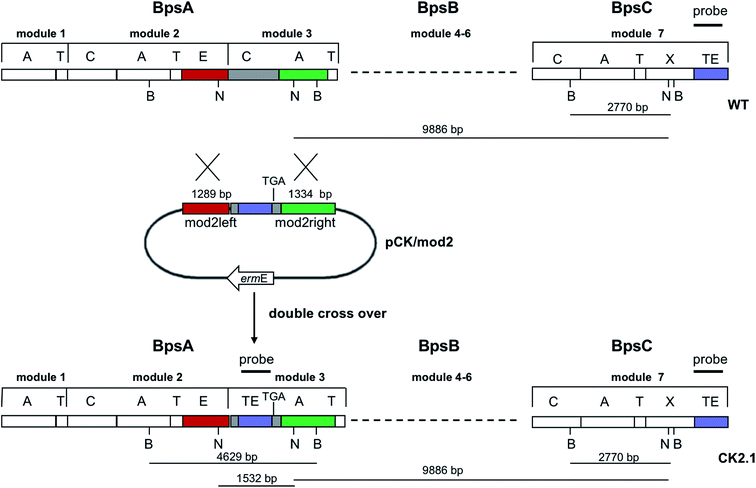 | ||
| Fig. 7 Construction of the mutant CK2.1. Schematic representation of the insertion of a TE-encoding DNA fragment into bpsA. Abbreviations for the domains see Fig. 1. WT, A. balhimycina wildtype; ermE, erythromycin resistance gene. Mod2left (1289 bp), containing the E-domain (red) and an N-terminal part of the C-domain (grey), and mod2right (1334 bp) including the A-domain (green) and a C-terminal part of the C-domain (grey), TE fragment (blue). To obtain the replacement mutant CK2.1 a double crossover via homologous recombination is required (see Results). Line: fragments after NcoI (N) or BamHI (B) restriction. | ||
The TE-domain of BpsC was amplified by PCR and inserted between the fragments mod2left and mod2right resulting in the plasmid pCKmod2. The non-replicative plasmid pCK/mod2 was used to transform the A. balhimycina wildtype strain by the direct transformation method. The plasmid selection marker conferring erythromycin resistance was used to identify clones in which a first homologous recombination event occurred. Ten erythromycin resistant clones were obtained. These clones were analysed in a bioassay with Bacillus subtilis ATCC6633, to check whether the integration of pCK/mod2 resulted in the loss of balhimycin production. Loss of balhimycin production would be the expected result of recombination via the chromosomal bpsA gene. Two of the ten clones lost the ability to produce an active antibiotic and the correct integration of pCKmod2 was verified by Southern Blot hybridisation; both clones turned out to be identical. The non-producing clone (CK1) showed the intended integration of the plasmid within the region coding for the C-domain. For the other eight erythromycin resistant clones that still produced balhimycin the crossover took place via the part of the bpsC gene which encodes the TE-domain at the end of BpsC.
To obtain the replacement mutant, a second homologous recombination in CK1 was required: since double crossover events happen at a low frequency in A. balhimycina, a “stress” treatment (see Experimental procedures) – whose application increases the probability of a second crossover event – was carried out with clone CK1. Following this protocol, 650 colonies were examined on R5 agar plates both in the presence and absence of erythromycin. Two of the “stressed” colonies lost the integrated plasmid as indicated by the lack of erythromycin resistance. These two clones, CK2.1 and CK2.2, were not able to produce an active antibiotic, indicating that the second crossover led to the disruption of the non-ribosomal peptide biosynthesis. The gene replacement and loss of pCK/mod2 was confirmed by Southern Blot analyses; a DIG-labelled PCR fragment of the TE-domain was used as a probe with total DNA of the two mutants CK2.1 and CK2.2 and the wildtype of A. balhimycina. The Southern Blot clearly showed two bands of the expected size in the case of the mutants while in the total DNA of A. balhimycina wildtype only a single band was visible (Fig. S2†). This result combined with the loss of balhimycin production showed that CK2.1 and CK2.2 are identical and confirmed the correct identity of the replacement mutants.
The mutant CK2.1 was fermented under production conditions and the culture broth was analysed by HPLC-ESI-MS. In accordance to the lack of antibiotic activity in the bioassays, no balhimycin was detected. In contrast, the search for biosynthetic intermediates resulted in signals that corresponded to Leu–OH–Cl-Tyr (Fig. S3†). Remarkably, the molecular mass of this dipeptide was found at two different retention times in the HPLC run, which suggests the presence of two diastereoisomers due to alternate stereochemistry of the second amino acid, β-hydroxytyrosine (Fig. S3†). Given that the epimerisation domain that converts (L)–OH–Tyr to (D)–OH–Tyr in the NRPS is still present, this implies that the inserted thioesterase domain is able to compete effectively with the E-domain for the peptidyl-PCP substrate to generate two diastereomers of the product dipeptide. In order to confirm the constitutional structures of these dipeptides, daughter ion spectra were generated and signals assigned. It was found that the main fragment signals of both diastereoisomers were identical indicating the same constitutional formulae (Fig. S4†). We further synthesised synthetic standards of the suspected Leu–OH–Cl-Tyr diastereomers and could thus confirm the identity of the products identified from the A. balhimycina mutant, with the major dipeptide peak corresponding to the dipeptide bearing the (L)–OH–Tyr residue (Fig. S5 and S6†). This result confirms that halogenation during GPA biosynthesis already takes place on module 2 during NRPS synthesis.
Discussion
The diversification of peptide scaffolds within NRPS biosynthesis provides a tremendous source of selectivity and function. Halogenation is an important example of one such modification, and the GPAs are an excellent example of the effects halogenation can have on compound activity, as the removal of chlorine atoms from GPAs leads to significant reduction in binding affinity for the dipeptide terminus of lipid II.14–16 Halogenase activity from different biosynthetic systems has been demonstrated for different types of substrates – either free in solution or carrier protein bound – with selectivity varying from one system to another.46 However, the timing and substrate selectivity of halogenation during GPA biosynthesis has previously been unclear. Whilst the activity of the vancomycin halogenase upon PCP-bound peptides has been reported, the use of mismatched peptide and PCP-domain within the substrates tested and the lack of cohesion with previous preliminary in vivo data led us to investigate this process further.40 In this work, we have been able to combine comprehensive results of both in vivo and in vitro experiments to show that GPA halogenase enzymes act during NRPS-mediated peptide assembly. Initially, through the in vitro characterisation of the halogenase Tcp21 from teicoplanin biosynthesis we could clearly demonstrate that halogenase activity was present with tyrosine only when the amino acid was presented in a PCP-bound form. Furthermore, we could show that the identity of the PCP-domain able to support tyrosine halogenation was not strictly correlated with the observed halogenation pattern of teicoplanin, which suggests that a combination of correct amino acid and PCP-domain is required for GPA halogenation during NRPS-catalysed peptide synthesis in vivo. Our results are in contrast to the results described by Schmartz et al.,40 in which the authors demonstrated the chlorination of both OH–Tyr residues on a linear hexapeptide intermediate attached to the PCP domain in module-6 of the NRPS during vancomycin assembly. Chlorination at this stage of GPA biosynthesis shows that this halogenase demonstrates the in vitro capability to modify two quite different sites within the hexapeptide-PCP substrate: this activity is challenging to reconcile with other examples of GPA halogenation (e.g. A40926,47 which displays chlorination at residues 3 and 6 of the peptide, or the significantly altered pattern found in the Type-V GPA complestatin).48 We therefore decided to explore our hypothesis that chlorination occurs on the PCP-bound amino acid using further in vitro and in vivo studies on a different GPA halogenase: the balhimycin halogenase BhaA. Previous in vivo gene disruption experiments of the balhimycin synthetase had revealed the presence of chlorinated GPA peptide precursors during NRPS-catalysed peptide synthesis.11 In order to further test whether the substrates of GPA halogenation are aminoacyl-PCPs, we constructed a mutant balhimycin producer strain that had been engineered to produce the minimal GPA peptide that could contain a chlorine atom – a dipeptide. Following the successful construction of the strain, analysis of the peptide products and comparison to authentic standards clearly revealed the presence and confirmed the identity of the chlorinated dipeptides produced. This result indicates that halogenation during GPA biosynthesis occurs during NRPS-mediated peptide biosynthesis: as neither Tcp21 nor BhaA were active upon peptide substrates, the halogenase activity observed during NRPS-catalysed peptide synthesis in vivo can be deduced as occurring upon PCP-bound amino acids, which matches the results of our in vitro experiments. The lack of activity of BhaA upon PCP-bound tyrosine is also consistent with the differences in the generation of OH–Tyr residues during balhimycin and teicoplanin biosynthesis. In balhimycin biosynthesis, the correct PCP-bound amino acid substrates for BhaA would be OH–Tyr and not Tyr, as OH–Tyr is incorporated directly into the precursor peptide by the balhimycin NRPS machinery.30,45 This is significantly different to teicoplanin biosynthesis, in which tyrosine is initially recognised and incorporated into the GPA peptide by the NRPS and that is followed by subsequent generation of OH–Tyr via the actions of a non-heme iron oxygenase.49 The results from BhaA in vitro experiments further demonstrate the substrate specificity of the GPA halogenase enzymes and underlines the importance of having the correct amino acid loaded onto the PCP to enable halogenation during GPA biosynthesis.Conclusion
In this work, we have obtained conclusive evidence that halogenation occurs during NRPS-mediated peptide assembly in GPA biosynthesis. By combining the results of in vitro and in vivo experiments we have been able to show that aminoacyl-PCP substrates are recognised by halogenase enzymes and that the pairing of both the PCP-domain and bound substrate is crucial to activity: this model (see also ref. 50) helps to explain why some GPAs (such as A40926) display halogenation at different residues47 or differing levels,51 whilst others (such as pekiskomycin)52 are lacking chlorination at residues that would anticipate to be halogenated based on similar GPAs.3 Our GPA halogenation model also aligns with results obtained from biosynthetic studies of other halogenated peptides produced by NRPS-systems (such as enduracidin/ramoplanin), where switching the halogenase enzyme in vivo leads to a product where the halogenation pattern is altered in spite of highly similar peptide structures.53,54 Our results suggest that the alteration of halogenase enzymes within GPA producer strains could well produce alternatively halogenated GPA products: given that halogen atoms can act as handles for chemical modification of GPAs55,56 and the synthetic challenge of producing such compounds,14,15 switching halogenase enzymes (such as has been done for other modification enzymes in GPA biosynthesis)57 would appear to be an important route that should be explored to enable the future diversification of GPA scaffolds.Experimental
In vivo experiments
PCR protocols for amplification of the fragments mod2left, mod2right and TE PCRs were performed on a Robo Cycler Gradient 40 thermocycler from Stratagene (La Jolla, CA, USA) with the Expand High Fidelity PCR System (Roche, Grenzach-Wyhlen, Germany). For the amplification of the fragments mod2left, mod2right and TE the following PCR conditions were used: initial denaturation (95 °C for 5 min), 30 cycles of denaturation (95 °C for 2 min), annealing (69 °C for 1 min), and polymerisation (72 °C for 1 min 30 s), an additional polymerisation step (72 °C for 10 min) at the end. The primers used were as follows: amplification of mod2left (1289 bp): Mod2leftP1, Mod2leftP2; for amplification of fragment mod2right (1334 bp): Mod2rightP1, Mod2rightP2, and for TE (831 bp): TE2left, TE2right.
In vitro experiments
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800g, 1 h). Supernatant was incubated with Ni-NTA beads (1 h, Macherey-Nagel) with gentle shaking and the beads were washed twice with NiNTA wash buffer (50 mM Tris·HCl, pH 7.4, 300 mM NaCl, 10 mM imidazole). Protein was eluted with NiNTA elution buffer (50 mM Tris·HCl, pH 7.4, 300 mM NaCl, 300 mM imidazole). Size exclusion chromatography (SEC) was performed using a Superose 12 10/300 GL column connected to an Äkta PURE system (GE Healthcare). Protein concentration was determined by spectroscopic method (A280, Nanodrop 2000c, Nanodrop) and protein concentrated using centrifugal concentrators (Sartorius). Protein purity was assessed by SDS-PAGE and concentrated proteins were aliquoted (30–100 μl), flash frozen and stored at −80 °C. Identity of the purified proteins was confirmed by MALDI-TOF MS peptide mass fingerprinting.
800g, 1 h). Supernatant was incubated with Ni-NTA beads (1 h, Macherey-Nagel) with gentle shaking and the beads were washed twice with NiNTA wash buffer (50 mM Tris·HCl, pH 7.4, 300 mM NaCl, 10 mM imidazole). Protein was eluted with NiNTA elution buffer (50 mM Tris·HCl, pH 7.4, 300 mM NaCl, 300 mM imidazole). Size exclusion chromatography (SEC) was performed using a Superose 12 10/300 GL column connected to an Äkta PURE system (GE Healthcare). Protein concentration was determined by spectroscopic method (A280, Nanodrop 2000c, Nanodrop) and protein concentrated using centrifugal concentrators (Sartorius). Protein purity was assessed by SDS-PAGE and concentrated proteins were aliquoted (30–100 μl), flash frozen and stored at −80 °C. Identity of the purified proteins was confirmed by MALDI-TOF MS peptide mass fingerprinting.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 790 M−1 cm−1 was used to determine protein concentration). YcnD: 4 ml NiNTA beads were incubated with the cleared lysate. Elution fraction was dialysed against 50 mM Tris·HCl, pH 7.4, 20 mM NaCl and subjected to anion exchange purification (Resource Q, GE Healthcare). Appropriate fractions were pooled and subjected to SEC (50 mM Tris·HCl, pH 7.4, 150 mM NaCl). Fractions containing monomeric YcnD were pooled and concentrated to 17 mg ml−1 (calculated ε = 36
790 M−1 cm−1 was used to determine protein concentration). YcnD: 4 ml NiNTA beads were incubated with the cleared lysate. Elution fraction was dialysed against 50 mM Tris·HCl, pH 7.4, 20 mM NaCl and subjected to anion exchange purification (Resource Q, GE Healthcare). Appropriate fractions were pooled and subjected to SEC (50 mM Tris·HCl, pH 7.4, 150 mM NaCl). Fractions containing monomeric YcnD were pooled and concentrated to 17 mg ml−1 (calculated ε = 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 910 M−1 cm−1 was used to determine protein concentration). Trx-PCP1, Trx-PCP2 and Trx-PCP6: 2 mM dithioerythritol (DTE) was added to cells and to all purification buffers. 5 ml NiNTA beads were added to the cleared lysate. NiNTA elution fraction was further purified by SEC (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 1.5 mM TCEP). Appropriate fractions were pooled and concentrated to approximately 15 mg ml−1 (calculated ε: Trx-PCP1: 22
910 M−1 cm−1 was used to determine protein concentration). Trx-PCP1, Trx-PCP2 and Trx-PCP6: 2 mM dithioerythritol (DTE) was added to cells and to all purification buffers. 5 ml NiNTA beads were added to the cleared lysate. NiNTA elution fraction was further purified by SEC (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 1.5 mM TCEP). Appropriate fractions were pooled and concentrated to approximately 15 mg ml−1 (calculated ε: Trx-PCP1: 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 234.4 M−1 cm−1, Trx-PCP2: 22
234.4 M−1 cm−1, Trx-PCP2: 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 607 M−1 cm−1 and Trx-PCP6: 20
607 M−1 cm−1 and Trx-PCP6: 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970 M−1 cm−1 were used to determine protein concentration). PanK: 4 ml NiNTA beads were added to the cleared lysate. NiNTA elution fraction was further purified by SEC (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2). Appropriate fractions were concentrated to 13 mg ml−1 (ε = 45
970 M−1 cm−1 were used to determine protein concentration). PanK: 4 ml NiNTA beads were added to the cleared lysate. NiNTA elution fraction was further purified by SEC (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2). Appropriate fractions were concentrated to 13 mg ml−1 (ε = 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 380 M−1 cm−1 was used to determine protein concentration). PPAT: 4 ml NiNTA beads were added to the cleared lysate. Elution fraction was dialysed against 50 mM Tris·HCl, pH 8.0, 150 mM NaCl and further purified by SEC (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2). Appropriate fractions were concentrated to 20 mg ml−1 (ε = 8480 M−1 cm−1 was used to determine protein concentration). GB1-PCP7tei and GB1-PCP7cep were purified according to Haslinger and Peschke et al.36
380 M−1 cm−1 was used to determine protein concentration). PPAT: 4 ml NiNTA beads were added to the cleared lysate. Elution fraction was dialysed against 50 mM Tris·HCl, pH 8.0, 150 mM NaCl and further purified by SEC (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2). Appropriate fractions were concentrated to 20 mg ml−1 (ε = 8480 M−1 cm−1 was used to determine protein concentration). GB1-PCP7tei and GB1-PCP7cep were purified according to Haslinger and Peschke et al.36
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) for 1.5 hours with gentle shaking, the peptide precipitated by addition of cold diethyl ether, pelleted using centrifugation before being taken up in a solution of 1
3) for 1.5 hours with gentle shaking, the peptide precipitated by addition of cold diethyl ether, pelleted using centrifugation before being taken up in a solution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 MeCN/water and lyophilised to afford a white powder. Analysis was performed using an XBridge BEH300 C18 column (5 μm, 4.6 × 250 mm) at 1 ml min−1 (gradient: 5% MeCN for 4 minutes, 5–75% MeCN in 21 minutes, tR = 12.4 minutes). HRMS (ESI): m/z calculated for C28H37N4O8S [M − H]− 589.2338, found 589.2332 (Δm = 1 ppm).
1 MeCN/water and lyophilised to afford a white powder. Analysis was performed using an XBridge BEH300 C18 column (5 μm, 4.6 × 250 mm) at 1 ml min−1 (gradient: 5% MeCN for 4 minutes, 5–75% MeCN in 21 minutes, tR = 12.4 minutes). HRMS (ESI): m/z calculated for C28H37N4O8S [M − H]− 589.2338, found 589.2332 (Δm = 1 ppm).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 MeCN/water and lyophilised to afford a white powder. This was purified by preparative HPLC on a Waters Xbridge BEH300 Prep C18 column (5 μm, 19 × 150 mm) at 20 ml min−1 (LCMS-2020, Shimadzu, gradient: 5% MeCN (acetonitrile) for 5 minutes, 5–35% MeCN in 30 minutes). H-(D)-Hpg-(D)-ClTyr–NH–Me (0.8 mg; yield% = 4.2%); MS (ESI): m/z calculated for C28H37N4O8S [M + H]+ 378.11, found 378.10. H-(D)-Hpg-(L)-ClTyr–NH–Me (1.2 mg; yield% = 6.3%); MS (ESI): m/z calculated for C28H37N4O8S [M + H]+ 378.11, found 378.10. HPLC analysis was performed using an XBridge BEH300 C18 column (5 μm, 4.6 × 250 mm) at 1 ml min−1 (gradient: 5% MeCN for 4 minutes, 5–65% MeCN in 30 minutes, (D,D)-dipeptide tR = 9.85 minutes, (D,L)-dipeptide tR = 10.20 minutes).
1 MeCN/water and lyophilised to afford a white powder. This was purified by preparative HPLC on a Waters Xbridge BEH300 Prep C18 column (5 μm, 19 × 150 mm) at 20 ml min−1 (LCMS-2020, Shimadzu, gradient: 5% MeCN (acetonitrile) for 5 minutes, 5–35% MeCN in 30 minutes). H-(D)-Hpg-(D)-ClTyr–NH–Me (0.8 mg; yield% = 4.2%); MS (ESI): m/z calculated for C28H37N4O8S [M + H]+ 378.11, found 378.10. H-(D)-Hpg-(L)-ClTyr–NH–Me (1.2 mg; yield% = 6.3%); MS (ESI): m/z calculated for C28H37N4O8S [M + H]+ 378.11, found 378.10. HPLC analysis was performed using an XBridge BEH300 C18 column (5 μm, 4.6 × 250 mm) at 1 ml min−1 (gradient: 5% MeCN for 4 minutes, 5–65% MeCN in 30 minutes, (D,D)-dipeptide tR = 9.85 minutes, (D,L)-dipeptide tR = 10.20 minutes).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.004 ratio of PCP domain, CoA-Tyr and Sfp R4-4 (ref. 65) respectively (10 min, RT, 25 mM Tris, pH 7.4, 5 mM MgCl2, 1 mM DTE). For hexa- and heptapeptide-CoAs41,42 ratio of 1
0.004 ratio of PCP domain, CoA-Tyr and Sfp R4-4 (ref. 65) respectively (10 min, RT, 25 mM Tris, pH 7.4, 5 mM MgCl2, 1 mM DTE). For hexa- and heptapeptide-CoAs41,42 ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 was used (1 hour, 30 °C). Dipeptide substrates were loaded to Trx-PCP2-domain in a modified one pot loading reaction.65,66 100 μM Trx-PCP2, 2.5 μM PanK and 300 μM pantetheine-dipeptide were mixed in loading buffer (100 mM Tris·HCl, pH 7.4, 5 mM MgCl2, 1 mM DTE). Reaction was started by addition on 1 mM ATP (5 minutes, RT). 10 μM PPAT were added and reaction incubated for another 10 minutes followed by addition of 0.1 U ml−1 of alkaline phosphatase (10 minutes, M0290, Roche). Finally, the substrate was loaded to Trx-PCP2 domain using 3 μM Sfp R4-4 (15 minutes).
10 was used (1 hour, 30 °C). Dipeptide substrates were loaded to Trx-PCP2-domain in a modified one pot loading reaction.65,66 100 μM Trx-PCP2, 2.5 μM PanK and 300 μM pantetheine-dipeptide were mixed in loading buffer (100 mM Tris·HCl, pH 7.4, 5 mM MgCl2, 1 mM DTE). Reaction was started by addition on 1 mM ATP (5 minutes, RT). 10 μM PPAT were added and reaction incubated for another 10 minutes followed by addition of 0.1 U ml−1 of alkaline phosphatase (10 minutes, M0290, Roche). Finally, the substrate was loaded to Trx-PCP2 domain using 3 μM Sfp R4-4 (15 minutes).
Excess of substrate was removed after all loading reactions in centrifugal concentrators using serial concentrating-diluting steps (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MWCO, 0.5 ml, Merck Millipore). Loaded PCP domains were used as quickly as possible for assays to prevent substrate hydrolysis.
000 MWCO, 0.5 ml, Merck Millipore). Loaded PCP domains were used as quickly as possible for assays to prevent substrate hydrolysis.
| Compound | SIM | MRM |
|---|---|---|
| Tyr | 182 ⇒ 91 | |
| Me–NH–Tyr | 195 ⇒ 91 | |
| Cl-Tyr | 216 ⇒ 134 | |
| MeNH–Cl–Tyr | 229 ⇒ 170 | |
| Tyr–Hpg | 331 (+) | |
| MeNH–Tyr–Hpg | 344 (+) | |
| Cl–Tyr–Hpg | 365 | |
| MeNH–Cl–Tyr–Hpg | 378 | |
| Type I hexapeptide (V6P) | 868 (−) | |
| MeNH–V6P | 881 (−) | |
| Cl–V6P | 902 (−) 904 (+) | |
| MeNH–Cl–V6P | 917 (+) | |
| Di-Cl-V6P | 938 (+), 936 (−) | |
| MeNH-di-Cl-V6P | 950 (+), 949 (−) | |
| Type I heptapeptide (V7P) | 1017 (−) | |
| MeNH-T7P | 1030 (−) | |
| Cl–V7P | 1053 (+), 1051 (−) | |
| MeNH–Cl–V7P | 1066 (+), 1064 (−) | |
| Di-Cl-V7P | 1087 (+), 1085 (−) | |
| MeNH-di-Cl-V7P | 1100 (+), 1098 (−) | |
| Type IV heptapeptide (T7P) | 1088.5 (−) | |
| MeNH-T7P | 1101.4 (−) | |
| Cl-T7P | 1124.4 (+), 1122.5 (−) | |
| MeNH–Cl-T7P | 1137.4 (+), 1135.5 (−) | |
| Di-Cl-T7P | 1158.4 (+), 1156.4 (−) | |
| MeNH-di-Cl-T7P | 1171 (+), 1169 (−) |
Acknowledgements
G. Stier (BZH-Heidelberg) for the thioredoxin fusion protein vector; Prof. P. Macheroux (TU Graz) for the ycnD expression plasmid, J. Yin (University of Chicago) for the R4-4 Sfp expression plasmid; D. Iftime and A. Kulik for fermentation of the mutant strain; Alexa Koch and Philipp Walch for assistance in cloning and protein preparation; to Clara Brieke for providing the hexapeptide-CoAs and heptapeptide-CoAs; Melanie Müller for mass spectral analysis; and to Christopher Roome for IT support. T. K. is grateful for the support of the Deutsche Akademischer Austausch Dienst (DAAD Graduate School Scholarship Program), M. J. C. is grateful for the support of the Deutsche Forschungsgemeinschaft (Emmy-Noether Program, CR 392/1-1), Monash University and the EMBL Australia program. This research was supported under Australian Research Council's Discovery Projects funding scheme (project number DP170102220) to MJC. This work has further been supported by the Universities Australia – DAAD 2016 Australia – Germany Joint Research Co-operation Scheme (Award ID 16679401) jointly awarded to ES and MJC. The Deutsche Forschungsgemeinschaft (DFG) supported ES and WW by the SFB 766 program TP-A03.References
- E. A. Felnagle, E. E. Jackson, Y. A. Chan, A. M. Podevels, A. D. Berti, M. D. McMahon and M. G. Thomas, Mol. Pharm., 2008, 5, 191–211 CrossRef CAS PubMed.
- R. D. Süssmuth and A. Mainz, Angew. Chem., Int. Ed., 2017, 56, 3770–3821 CrossRef PubMed.
- R. S. Al Toma, C. Brieke, M. J. Cryle and R. D. Süssmuth, Nat. Prod. Rep., 2015, 32, 1207–1235 RSC.
- A. M. Gulick, ACS Chem. Biol., 2009, 4, 811–827 CrossRef CAS PubMed.
- G. H. Hur, C. R. Vickery and M. D. Burkart, Nat. Prod. Rep., 2012, 29, 1074–1098 RSC.
- U. Linne and M. A. Marahiel, Biochemistry, 2000, 39, 10439–10447 CrossRef CAS PubMed.
- N. M. Gaudelli and C. A. Townsend, Nat. Chem. Biol., 2014, 10, 251–258 CrossRef CAS PubMed.
- L. Ray, K. Yamanaka and B. S. Moore, Angew. Chem., Int. Ed., 2016, 55, 364–367 CrossRef CAS PubMed.
- M. E. Horsman, T. P. A. Hari and C. N. Boddy, Nat. Prod. Rep., 2016, 33, 183–202 RSC.
- G. Yim, M. N. Thaker, K. Koteva and G. Wright, J. Antibiot., 2014, 67, 31–41 CrossRef CAS PubMed.
- D. Bischoff, B. Bister, M. Bertazzo, V. Pfeifer, E. Stegmann, G. J. Nicholson, S. Keller, S. Pelzer, W. Wohlleben and R. D. Süssmuth, ChemBioChem, 2005, 6, 267–272 CrossRef CAS PubMed.
- E. Stegmann, H.-J. Frasch and W. Wohlleben, Curr. Opin. Microbiol., 2010, 13, 595–602 CrossRef CAS PubMed.
- K. C. Nicolaou, C. N. C. Boddy, S. Braese and N. Winssinger, Angew. Chem., Int. Ed., 1999, 38, 2096–2152 CrossRef PubMed.
- J. R. Pinchman and D. L. Boger, J. Med. Chem., 2013, 56, 4116–4124 CrossRef CAS PubMed.
- J. R. Pinchman and D. L. Boger, Bioorg. Med. Chem. Lett., 2013, 23, 4817–4819 CrossRef CAS PubMed.
- C. M. Harris, R. Kannan, H. Kopecka and T. M. Harris, J. Am. Chem. Soc., 1985, 107, 6652–6658 CrossRef CAS.
- O. Puk, P. Huber, D. Bischoff, J. Recktenwald, G. Jung, R. D. Süssmuth, K.-H. van Pée, W. Wohlleben and S. Pelzer, Chem. Biol., 2002, 9, 225–235 CrossRef CAS PubMed.
- J. Pootoolal, M. G. Thomas, C. G. Marshall, J. M. Neu, B. K. Hubbard, C. T. Walsh and G. D. Wright, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 8962–8967 CrossRef CAS PubMed.
- E. Yeh, L. C. Blasiak, A. Koglin, C. L. Drennan and C. T. Walsh, Biochemistry, 2007, 46, 1284–1292 CrossRef CAS PubMed.
- C. Dong, S. Flecks, S. Unversucht, C. Haupt, K.-H. van Pée and J. H. Naismith, Science, 2005, 309, 2216–2219 CrossRef CAS PubMed.
- S. Keller, T. Wage, K. Hohaus, M. Hölzer, E. Eichhorn and K.-H. van Pée, Angew. Chem., Int. Ed., 2000, 39, 2300–2302 CrossRef CAS PubMed.
- E. Yeh, S. Garneau and C. T. Walsh, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 3960–3965 CrossRef CAS PubMed.
- S. A. Shepherd, C. Karthikeyan, J. Latham, A.-W. Struck, M. L. Thompson, B. R. K. Menon, M. Q. Styles, C. Levy, D. Leys and J. Micklefield, Chem. Sci., 2015, 6, 3454–3460 RSC.
- P. C. Dorrestein, E. Yeh, S. Garneau-Tsodikova, N. L. Kelleher and C. T. Walsh, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 13843–13848 CrossRef CAS PubMed.
- S. Lin, S. G. Van Lanen and B. Shen, J. Am. Chem. Soc., 2007, 129, 12432–12438 CrossRef CAS PubMed.
- B. Hadatsch, D. Butz, T. Schmiederer, J. Steudle, W. Wohlleben, R. Süssmuth and E. Stegmann, Chem. Biol., 2007, 14, 1078–1089 CrossRef CAS PubMed.
- D. Bischoff, S. Pelzer, A. Holtzel, G. J. Nicholson, S. Stockert, W. Wohlleben, G. Jung and R. D. Sussmuth, Angew. Chem., Int. Ed., 2001, 40, 1693–1696 CrossRef CAS PubMed.
- D. Bischoff, S. Pelzer, B. Bister, G. J. Nicholson, S. Stockert, M. Schirle, W. Wohlleben, G. Jung and R. D. Sussmuth, Angew. Chem., Int. Ed., 2001, 40, 4688–4691 CrossRef CAS PubMed.
- R. D. Sussmuth, S. Pelzer, G. Nicholson, T. Walk, W. Wohlleben and G. Jung, Angew. Chem., Int. Ed., 1999, 38, 1976–1979 CrossRef CAS.
- O. Puk, D. Bischoff, C. Kittel, S. Pelzer, S. Weist, E. Stegmann, R. D. Süssmuth and W. Wohlleben, J. Bacteriol., 2004, 186, 6093–6100 CrossRef CAS PubMed.
- S. Uhlmann, R. D. Süssmuth and M. J. Cryle, ACS Chem. Biol., 2013, 8, 2586–2596 CrossRef CAS PubMed.
- K. Haslinger, C. Brieke, S. Uhlmann, L. Sieverling, R. D. Süssmuth and M. J. Cryle, Angew. Chem., Int. Ed., 2014, 53, 8518–8522 CrossRef CAS PubMed.
- P. C. Schmartz, K. Wölfel, K. Zerbe, E. Gad, E. S. El Tamany, H. K. Ibrahim, K. Abou-Hadeed and J. A. Robinson, Angew. Chem., Int. Ed., 2012, 51, 11468–11472 CrossRef CAS PubMed.
- V. Ulrich, M. Peschke, C. Brieke and M. Cryle, Mol. BioSyst., 2016, 12, 2992–3004 RSC.
- M. Peschke, K. Haslinger, C. Brieke, J. Reinstein and M. Cryle, J. Am. Chem. Soc., 2016, 138, 6746–6753 CrossRef CAS PubMed.
- K. Haslinger, M. Peschke, C. Brieke, E. Maximowitsch and M. J. Cryle, Nature, 2015, 521, 105–109 CrossRef CAS PubMed.
- C. Brieke, M. Peschke, K. Haslinger and M. J. Cryle, Angew. Chem., Int. Ed., 2015, 54, 15715–15719 CrossRef CAS PubMed.
- M. Peschke, M. Gonsior, R. D. Süssmuth and M. J. Cryle, Curr. Opin. Struct. Biol., 2016, 41, 46–53 CrossRef CAS PubMed.
- M. Peschke, C. Brieke, R. J. A. Goode, R. B. Schittenhelm and M. J. Cryle, Biochemistry, 2017, 56, 1239–1247 CrossRef CAS PubMed.
- P. C. Schmartz, K. Zerbe, K. Abou-Hadeed and J. A. Robinson, Org. Biomol. Chem., 2014, 12, 5574–5577 CAS.
- C. Brieke, V. Kratzig, K. Haslinger, A. Winkler and M. J. Cryle, Org. Biomol. Chem., 2015, 13, 2012–2021 CAS.
- C. Brieke, M. Peschke, K. Haslinger and M. J. Cryle, Angew. Chem., Int. Ed., 2015, 54, 15715–15719 CrossRef CAS PubMed.
- S. Pelzer, W. Reichert, M. Huppert, D. Heckmann and W. Wohlleben, J. Biotechnol., 1997, 56, 115–128 CrossRef CAS PubMed.
- S. Pelzer, R. Süssmuth, D. Heckmann, J. Recktenwald, P. Huber, G. Jung and W. Wohlleben, Antimicrob. Agents Chemother., 1999, 43, 1565–1573 CAS.
- J. Recktenwald, R. Shawky, O. Puk, F. Pfennig, U. Keller, W. Wohlleben and S. Pelzer, Microbiology, 2002, 148, 1105–1118 CrossRef CAS PubMed.
- V. Weichold, D. Milbredt and K.-H. van Pée, Angew. Chem., Int. Ed., 2016, 55, 6374–6389 CrossRef CAS PubMed.
- M. Sosio, S. Stinchi, F. Beltrametti, A. Lazzarini and S. Donadio, Chem. Biol., 2003, 10, 541–549 CrossRef CAS PubMed.
- H.-T. Chiu, B. K. Hubbard, A. N. Shah, J. Eide, R. A. Fredenburg, C. T. Walsh and C. Khosla, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 8548–8553 CrossRef CAS PubMed.
- S. Stinchi, L. Carrano, A. Lazzarini, M. Feroggio, A. Grigoletto, M. Sosio and S. Donadio, FEMS Microbiol. Lett., 2006, 256, 229–235 CrossRef CAS PubMed.
- W. Wohlleben, E. Stegmann and R. D. Süssmuth, Methods Enzymol., 2009, 458, 459–486 CAS.
- F. Beltrametti, A. Lazzarini, C. Brunati, A. Marazzi, S. Jovetic, E. Selva and F. Marinelli, J. Antibiot., 2003, 56, 773–782 CrossRef CAS PubMed.
- M. N. Thaker, W. Wang, P. Spanogiannopoulos, N. Waglechner, A. M. King, R. Medina and G. D. Wright, Nat. Biotechnol., 2013, 31, 922–927 CrossRef CAS PubMed.
- X. Yin, Y. Chen, L. Zhang, Y. Wang and T. M. Zabriskie, J. Nat. Prod., 2010, 73, 583–589 CrossRef CAS PubMed.
- J.-S. Chen, M. Su, L. Shao, Y.-X. Wang, H.-M. Lin and D.-J. Chen, Appl. Microbiol. Biotechnol., 2016, 100, 289–298 CrossRef CAS PubMed.
- T. J. Wadzinski, K. D. Gea and S. J. Miller, Bioorg. Med. Chem. Lett., 2016, 26, 1025–1028 CrossRef CAS PubMed.
- Y. Nakama, O. Yoshida, M. Yoda, K. Araki, Y. Sawada, J. Nakamura, S. Xu, K. Miura, H. Maki and H. Arimoto, J. Med. Chem., 2010, 53, 2528–2533 CrossRef CAS PubMed.
- G. Yim, W. Wang, M. N. Thaker, S. Tan and G. D. Wright, ACS Infect. Dis., 2016, 2, 642–650 CrossRef CAS PubMed.
- W. O. Bullock, J. M. Fernandez and J. M. Short, BioTechniques, 1987, 5, 376–378 CAS.
- S. R. Nadkarni, M. V. Patel, S. Chatterjee, E. K. Vijayakumar, K. R. Desikan, J. Blumbach, B. N. Ganguli and M. Limbert, J. Antibiot., 1994, 47, 334–341 CrossRef CAS PubMed.
- J. Sambrook and D. W. Russel, Molecular Cloning - A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York, 2001 Search PubMed.
- T. Kieser, M. J. Bibb, M. J. Buttner, K. F. Chater and D. A. Hopwood, Practical Streptomycetes Genetics, The John Innes Foundation, Norwich, 2000 Search PubMed.
- C. J. Thompson, J. M. Ward and D. A. Hopwood, J. Bacteriol., 1982, 151, 668–677 CAS.
- J. Bogomolovas, B. Simon, M. Sattler and G. Stier, Protein Expression Purif., 2009, 64, 16–23 CrossRef CAS PubMed.
- A. Morokutti, A. Lyskowski, S. Sollner, E. Pointner, T. B. Fitzpatrick, C. Kratky, K. Gruber and P. Macheroux, Biochemistry, 2005, 44, 13724–13733 CrossRef CAS PubMed.
- M. Sunbul, N. J. Marshall, Y. Zou, K. Zhang and J. Yin, J. Mol. Biol., 2009, 387, 883–898 CrossRef CAS PubMed.
- K. M. Clarke, A. C. Mercer, J. J. La Clair and M. D. Burkart, J. Am. Chem. Soc., 2005, 127, 11234–11235 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Southern blot analyses of wildtype and the truncation mutants CK2.1 and CK2.2; LC-MS analysis of dipeptides from in vivo expression experiments together with authentic standards; SDS-PAGE and spectral analysis of Tcp21 and BhaA halogenase enzymes. See DOI: 10.1039/c7sc00460e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |