Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The in vivo hepato-recovery effects of the polyphenol-rich fermented food Xeniji™ on ethanol-induced liver damage
Noraisyah Zulkawi†
a,
Kam Heng Ng†a,
Rizi Zamberi†bc,
Swee Keong Yeap d,
Indu Bala Jaganathc,
Dilan Satharasinghee,
Chean Yeah Yongb,
Anisah Binti Jamaluddinc,
Sheau Wei Tanb,
Wan Yong Hof,
Noorjahan Banu Alitheenbg and
Kamariah Long
d,
Indu Bala Jaganathc,
Dilan Satharasinghee,
Chean Yeah Yongb,
Anisah Binti Jamaluddinc,
Sheau Wei Tanb,
Wan Yong Hof,
Noorjahan Banu Alitheenbg and
Kamariah Long *c
*c
aElken Sdn Bhd, 20, Bangunan Elken, Jalan 1/137C, Batu 5, Jalan Kelang Lama, 58000 Kuala Lumpur, Malaysia
bInstitute of Bioscience, Universiti Putra Malaysia, Serdang, Selangor, Malaysia
cMalaysian Agricultural Research and Development Institute (MARDI), Serdang, Selangor 43400, Malaysia. E-mail: amai@mardi.gov.my; Tel: +60-3-89537238
dXiamen University Malaysia, Jalan Sunsuria, Bandar Sunsuria, 43900 Sepang, Selangor Darul Ehsan, Malaysia
eDepartment of Basic Veterinary Sciences, Faculty of Veterinary Medicine & Animal Science, University of Peradeniya, Peradeniya 20400, Sri Lanka
fSchool of Biomedical Sciences, The University of Nottingham Malaysia Campus, Jalan Broga, 43500 Semenyih, Selangor, Malaysia
gDepartment of Cell and Molecular Biology, Faculty of Biotechnology and Biomolecular Science, Universiti Putra Malaysia, Serdang, Selangor 43400, Malaysia
First published on 3rd August 2017
Abstract
Xeniji is a health food product produced by bacterial lactic acid fermentation. Although fermented foods, including Xeniji, have been widely consumed for various health purposes, including as an antioxidant to improve liver disease, their polyphenol content and in vivo hepato-recovery effects have yet to be evaluated. This study aims to evaluate the polyphenol content of Xeniji and its in vivo hepato-recovery effect on ethanol-induced liver damage. In this study, the polyphenolic acids of Xeniji are quantified by liquid chromatography-mass spectrometry (LCMS). In addition, the recovery effect of Xeniji on ethanol-induced liver damage in mice is evaluated by assessing the serum liver enzyme profile, liver inflammation, liver oxidative stress, and the level of CYP2E1. Xeniji is recorded to contain a high concentration of caffeoylquinic acid and sakuranetin, based on the LCMS-MS quantification results. In terms of the in vivo hepato-recovery study, intake of ethanol induces substantial liver damage indicated by an increase of the serum liver enzyme profile, liver inflammation, liver oxidative stress and the level of CYP2E1. On the other hand, Xeniji promotes recovery from ethanol-induced liver damage by restoring antioxidant levels, enhancing the metabolism of ethanol in the liver and suppressing inflammation in a dosage-dependent manner. Xeniji is a fermented functional food that possesses hepato-recovery effects on ethanol-induced liver damage.
1 Introduction
Ethanol is a natural product that is present in certain beverages that have been commonly consumed by human beings for thousands of years.1 Although it is a common beverage ingredient, ethanol abuse has been identified as one of the major causes of liver disease, because it contributes to the generation of oxidative stress in the liver.2 The liver is an organ with a wide range of functions, including the detoxification of metabolites, drugs, and xenobiotics.2 Cytochrome P450, ethanol dehydrogenase (ADH) and catalase in the liver are the major enzymes that metabolise ethanol to acetaldehyde.2 Subsequently, acetaldehyde is metabolised to acetate by aldehyde dehydrogenase.2 During ethanol metabolism, excessive reactive oxygen species (ROS) are generated as a by-product, leading to a disturbance of the homeostasis of antioxidant defences in the liver.3 Exhaustion of antioxidants in the liver contributes to lipid peroxidation, indicated by the formation of compounds such as malondialdehyde (MDA).3 The presence of MDA and acetaldehyde also leads to the formation of a stable MDA-acetaldehyde-protein adduct (MAA), which induces a pro-inflammatory response in the liver.3 Both oxidative stress and chronic inflammation consequently promote liver damage, as indicated by an increased serum liver enzyme profile and liver cell death.4 Chronic ethanol abuse may subsequently be associated with morbidity and mortality.2The use of drugs in the treatment of ethanol-induced liver damage is still limited by the complications associated with their long-term use.4 Thus, the use of natural products with known bioactive agents and better safety profiles has been proposed as a better alternative for treating liver disease.4 Previous studies have shown that administration of herbal foods that are rich in antioxidants, including polyphenols, was able to promote recovery from ethanol-induced liver damage by restoring liver antioxidant levels and suppressing liver inflammation.1 Fermented foods have been identified as a good source of dietary antioxidants.5 Besides prolonging the shelf-life of foods and removing anti-nutrients, fermentation using lactic acid bacteria6 and yeast7 has also been reported to have benefits including enhancing the polyphenol and vitamin content of foods.8 There are various lactic acid bacteria fermented food products that are consumed as health supplements.9 For example, plant-based pastes produced by fermenting fruits, vegetables, wild herbs, mushrooms, seaweed and cereals with lactic acid bacteria have been reported to be rich in amino acids, vitamins and organic acids.9 In addition, they also possess various in vitro bioactivities, including antioxidant, anti-hypertensive, antibacterial, anti-inflammatory and anti-tyrosinase effects.9 Besides plant-based pastes, Xeniji is another lactic acid bacteria fermented food product that is rich in β-carotene, polyphenols, citric acid and essential amino acids, contributed by the lactic acid bacteria fermentation of the plant-based ingredients (Table 1).10 These nutrients have contributed to enhanced antioxidant activity and immunity in normal healthy mice in vivo without causing acute or subchronic toxicity.10 The nutritional value of Xeniji, particularly its high total antioxidant capacity and enhancement of in vivo antioxidant activity,10 shows its potential as a functional food to overcome alcohol-induced liver disease. However, the phenolic acid profile and the benefits of these nutrients, particularly for liver protection effects, have not been evaluated, although Xeniji is widely consumed as a health supplement.10 Thus, the phenolic acid content of Xeniji was quantified by LC-MS. In addition, the hepato-recovery effect of Xeniji on ethanol-induced liver damage in mice was also tested in this study.
| Material name | Content | |
|---|---|---|
| Sugar | Brown sugar, galacto-oligosaccharide (GOS) and oligosaccharide | 67.2% |
| Fruits | Prunus domestica L. (Prune), Fragaria x ananassa (Strawberry), Malus domestica (Apple), Vitis pione (Grape), Prunus persica (Peach), Citrus unshiu (Mandarin orange), Mulberry, Cherry blossom paste, Citrus junos (Yuzu), Diospyros kaki (Persimmon), Actinidia chinensis (Kiwi), Fortunella japonica (Kumquat), Citrus limon (Lemon), Vaccinium corymbosum (Blueberry), Myrica rubra (Artubus), Pyrus pyrifola (Pear), Prunus mume (Ume), Citrus iyo (Iyo-orange), Ficus carica (Fig), Rubus buergeri (Raspberry) and Rubus fruticosus (Blackberry) | 18.0% |
| Vegetables and wild herbs | Angelica keiskei (Folium) (Angelica keiskei leaf powder), Perilla frutescens (Perilla), Cucurbita maxima (Pumpkin), Raphanus sativus (Japanese radish), Spinacia oleracea (Spinach), Daucus carota var. Sativus (Carrot), Brassica oleracea, Acephala (Kale), Hordeum vulgare L. (Barley grass), Corchorus olitorius (Jew's mallow), Lycopersicon esculentum (Tomato), Cucumis sativus (Cucumber), Plantago asiatica (Plantain), Sasa veitchii (Stripped bamboo), Equisetum arvense (Field horsetail), Eriobotrya japonica (Loquat leaf), Brassica oleracea var. Capitata (Cabbage), Solanum melongena (Eggplant), Apium graveolens var. Dulce (Celery), Capsicum annuum (Sweet pepper), Momordica charantia (Bitter melon), Brassica rapa chinensis (Bok Choi), Nelumbo nucifera (Radix) (Lotus root), Curcuma longa (Turmeric), Brassica oleracea var. Italica (Broccoli), Zingiber officinale (Ginger), Petroselinum crispum cv (Parsley), Asparagus officinalis var. Altilis (Asparagus) and Oenanthe stolonifera (Japanese Parsley) | 7.4% |
| Mushrooms | Ganoderma lucidum (Reishi), Lentinula edodes (Shiitake mushroom), Auricularia polytricha (Jew's ear), Grifola frondosa (Maitake mushroom) | 1.3% |
| Seaweed | Ascophyllum nodosum (Kelp), Laminaria japonica Areschoug (Kombu), Undaria pinnatifida suringer (Wakame), Fucus evanescens (Fucus), Sargassum fusiforme setchell (Hijiki) | 1.6% |
| Pulse and cereals | Glycine max (Soybean), Theobroma cacao (Cocoa), Zea mays L. (Sweet Corn), Oryza sativa (Rice) | 4.4% |
| Lactic acid bacteria species | Lactobacillus brevis, Lactobacillus casei, Lactobacillus curvatus, Lactobacillus paracasei, Lactobacillus pentosus, Lactobacillus plantarum, Lactococcus lactis, Leuconostoc mesenteroides, Pediococcus acidilactici, Pediococcus pentosaceus | 0.1% |
2 Materials and methods
2.1 Preparation of Xeniji
Commercially available Xeniji™ was provided by Elken Sdn Bhd, Malaysia. It was prepared by fermenting multiple food ingredients using commercial lactic acid bacteria (Table 1)10 for 3 years and 6 months. Xeniji (10 ml) was weighed, mixed with 20 ml of methanol, vortexed for 15 minutes, centrifuged at 500 × g for 5 minutes and filtered with a 0.45 μm syringe filter (Merck Millipore, USA) to separate the supernatant from the residue. The same procedure was repeated twice with 10 ml of methanol. All the supernatants were mixed and stored at 4 °C. Prior to analysis, the supernatants were dried using a vacuum concentrator yielding a dry crude methanol extract.2.2 LC-MS quantification of phenolic acids
The crude Xeniji extract was reconstituted at 10 mg ml−1 and subjected to LCMS-MS in triplicate. The injection volume and the concentration of the extract were kept constant for quantification purposes. For optimal separation of phytochemicals, a high-performance liquid chromatography (HPLC) system (Agilent 1200 series) with an Agilent 1100LC Binary Pump set at 5801 psi was used with a Thermo-hyperseal Gold C18 column (150 × 4.6 mm). The reverse phase used was a C-18, 150 mm × 4.6 mm i.d. 5 μm particle size Thermo Hypersil GOLD column (Thermo Scientific, UK), while the composition of the mobile phase used was 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) with the following gradient for solvent B: 5% (5 min), 5–90% (30 min), 5% (5 min), at a flow rate of 1 ml min−1. The total injection volume was 20 μl and detection was performed at 280 and 360 nm with the column oven set at 28 °C. For phytochemical identification and quantification, a MS system was used to ionise the samples that were eluted from HPLC to generate charged molecules. In brief, a 3200 Qtrap ABsciex LCMS-MS was used with the following set parameters: (i) injection volume: 20 μl; (ii) range of ion detection: 50–1200 mg; (iii) scan type: EMS; (iv) ion source: turbo spray; (v) polarity: negative; (vi) voltage: −4500 V; (vii) scan rate: 1000 amu s−1; (viii) source temperature: 550 °C. These charged molecules generated an isotopic signature through the mass-to-charge ratios, which was then used to identify the polyphenols present in the samples.2.3 Animals
Male inbred Balb/c mice (n = 30, 6 weeks old) were purchased from the animal house at the Institute of Bioscience, Universiti Putra Malaysia (UPM). The animals were acclimatised for 2 weeks under standard conditions at 22 °C, with a 12 hour day/night light cycle, and were fed with distilled water and standard pellets ad libitum before starting the experiment. This work was approved by the Institutional Animal Care and Use Committee (IACUC), UPM (R045/2016) and was performed in accordance with the Guide for the Care and Use of Laboratory Animals prepared by the Institutional Animal Care and Use Committee (IACUC), UPM (Malaysia).2.4 Experimental design
The mice were randomly divided into 6 groups as stated below under standard conditions at 22 °C, with a 12 hour day/night light cycle, and were fed with distilled water and standard pellets ad libitum. Mice from groups 2 to 6 were fed with 5 g kg−1 body weight of ethanol (in 100 μl) once daily every afternoon using oral gavage for 21 days.11 Starting from day 7, the mice were orally fed with silybin or a different concentration of Xeniji using oral gavage every morning for 14 days as stated below. The time interval between the treatment and ethanol administration was 6 hours.Group 1 (n = 5): buffer only, healthy mice.
Group 2 (n = 5): ethanol (5 g kg−1) challenged + buffer.
Group 3 (n = 5): ethanol (5 g kg−1) challenged + silymarin (50 mg kg−1 BW).
Group 4 (n = 5): ethanol (5 g kg−1) challenged + Xeniji 0.1 g kg−1 BW.
Group 5 (n = 5): ethanol (5 g kg−1) challenged + Xeniji 1.0 g kg−1 BW.
Group 6 (n = 5): ethanol (5 g kg−1) challenged + Xeniji 2.0 g kg−1 BW.
At the end of the experimental period, all of the mice were anesthetized with isoflurane, and euthanized by cervical dislocation. Liver and serum samples were collected and subjected to the following assays.
2.5 Serum biomarker assays
Sera were collected for quantification of the following enzyme markers: aspartate aminotransferase (AST), alanine aminotransferase (ALT) and alkaline phosphatase (ALP). The assays were performed in a biochemical analyser (Hitachi 902 Automatic Analyser; Hitachi, Japan) with adapted reagents from Roche (Germany).2.6 Protein extraction
Total protein was extracted from 30 mg of fresh liver tissue using RIPA buffer (50 mM Tris pH 8, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) supplemented with a phosphatase inhibitor cocktail (Roche, Canada). The protein concentration was quantified using a Bradford protein assay (Life technology, USA) according to the manufacturer's protocol.2.7 Liver western blot analysis of CYP2E1
CYP2E1 protein expression levels were determined by western blotting using anti-cytochrome P450 2E1 rabbit polyclonal antibody (ab28146; Abcam, USA) with anti-β-actin mouse monoclonal antibody (ab8226; Abcam, USA) as a housekeeping control and Odyssey pre-stained molecular weight marker (LICOR, USA). Briefly, 100 μg of extracted protein was separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to a PVDF membrane (LICOR, USA) using a Pierce Fast Semi-Dry Blotter (Pierce, USA). The membrane was blocked with 1% Odyssey blocking buffer (LICOR, USA) for 2 h, washed with TBST buffer (1.5 M NaCl, 0.5 M Tris, pH 7.5) three times, and incubated with anti-cytochrome P450 2E1 rabbit polyclonal antibody (ab28146; Abcam, USA) and anti-β-actin mouse monoclonal antibody (ab8226; Abcam, USA). Next, the membrane was incubated with an IRDye 800CW Donkey anti-Rabbit IgG secondary antibody and IRDye 680RD Goat anti-Mouse IgG secondary antibody (LICOR, USA). The near-infrared signals were detected using a LICOR Odyssey Classic machine (LICOR, USA). The density results were analysed using Image Studio Software (LICOR, USA).2.8 Liver ADH and ALDH activity assays
Extracted and standardized protein was used for the quantification of ADH and ALDH activity. ADH and ALDH were quantified using an alcohol dehydrogenase activity assay kit and an aldehyde dehydrogenase activity colorimetric assay kit (Sigma, USA) according to the manufacturer's protocol.2.9 Liver antioxidant levels
The liver was excised from each mouse, washed with ice-cold phosphate buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), and weighed before being divided into several parts. One gram of the liver was mashed using a 0.2 μm cell strainer (SPL Life Sciences, China) and a syringe rubber plunger in 10 ml of cold phosphate-buffered saline (PBS) to obtain a liver homogenate. The liver homogenate was used to determine the levels of the ferric reducing ability of plasma (FRAP), superoxide dismutase (SOD), malondialdehyde (MDA), and nitric oxide (NO) according to previously described methods,1 while the GSH and ROS levels were determined with a Glutathione Assay Kit (Sigma-Aldrich, USA) and ROS detection kit (Cell Biolabs, USA) according to the manufacturers' protocols.2.10 Serum cytokine quantification
Serum collected from all mice was subjected to quantification of IL1β, IL-6 and TNF-α using ELISA kits (Biolegend, USA) according to the manufacturer's protocol.2.11 Liver gene expression using quantitative reverse transcriptase PCR (qRT-PCR)
The extracted livers (three biological replicates with three technical replicates) were stored in RNAlater solution (Life Technologies, USA) to preserve the RNA. The RNA was then extracted using an RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's protocol. Total RNA (1 μg) was reverse-transcribed to first-strand cDNA using a NEXscript cDNA synthesis kit (NEX Diagnostics, Korea) according to the manufacturer's protocol. Quantitative real-time PCR was performed with NEXpro qPCR Evagreen Master Mix (NEX Diagnostics, Korea). Primers for the following targets were designed and used: iNOS, NFkB,12 Nrf2, NQO-1, GSTA2, GCLM, and β-actin (Table 2).13 Quantitative PCR (qPCR) was performed using an Eco Real Time PCR system (Illumina, USA) with the following steps: 95 °C for 2 min, 40 cycles of 95 °C for 10 s, 60 °C for 45 s, and acquisition of the fluorescent signal. The specificity of the real-time PCR was confirmed by a melt curve analysis. Standard curves for each gene were generated to obtain the PCR efficiency for each primer. The expression of target genes in the treatment groups and control group was normalised using β-actin and the fold change in the expression of each target gene was calculated with Eco 48 Software (Illumina, USA) using the efficiency-corrected method.| Gene | Primer sequence (5′-3′) | |
|---|---|---|
| Forward | Reverse | |
| Nrf-2 | AGGACATGGAGCAAGTTTGG | TTCTTTTTCCAGCGAGGAGA |
| NQO1 | GGTAGCGGCTCCATGTACTC | CATCCTTCCAGGATCTGCAT |
| GCLM | AATCAGCCCCGATTTAGTCAGG | CCAGCTGTGCAACTCCAAGGAC |
| GSTA2 | CGCCACCAAATATGACCTCT | CCTGTTGCCCACAAGGTAGT |
| iNOS | GCACCGAGATTGGAGTTC | TAGTTACACCGACACGAG |
| NF-kB | CATTCTGACCTTGCCTATCT | TCTTACCTGTCTTGTCGTC |
| β-Actin | TCCTTCCTGGGCATGGAG | AGGAGGAGCAATGATCTTGATCTT |
2.12 Statistics
Means and standard deviations for three biological replicates, each with three technical replicates, were calculated using Excel for all measured parameters. One-way analysis of variance (ANOVA) was followed by a post-hoc Duncan test using SPSS version 20 and the results were used to compare groups for all assays with a single factor. Differences were considered significant at p < 0.05.3 Results
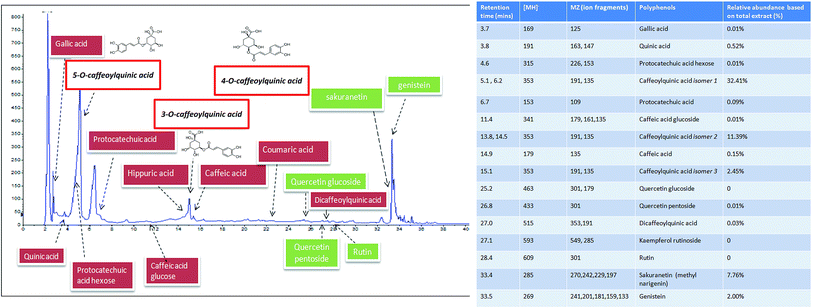
3.1 Phenolic acid profile of Xeniji
Based on LC-MS quantification, the most abundant polyphenols in Xeniji were 5-O-caffeoylquinic acid (32.41%), 3-O-caffeoylquinic acid (11.39%), 4-O-caffeoylquinic acid (2.45%), sakuranetin (7.76%) and quinic acid (0.52%). The other detected polyphenols were present at a level of less than 0.20% (Fig. 1).3.2 Serum biomarkers
Untreated ethanol-challenged mice had a higher level of serum ALT, ALP and AST enzymes (Table 3). On the other hand, Xeniji treatment significantly reduced these serum liver enzyme levels. Notably, administration of Xeniji at the dose of 2 g kg−1 body weight reduced the serum ALT, ALP and AST enzyme levels to close to those of healthy normal mice.| ALT (U/L) | ALP (U/L) | AST (U/L) | |
|---|---|---|---|
| Normal | 64.75 ± 5.80* | 91.67 ± 6.53* | 112.73 ± 6.26* |
| Untreated | 132.70 ± 1.38 | 151.33 ± 8.13 | 389.71 ± 2.86 |
| Silymarin | 88.20 ± 5.16* | 119.67 ± 14.00* | 398.51 ± 7.43 |
| Xeniji 0.1 | 81.40 ± 7.31* | 140.33 ± 7.51* | 301.99 ± 6.95* |
| Xeniji 1.0 | 80.10 ± 0.80* | 100.67 ± 2.02* | 282.28 ± 6.14* |
| Xeniji 2.0 | 78.53 ± 6.22* | 77.67 ± 9.78* | 82.73 ± 2.63* |
3.3 Liver histopathological analysis
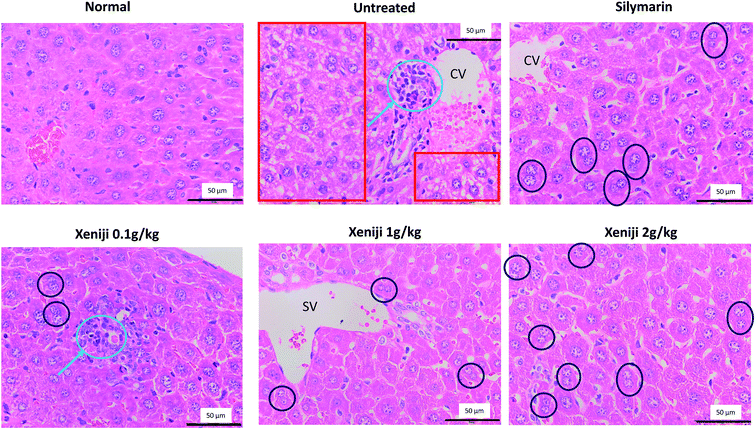
In the livers of normal mice, no abnormal appearance or histopathological change was observed. Unlike the normal mice, the livers of the untreated mice challenged with ethanol were observed to exhibit hepatocyte injury indicated by ballooning. In addition, inflammatory cell infiltration in the central vein was observed in the liver histology of the untreated and Xeniji 0.1 g kg−1 body weight-treated ethanol-challenged mice. On the other hand, the silymarin and Xeniji 1.0 and 2.0 g kg−1 body weight-treated mice were observed to exhibit binuclear hepatocytes (BNs), indicating that the treatments supported recovery from ethanol-induced liver damage by promoting the proliferation of hepatocytes (Fig. 2).3.4 Liver cytochrome P450 2E1 (CYP2E1) levels
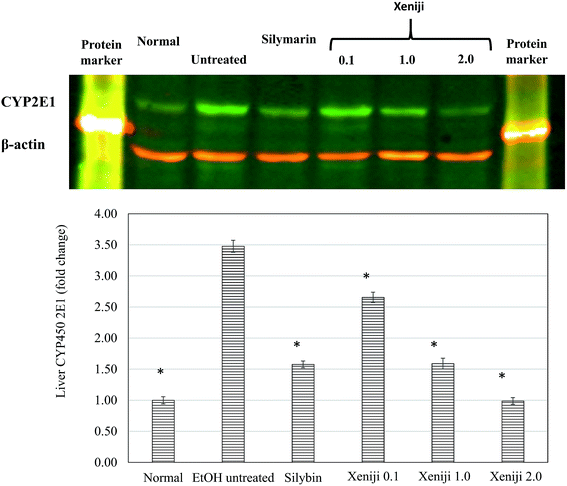
Based on the western blot analysis, Xeniji and silymarin-treated mice exhibited lower levels of CYP2E1 protein in the liver as compared to untreated ethanol-challenged mice in a dosage-dependent manner (Fig. 3).3.5 Liver ethanol dehydrogenase (ALDH) and aldehyde dehydrogenase (ADH) activities
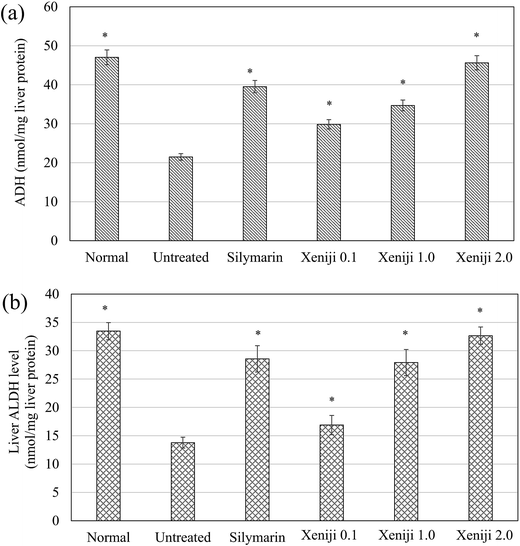
Untreated ethanol-challenged mice were observed to exhibit low levels of liver ADH (Fig. 4a) and ALDH (Fig. 4b) activity as compared to the normal, healthy control mice. In contrast, Xeniji and silymarin were able to restore the ADH (Fig. 4a) and ALDH (Fig. 4b) activities in the livers of the mice challenged with ethanol in a dosage-dependent manner.3.6 Liver antioxidant level
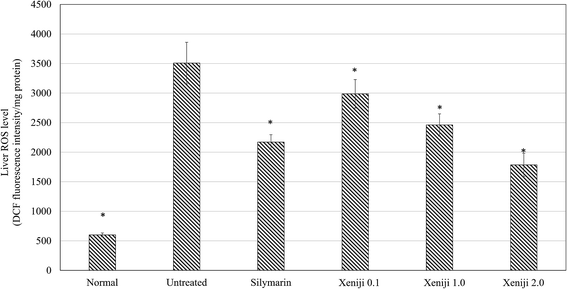
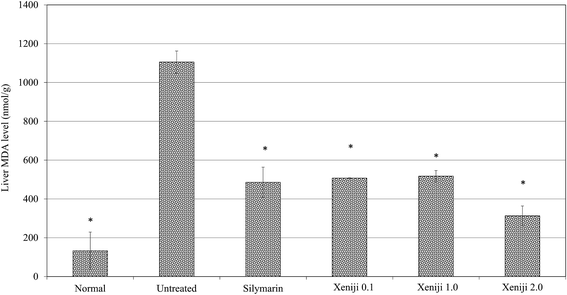
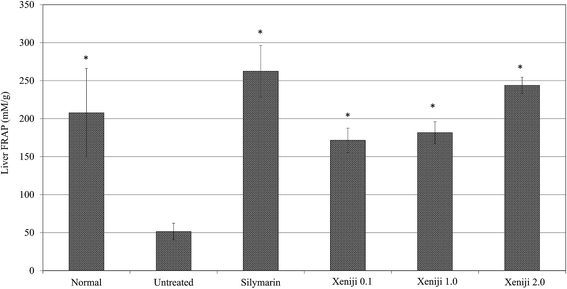
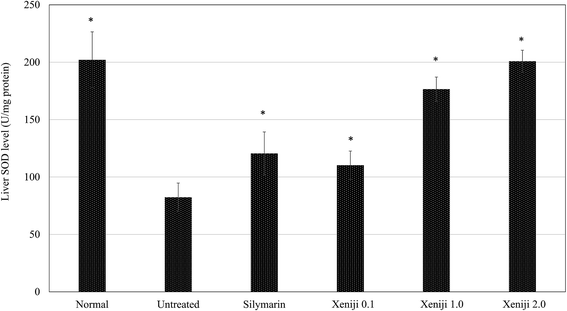
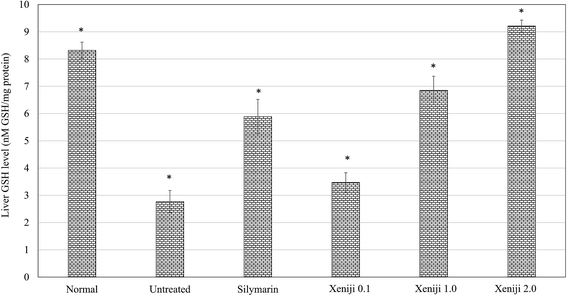
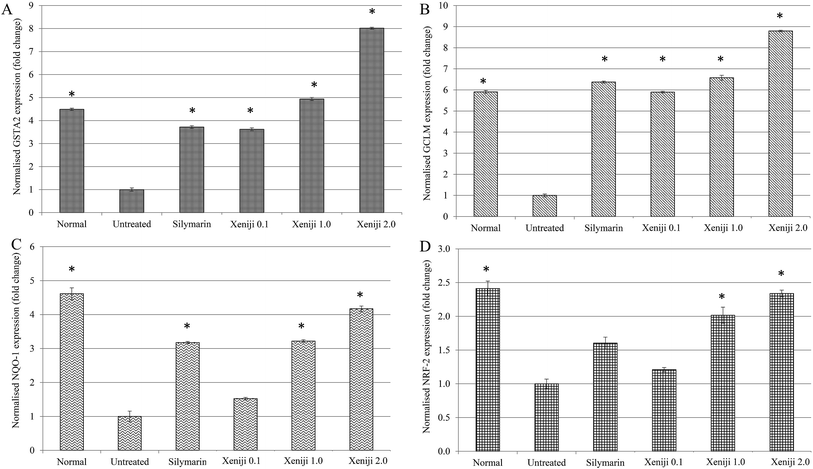
In this study, ethanol treatment caused reactive oxygen species (ROS) (Fig. 5) and lipid peroxidation malondialdehyde (MDA) (Fig. 6) accumulation associated with the down-regulation of the ferric reducing ability of plasma (FRAP), the total antioxidant capacity (Fig. 7), superoxide dismutase enzyme (SOD) activity (Fig. 8), and the glutathione (GSH) level (Fig. 9) in the livers of the untreated mice. On the other hand, the levels of ROS (Fig. 5) and MDA (Fig. 6) in the livers of the Xeniji and silymarin-treated mice were significantly reduced as compared to in the livers of the untreated mice. In addition, Xeniji and silymarin also restored the total antioxidant capacity (Fig. 7), SOD activity (Fig. 8) and GSH level (Fig. 9), which were depleted by the ROS generated from ethanol metabolism. Overall, amelioration of the antioxidant activity by Xeniji treatment was found to occur in a dosage-dependent manner.To understand the activation of antioxidant mechanisms by Xeniji, the regulation of nuclear factor-like 2 (Nrf2), glutamate-cysteine ligase modifier (GCLM), glutathione S-transferase alpha 2 (GSTA2) and NAD(P)H dehydrogenase (quinone 1) (NQO1) gene expression was evaluated by quantitative real time PCR (qRT-PCR) and the results are presented in Fig. 10. Only differential expressions with normalised fold changes of more than two as compared to the untreated group were considered significant. Expression of all evaluated genes was significantly higher (>2 fold) in the livers of normal mice than in untreated ethanol-challenged mice. In the mice treated with silymarin and 0.1 g kg−1 body weight of Xeniji, only the expression of GCLM, GSTA2 and NQO1 was significantly altered (>2 fold) when compared to the untreated ethanol-challenged mice. However, the expression of all evaluated genes in the liver was significantly greater (>2 fold) in the 1 and 2 g kg−1 body weight Xeniji-treated mice.
3.7 Pro-inflammatory cytokine level in the serum and nitric oxide level in the liver
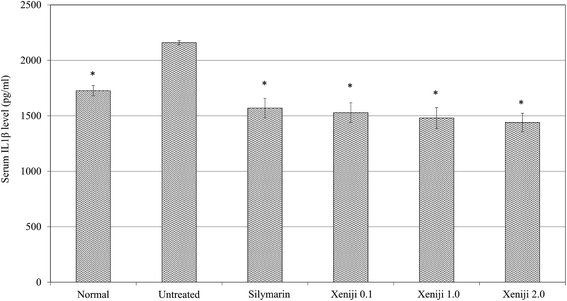
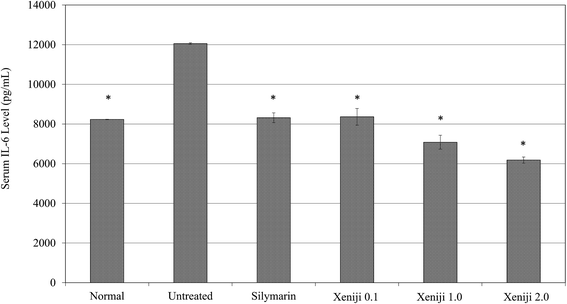
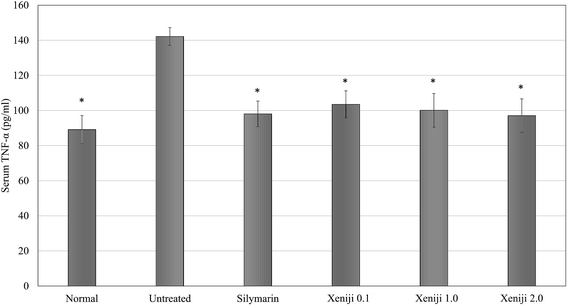
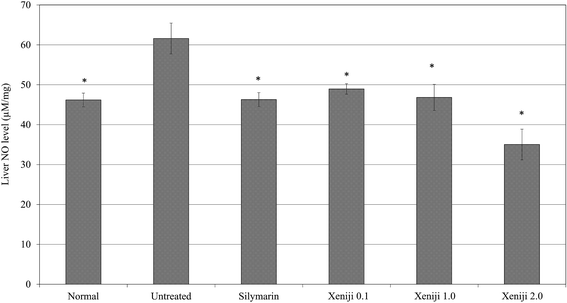
In terms of the anti-inflammatory effect of Xeniji, the levels of proinflammatory cytokines in the serum were evaluated using ELISA, while the levels of reactive nitrogen species and nitric oxide (NO) in the liver were evaluated using the Griess assay. The serum levels of IL-1β, IL-6 and TNF-α were higher in the untreated mice as compared to the normal, healthy group. Similar to the silymarin treatment, Xeniji significantly (p < 0.05) reduced the serum levels of the IL-1β (Fig. 11), IL-6 (Fig. 12) and TNF-α (Fig. 13) cytokines, but not in a dosage-dependent manner. In terms of the nitric oxide level in the liver, normal mice and all of the treated mice maintained significantly lower levels of NO in the liver as compared to the untreated mice. Only Xeniji at 2.0 g kg−1 body weight resulted in a significantly lower level of NO in the liver as compared to all other groups (Fig. 14).4 Discussion
Although ethanol is a common beverage, its chronic excessive use leads to various liver diseases including ethanol hepatitis, ethanolic cirrhosis and liver cancer.14 To date, ethanol-related liver disease remains an important public health problem worldwide.14 Ethanol has been identified as a hepatotoxin, and is the second leading cause of liver cirrhosis after the hepatitis C virus.14 Nutritional therapy is the first-line defence for ethanolic liver disease15 and a combination of pharmaceutical drugs with natural agents from food and herbs has been proposed as the optimal management for this disease.16 Thus, food ingredients, particularly those rich in antioxidant phenolic acids, have been proposed as potential functional foods for liver disease.17 The present investigation demonstrated that Xeniji promoted recovery from ethanol-induced liver damage in a dosage-dependent manner by restoring antioxidants and suppressing inflammation. Previous studies have reported that a plant-based paste fermented for 3 years was detected to have a high total phenol content, which contributed to its SOD-like antioxidant activity.9 Xeniji is a plant-based fermented food that contains high levels of total phenolics, citric acid, β-carotene and essential amino acids.10 These active nutrients enhance the in vivo antioxidant level in healthy mice without causing toxic side effects.10 In this study, the polyphenol profile that contributed to the total phenolic content of Xeniji was quantified by LC-MS-MS. The most abundant polyphenols in Xeniji were 5-O-caffeoylquinic acid, followed by 3-O-caffeoylquinic acid and sakuranetin. 5-O-Caffeoylquinic acid and 3-O-caffeoylquinic acid are two phenolic acids that have been previously reported to induce various bioactivities including antioxidant, antibacterial, antitumor, antihistaminic18 and antiobesity19 effects. In addition, both 5-O-caffeoylquinic acid and 3-O-caffeoylquinic acid were also reported to have an in vitro liver protective effect against carbon tetrachloride (CCl4) challenges.20 Sakuranetin is a flavonoid that has been previously reported to have antioxidant21 and anti-inflammatory effects,21–23 which may contribute to the in vitro anti-inflammatory effect of Xeniji.9 The presence of these polyphenols contributed to the antioxidant effect of Xeniji10 and provided evidence that Xeniji may possess a hepato-recovery effect.Ethanol-induced liver damage is always attributed to the elevation of serum liver enzyme levels including ALT, ALP and AST,1 which was observed in the untreated mice. Among the serum liver enzymes, ALT is a liver-specific enzyme that leaks into the blood from the cytoplasm of hepatic cells after causing cell damage. In addition, the leakage of AST and ALP into the blood also indicates muscle injury and abnormal hepatic cell function, respectively.24 In the Xeniji-treated mice, the serum levels of ALT, ALP and AST were significantly reduced in a dosage-dependent manner. This result was supported by the liver histopathology analysis in which the Xeniji-treated mice were not observed to exhibit hepatocyte ballooning, but were noticed to have a higher number of binuclear hepatocytes. Hepatocyte ballooning, which is a common histological feature indicating the apoptotic cell death of hepatocytes in chronic ethanol-induced liver damage, was found to be positively correlated with an increase in serum liver enzyme markers.25 On the other hand, binuclear hepatocytes are always used to show liver regeneration as they indicate the division of liver cells.26 Both serum liver enzyme and liver histopathology studies show that Xeniji, especially at a higher concentration, ameliorates the liver damage induced by ethanol in mice. Subsequently, regulation of ethanol metabolism, oxidative stress and inflammation in the liver by Xeniji were further evaluated to understand how Xeniji ameliorates ethanol-induced liver damage.
Ethanol can be metabolised in the liver by alcohol dehydrogenase (ADH) and cytochrome P450 2E1 (CYP2E1) to acetaldehyde. Subsequently, acetaldehyde is further metabolised to acetate by aldehyde dehydrogenase (ALDH).27 Chronic ethanol exposure is known to induce CYP2E1,28 which leads to overproduction of reactive oxygen species (ROS).29 An excessive level of ROS produced by the activation of CYP2E1 depletes the antioxidants and promotes lipid peroxidation in the liver3 as observed in the untreated ethanol-challenged mice. In addition, overproduction of ROS leads to reduced levels of ADH and ALDH.29,30 Activation of CYP2E1 and suppression of ADH/ALDH enzyme activity contributes to accumulation of ROS, lipid peroxidation, and carcinogenic acetaldehyde, which causes hepatocyte cell death,29 as was seen in the histopathology observation and serum liver enzyme quantification of the untreated ethanol-challenged mice. Accumulation of acetaldehyde also contributes to oxidative stress by promoting GSH utilisation, which subsequently leads to depletion of antioxidants in the liver.31 Xeniji treatment significantly increased the efficiency of ethanol metabolism to produce acetate by restoring the enzyme activities of both ADH and ALDH in the liver tissue. The increased efficiency of ADH and ALDH contributes to reduced dependency on the CYP2E1 protein,28 which subsequently prevents accumulation of ROS and MDA caused by activation of the CYP2E1 enzyme in the liver,29 as observed in the livers of the Xeniji-treated mice. On the other hand, polyphenols in Xeniji may also directly suppress the CYP2E1 activity, as recent reports have proposed that polyphenols can suppress the CYP450 mechanism by blocking the hepatic organic anion-transporting polypeptide to inhibit transportation of toxins into hepatocytes, which subsequently reduces the production of ROS.32
Oxidative stress caused by the induction of CYP2E1 is one of the key mechanisms that depletes antioxidants and drives liver damage.29 Previous studies have reported that polyphenols with antioxidant capacity can directly suppress the oxidative stress in vivo through overexpression of the Nrf2 pathway, which restores the level of glutathione (GSH).33 Xeniji, which is rich in polyphenols, was found to enhance the antioxidant levels of healthy mice, indicated by the upregulation of the GCLM and GSTA2 genes that subsequently contribute to increasing the level of GSH in the liver.10 Besides suppression of CYP2E1, Xeniji also suppresses the ethanol-induced ROS by restoring the expression of Nrf2 pathway-related genes, i.e. Nrf2, NQO1, GSTA2 and GPX2 genes in the liver. Nrf2 is a transcription factor that regulates the synthesis of GSH peptide, heme oxygenase (HO-1) enzyme, and NAD(P)H quinone dehydrogenase 1 (NQO1) enzymes.34 GCLM is the gene encoding the enzyme glutamate cysteine ligase that catalyses the synthesis of GSH, while GSTA2 is the gene encoding the enzyme GPX2 for the utilization of GSH.35 Phenolic antioxidants have been reported as inducers of Nrf2 antioxidant mechanisms.36 Xeniji, which is rich in phenolic acids including 5-O-caffeoylquinic acid, 3-O-caffeoylquinic acid, and sakuranetin, activated the expression of Nrf-2, NQO1, GCLM and GSTA2, which subsequently restored the GSH levels in the liver. The enhanced levels of the liver GSH peptide and antioxidant SOD enzyme help to neutralise the ROS and lipid peroxidation in the liver.33
Ethanol-induced liver damage is always observed with chronic inflammation.33 Accumulation of ROS during ethanol metabolism promotes the expression of the pro-inflammatory mediator NF-kB, which subsequently activates the expression of inducible nitric oxide synthase (iNOS) and pro-inflammatory cytokines including IL-1β, IL-6 and TNF-α.31,33 IL-1β, IL-6 and TNF-α have been identified as major inflammatory-related cytokines that are over-expressed and promote the production of nitric oxide in the liver.14,15 Over-expression of pro-inflammatory cytokines subsequently recruits inflammatory immune cells such as hepatic macrophages, which were observed in the histopathological analysis of the livers from untreated ethanol-challenged mice (Fig. 2). Subsequently, the infiltrated inflammatory immune cells promote the progression of liver inflammation,37 activate iNOS expression and subsequently promote the production of nitric oxide,38,39 which reacts with superoxides to produce reactive peroxynitrite.40 In addition, ethanol was found to switch the signalling pathway of TNF-α from the induction of proliferation to cytotoxic killing. Thus, besides ROS, both NO and TNF-α also contribute to the death of hepatocytes.31 A previous in vitro study has reported that Xeniji possesses anti-inflammatory effects.9 In this study, mice treated with Xeniji were observed to exhibit lower levels of serum proinflammatory cytokines, downregulation of NF-kB and iNOS gene expression, and lower levels of NO, which may be attributed to the sakuranetin. Previous studies have reported that sakuranetin possesses anti-inflammatory effects by inhibiting the NF-kB pathway.21 The anti-inflammatory effect of the phenolic acids of Xeniji helps to prevent the formation of reactive peroxynitrite and TNF-α, which induce apoptosis of hepatic cells.
5 Conclusion
Xeniji, which is rich in caffeoylquinic acid and sakuranetin, is able to promote recovery from ethanol-induced liver damaged in mice by improving the metabolism of ethanol, restoring antioxidants, and suppressing ethanol-induced inflammation in the liver in a dosage-dependent manner. Further studies shall focus on the detailed mechanism activated by Xeniji, particularly the major phenolic acids caffeoylquinic acid and sakuranetin, to protect against alcohol-induced liver damage.Conflict of interest
NZ and NKH are from Elken Sdn Bhd, Malaysia, which funded this study. Other authors of this article declare no conflict of interest.Acknowledgements
Dr Kamariah Long would like to thank Elken Sdn Bhd for awarding a research grant.References
- W. Y. Ho, S. K. Yeap, C. L. Ho, R. A. Rahim and N. B. Alitheen, Evid. base. Compl. Alternative Med., 2012, 2012, 417953 Search PubMed.
- K. K. Bhopale, H. Wu, P. J. Boor, V. L. Popov, G. A. S. Ansari and B. S. Kaphalia, Ethanol, 2006, 39, 179–188 CAS.
- D. J. Tuma, Free Radical Biol. Med., 2002, 32, 303–308 CrossRef CAS PubMed.
- A. Bishayee, A. S. Darvesh, T. Politis and R. McGory, Liver Int., 2010, 30, 1103–1114 CrossRef CAS PubMed.
- E. M. Selhub, A. C. Logan and A. C. Bested, J. Physiol. Anthropol., 2014, 33, 2 CrossRef PubMed.
- J. A. Curiel, D. Pinto, B. Marzani, P. Filannino, G. A. Farris, M. Gobbetti and C. G. Rizzello, Microb. Cell Fact., 2015, 14, 67 CrossRef PubMed.
- S. H. Eom, D. S. Lee, Y. M. Kang, K. T. Son, Y. J. Jeon and Y. M. Kim, Appl. Biochem. Biotechnol., 2013, 171, 569–582 CrossRef CAS PubMed.
- F. O. Adetuyi and T. A. Ibrahim, Niger. Food J., 2014, 32, 128–137 CrossRef.
- S. Kuwaki, N. Nakajima, H. Tanaka and K. Ishihara, Biochem. Insights, 2012, 5, 21–29 CrossRef CAS PubMed.
- N. Zulkawi, K. H. Ng, R. Zamberi, S. K. Yeap, D. Satharasinghe, I. B. Jaganath, A. B. Jamaluddin, S. W. Tan, W. Y. Ho, N. B. Alitheen and K. Long, BMC Complementary Altern. Med., 2017, 17, 344 CrossRef PubMed.
- A. Srivastava and T. Shivanandappa, Hepatol. Res., 2006, 35, 267–275 CrossRef PubMed.
- N. E. Mohamad, S. K. Yeap, K. L. Lim, H. M. Yusof, B. K. Beh, S. W. Tan, W. Y. Ho, S. A. Sharifuddin, A. Jamaluddin, K. Long, N. M. A. N. A. Rahman and N. B. Alitheen, Chin. Med., 2015, 10, 3 CrossRef PubMed.
- G. Karthivashan, P. Arulselvan, S. W. Tan and S. Fakurazi, J. Funct. Foods, 2015, 17, 115–126 CrossRef CAS.
- A. K. Singal and B. S. Anand, Clin. Liver Dis., 2013, 2, 53–56 CrossRef.
- T. H. Frazier, Ther. Adv. Gastroenterol., 2011, 4, 63–81 CrossRef PubMed.
- M. S. Kim, M. Ong and X. Qu, World J. Gastroenterol., 2016, 22, 8–23 CrossRef CAS PubMed.
- K. B. Pandey and S. I. Rizvi, Oxid. Med. Cell. Longevity, 2009, 2, 270–278 CrossRef PubMed.
- Y. Miyamae, M. Kurisu, J. Han, H. Isoda and H. Shigemori, Chem. Pharm. Bull., 2011, 59, 502–507 CrossRef CAS PubMed.
- K. Huang, X. C. Liang, Y. L. Zhong, W. Y. He and Z. Wang, J. Sci. Food Agric., 2015, 95, 1903–1910 CrossRef CAS PubMed.
- S. Kan, M. W. Cheung, Y. Zhou and W. S. Ho, J. Food Sci., 2014, 79, C147–C154 CrossRef CAS PubMed.
- L. Taguchi, N. M. Pinheiro, C. R. Olivo, A. Choqueta-Toledo, S. S. Grecco, F. D. Lopes, L. C. Caperuto, M. A. Martins, I. F. Tiberio, N. O. Camara, J. H. Lago and C. M. Prado, Respir. Res., 2015, 16, 79 CrossRef PubMed.
- A. C. Toledo, C. P. Sakoda, A. Perini, N. M. Pinheiro, R. M. Maqalhaes, S. Grecco, I. F. Tiberio, N. O. Camara, M. A. Martins, J. H. Lago and C. M. Prado, Br. J. Pharmacol., 2013, 168, 1736–1749 CrossRef CAS PubMed.
- K. Y. Kim and H. Kang, Evid. base. Compl. Alternative Med., 2016, 2016, 9824203 Search PubMed.
- F. F. Madkour and M. M. Abdel-Daim, Indian J. Pharm. Sci., 2013, 75, 642–648 Search PubMed.
- V. J. Lavallard, S. Bonnafous, S. Patouraux, M. C. Saint-Paul, D. Rousseau, R. Anty, Y. L. Marchand-brustel, A. Tran and P. Gual, PLoS One, 2011, 6, e17599 CAS.
- Y. Miyaoka and A. Miyajima, Cell Div., 2013, 8, 8 CrossRef CAS PubMed.
- M. Setshedi, J. R. Wands and S. M. Monnte, Oxid. Med. Cell. Longevity, 2010, 3, 178–185 CrossRef PubMed.
- Z. Zhou, L. Wang, Z. Song, J. T. Saari, C. J. McClain and Y. J. Kang, Am. J. Pathol., 2005, 166, 1681–1690 CrossRef CAS PubMed.
- M. J. Bak, V. L. Truong, S. Y. Ko, X. N. G. Nguyen, P. Ingkasupart, M. Jun, J. Y. Shin and W. S. Jeong, Int. J. Mol. Sci., 2016, 17, 758 CrossRef PubMed.
- G. Wu, J. Yang, S. Lin, Y. Feng, Q. Yang, Q. Lv and J. Hu, Adv. Exp. Med. Biol., 2013, 776, 21–28 CrossRef CAS PubMed.
- J. I. Beier and C. J. McClain, Biol. Chem., 2010, 391, 1249–1264 CrossRef CAS PubMed.
- R. V. Priyadarsini and S. Nagini, Free Radical Res., 2012, 46, 41–49 CrossRef PubMed.
- A. Louvet and P. Mathurin, Nat. Rev. Gastroenterol. Hepatol., 2015, 12, 231–242 CrossRef PubMed.
- H. Y. Kay, J. W. Yang, T. H. Kim, D. Y. Lee, B. Kang, J. H. Ryu, R. Jeon and S. G. Kim, J. Nutr., 2010, 140, 1211–1219 CrossRef CAS PubMed.
- C. S. Weldy, I. P. Luttrell, C. C. White, V. Morgan-Stevenson, T. K. Bammler, R. P. Beyer, Z. Afsharinejad, F. Kim, K. Chitaley and T. J. Kavanagh, Free Radical Biol. Med., 2012, 53, 1264–1278 CrossRef CAS PubMed.
- Q. Ma, Annu. Rev. Pharmacol. Toxicol., 2013, 53, 401–426 CrossRef CAS PubMed.
- R. Zamora, Y. Vodovotz and T. R. Billiar, Mol. Med., 2000, 6, 347–373 CAS.
- J. Kato, Y. Mogi, Y. Kohgo, R. Takimoto, M. Kobune, H. Hisai, T. Nakamura, K. Takada and Y. Niitsu, J. Gastroenterol., 1998, 33, 855–859 CrossRef CAS PubMed.
- N. B. D. El-Guindy, W. J. Villiers and D. E. Doherty, Ethanol, 2007, 41, 335–345 Search PubMed.
- N. Karunaweera, R. Raju, E. Gyengesi and G. Munch, Front. Mol. Neurosci., 2015, 8, 24 Search PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |