DOI:
10.1039/C7RA04008C
(Paper)
RSC Adv., 2017,
7, 27796-27806
Investigating the interaction mechanism of fluorescent whitening agents to human serum albumin using saturation transfer difference-NMR, multi-spectroscopy, and docking studies†
Received
8th April 2017
, Accepted 19th May 2017
First published on 25th May 2017
Abstract
In this study, 2,2′-(4,4′-biphenylylenebisvinylene)bisbenzenesulfonicacid (CBS-X) and its disodiumsalt (CBS) were used as model compounds to investigate the interaction mechanism between 4,4′-distyrylbiphenyl based fluorescent whitening agents (DSBP-FWAs) and human serum albumin (HSA) through various techniques, including 1H saturation transfer difference nuclear magnetic resonance (1H STD-NMR), fluorescence studies, UV-vis absorption, Fourier transform infrared (FT-IR) spectroscopy, circular dichroism (CD) spectroscopy, and molecular docking. The 1H STD-NMR analyses indicated that CBS and CBS-X can bind to HSA at the favored Sudlow's sites II and I, respectively. Fluorescence emission spectra showed that CBS and CBS-X quenches HSA fluorescence through a dynamic mechanism, and this was further verified by fluorescence lifetime experiments and UV-vis absorption. Moreover, the effective binding constant values of the two compounds at the same temperature decreased in the order CBS > CBS-X. Furthermore, the energy transfer efficiency for CBS and CBS-X were 50.5% and 40.6%, respectively. Thermodynamic analyses indicated that the binding of CBS and CBS-X with HSA are both primarily controlled by hydrophobic forces. FT-IR and CD spectroscopy provided complementary information on the micro-environmental and conformational changes of HSA with the additions of CBS and CBS-X. Molecular docking further confirmed the NMR and spectroscopic results. Overall, the comparative studies on the interaction mechanism of CBS and CBS-X when binding to HSA may provide useful information for evaluating their effects on the human body.
1. Introduction
Fluorescent whitening agents (FWAs) are fluorescent dyes that can absorb ultraviolet light (usually 340–370 nm) and emit blue light (typically 420–470 nm). They are extensively applied in detergents, papers, textiles, coatings, and plastic products to eliminate the yellowish and visually increase the whiteness and brightness of products.1–4 2,2′-(4,4′-Biphenylylenebisvinylene)bisbenzenesulfonicacid (CBS-X) and its disodiumsalt 2,2′-(4,4′-biphenylylenedivinylene)di-benzenesulfonicacidisodiumsalt (CBS) (Fig. 1) are 4,4′-distyrylbiphenyl based fluorescent whitening agents (DSBP-FWAs) that can modify the cotton, silk, wool, and polyester fabrics with a higher whiteness level.5–8 However, FWAs can easily enter the human circulatory system through skin, food, and water. Moreover, FWAs are structurally stable and difficult to metabolize and can accumulate in the body and endanger human health.9 Furthermore, DSBP-FWAs can irritate skin and mucous membranes.10,11 CBS and CBS-X can cause potential toxicity during transmission to the human body. The toxic characteristics of CBS and CBS-X need to be explored because these substances cause concern in the modern times.
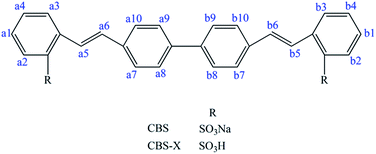 |
| | Fig. 1 Chemical structures of CBS and CBS-X. | |
Human serum albumin (HSA) is the most abundant carrier protein in the circulatory system and plays a major role in the transportation and disposition of many endogenous and exogenous compounds.12,13 HSA is a globular protein that contains 585 amino acids with three homologous helical domains (I, II, and III), with each domain comprising two subdomains (A and B).14,15 Sudlow's sites I and II are two major ligand binding sites of HSA, which located in subdomains IIA and IIIA, respectively. Given the high albumin concentration in the human blood plasma and its high binding affinity to various small molecules, HSA has been extensively used as a model protein to evaluate small molecular–protein systems.16,17 Studies on the interactions between dyes and HSA have received increasing attention and have significantly contributed to the understanding of the toxicity of dyes that is being transported to the human body.18–20 Thus, knowledge of the interaction mechanisms between CBS/CBS-X and HSA is important for us to understand the toxicity of DSBP-FWAs in detail.
Several methods, including isothermal titration calorimetry,21 electrophoretic techniques,22,23 and spectroscopic analyses,24,25 have been employed to explain the interaction mechanisms of small molecules with proteins. These methods have been proved reasonable and reliable. However, they cannot identify small molecule epitope and structure affinity even if these are highly laborious experimental protocols for clarifying interaction mechanism. In recent years, nuclear magnetic resonance (NMR) has been efficient in characterizing small molecule–protein interactions at the molecular level without prior knowledge of protein functions.26–29 Accordingly, saturation transfer difference (STD) is a popular small molecule-based NMR technique that can identify the small molecules involved in the structural information without protein labeling. In this study, 1H STD-NMR, multi-spectroscopic methods (fluorescence, UV-vis, FT-IR, and CD spectroscopy), and molecular docking were used to initially investigate the interactions of CBS and CBS-X with HSA and show the differences between CBS–HSA and CBS-X–HSA interactions. The binding affinity of the H proton in CBS and CBS-X with HSA and their possible binding sites were initially investigated using 1H STD-NMR. Moreover, multi-spectroscopic methods (fluorescence, UV-vis, FT-IR, and CD spectroscopy) were used to clarify the binding mechanism and conformational alternation of HSA. The molecular docking technique was performed to further examine the binding process. This study will provide insights into the molecule-based interactions between CBS and CBS-X and HSA.
2. Experimental
2.1. Materials
HSA without essential fatty acids was purchased from Sigma-Aldrich (Milwaukee, USA) and used without further purification. CBS (99%) was provided by Tianjin Heowns Biochem LLC (Tianjin, China), and CBS-X (98%) was provided by Huaxia Reagent Ltd. (Beijing, China). Warfarin, ibuprofen, deuterium oxide (D2O), and dimethyl sulfoxide-d6 (DMSO-d6) were purchased from J&K Scientific Ltd. (Beijing, China). Triple-distilled water was used to prepare the phosphate buffer solution (PBS) and stock solutions of CBS (2.0 mM) and CBS-X (2.0 mM). The stock solutions of HSA (20 μM) were prepared by dissolving solid HSA into 0.01 M PBS at pH 7.4 and storing at 273 K to 277 K. All other reagents were of analytical grade and used without further purification.
2.2. Instruments and operations
2.2.1. NMR experiments. All NMR experiments were conducted on a Varian Inova 700 MHz spectrometer operating at 298 K using the VNMRJ software (version 2.1B). The HSA (0.2 mM) solution was prepared in the 0.01 M PBS (pH 7.4). The CBS (40 mM), CBS-X (40 mM), warfarin (80 mM), and ibuprofen (80 mM) were directly dissolved in DMSO-d6. The NMR samples were prepared in 500 μL of 0.01 M PBS buffer containing 10 μM HSA and 400 μM ligand (molar ratio of HSA to ligand = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) at room temperature. Competition studies were performed with ibuprofen and warfarin, respectively. Competitive NMR samples, including identical HSA and ligand concentrations containing two different concentrations of warfarin (600 and 1200 μM) or ibuprofen (600 and 1200 μM), were analyzed. In the STD experiments, 50 ms Gaussian pulses were applied at an on-resonance saturation of −0.5 ppm and an off-resonance saturation of 34 ppm. All spectra were acquired with a sweep width of 8389.26 Hz, 256 transients, and acquisition time of 1 s. The total scan number was 1024 and typically used 16 ppm spectral widths for the 1H STD spectra. The subtraction was performed after every STD scan by phase cycling. Each sample was recorded with Watergate solvent suppression to obtain a standard 1H spectrum. All the spectra were processed and analyzed using the ACD/CNMR software (Advanced Chemistry Development, Inc., version 11.0).
40) at room temperature. Competition studies were performed with ibuprofen and warfarin, respectively. Competitive NMR samples, including identical HSA and ligand concentrations containing two different concentrations of warfarin (600 and 1200 μM) or ibuprofen (600 and 1200 μM), were analyzed. In the STD experiments, 50 ms Gaussian pulses were applied at an on-resonance saturation of −0.5 ppm and an off-resonance saturation of 34 ppm. All spectra were acquired with a sweep width of 8389.26 Hz, 256 transients, and acquisition time of 1 s. The total scan number was 1024 and typically used 16 ppm spectral widths for the 1H STD spectra. The subtraction was performed after every STD scan by phase cycling. Each sample was recorded with Watergate solvent suppression to obtain a standard 1H spectrum. All the spectra were processed and analyzed using the ACD/CNMR software (Advanced Chemistry Development, Inc., version 11.0).
2.2.2. Fluorescence measurements. Fluorescence measurements were conducted on a Cary Eclipse Fluorescence Spectrophotometer (Varian, CA, USA). The emission spectra of HSA with or without ligand (CBS or CBS-X) were recorded at 298, 304, and 310 K by setting a 280 nm excitation wavelength, 5/10 nm (excitation/emission) slit widths, and a 300–500 nm scanning range. In the measurements, HSA was diluted with 0.01 M PBS to obtain a final concentration of 2 μM. A series of samples was prepared by adding a ligand (CBS or CBS-X) stock solution to HSA to ensure that the final ligand concentrations increased from 0.2 μM to 1.4 μM with a gradient growth of 0.2 μM. In this study, all fluorescence intensities were corrected for the absorption of excited light and reabsorption of emitted light based on the following equation:| | |
Fcorr = Fobs × exp(Aex + Aem)/2
| (1) |
where Fcorr and Fobs are the corrected and observed fluorescence intensities, respectively. Aex and Aem are the absorption of the system at the excitation and emission wavelengths, respectively.The fluorescence lifetimes of the HSA and HSA–ligand (CBS or CBS-X) systems were measured at room temperature on a Horiba Jobin Yvon FluoroLog-TCSPC spectrofluorometer (HORIBA, France). The values of excitation wavelength (λex) and emission wavelength (λem) were fixed at 280 and 345 nm, respectively. Free HSA (2 μM) and a mixture of HSA and ligand (CBS or CBS-X) (1 : 0.5 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratios) were observed in sequence. Experimental data were analyzed using the tail-fitting method, and the qualities of the fits were assessed using χ2 values and residuals.
1 molar ratios) were observed in sequence. Experimental data were analyzed using the tail-fitting method, and the qualities of the fits were assessed using χ2 values and residuals.
2.2.3. UV-vis spectroscopy. The absorption spectra of HSA with or without ligand (CBS or CBS-X) and only ligands were scanned within 200–400 nm at 298 K using a TU1901 double beam UV-vis spectrophotometer (Purkinje General, Beijing, China). PBS (0.01 M) buffer was used as the reference solution. HSA concentration was maintained at 2 μM, and the ligand (CBS or CBS-X) concentrations were set at 1.4 μM. Moreover, the absorption spectra of CBS (2 μM) and CBS-X (2 μM) were scanned within 200–500 nm, respectively.
2.2.4. FT-IR spectroscopy. FT-IR measurements were performed using a Nicolet 6700 FT-IR spectrometer (Thermo Scientific, WI, USA) combined with a Smart OMNI-Sampler accessory. The spectra were obtained using the attenuated total reflection method with a resolution of 4 cm−1, 128 scans, and a range of 4000–600 cm−1 at 298 K. HSA concentration was fixed at 0.2 mM, whereas that of the ligand (CBS or CBS-X) was 0.14 mM.
2.2.5. CD spectroscopy. CD spectral measurements were conducted using an automatic recording spectrophotometer (Model 400, AVIV, USA) in a 10 mm path length cell at 298 K. Each spectrum was the average of three successive scans within the wavelength range of 190–280 nm. The HSA concentration was kept at 2 μM, and other samples were prepared by adding CBS or CBS-X to obtain the final ligand concentrations of 1 and 2 μM (i.e., 1 : 0.5 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratios).
1 molar ratios).
2.2.6. Molecular docking. Docking between HSA (as receptor) and CBS or CBS-X (as ligand) was simulated with the AutoDockTools-1.5.6 software and the AutoDock-4.2.6 program to identify potential binding sites. Both ligand and receptor molecules were pretreated prior to docking. The crystal structure of the free HSA was taken from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB), with PDB ID 1BM0. Chain B was deleted and all water molecules were removed, polar hydrogen and Kollman charges were added, and the AD4 atom type was assigned. The three-dimensional (3D) structure of CBS and CBS-X were generated using Chem3D Ultra 11.0, and energy was minimized using the “Discover Minimization” tool in Materials Studio 6.0. To explore all the binding sites of CBS and CBS-X in HSA, a blind docking simulation strategy was adopted by setting the grid box size to 126 Å × 126 Å × 126 Å along the X, Y, and Z axes, respectively, with 0.581 Å grid spacing. The center of the grid was set to the position of 27.236, −15.003, and 23.042. During the docking, Genetic Algorithm (GA) was selected to set the search parameters, and global optimization was conducted with 200 GA runs and 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 generations, as well as a medium number of 2
000 generations, as well as a medium number of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 energy evaluations. Other parameters were set as the default protocol settings. Subsequently, the lowest-energy docked conformation was applied to further locate the most suitable binding mode.
000 energy evaluations. Other parameters were set as the default protocol settings. Subsequently, the lowest-energy docked conformation was applied to further locate the most suitable binding mode.
3. Results and discussion
3.1. Identification of CBS and CBS-X binding to HSA by 1H STD-NMR
3.1.1. Binding of CBS and CBS-X to HSA. The NMR technique is a well-established tool for studying the interaction between small molecular compounds and biological macromolecules, particularly for drug screening and assessment in the pharmaceutical industry.20,30,31 As a new NMR-based screening method, the 1H STD-NMR technique exploits the nuclear Overhauser effect (NOE) transference from the macromolecule to the ligand. When a macromolecule is saturated by a selective radio-frequency irradiation, this saturation will spread within the entire macromolecule via spin diffusion and be transferred to the bound ligand through intermolecular NOEs.30 During the transfer, the small molecule hydrogen that is in close contact with the macromolecule receives the most intense magnetization transfer and provides the most intense NMR signals. Therefore, the spectrum data obtained with STD are different spectrum data and exhibits only the signals of the bound ligand.32,33 The 1H STD-NMR technique is effective for identifying binding epitopes by calculating the relative STD (%) signals.30 For the relative STD (%) signals, the STD integral intensities of each proton of a small molecule are calculated with the strongest STD effect, which is assigned with a value of 100% as reference.The reference 1H NMR spectrum and the corresponding 1H STD-NMR spectra of CBS–HSA and CBS-X–HSA systems are displayed in Fig. 2. The detailed 1H chemical shift values of CBS and CBS-X are shown in Fig. 2(a) and (c), respectively. The difference in the structures of CBS and CBS-X are only the sodium sulfonate and sulfonic acid groups (Fig. 1). Furthermore, the 1H of the sulfonic acid group would be replaced by D2O in NMR experiments, and this caused the 1H NMR spectrum of CBS and CBS-X to be the same in this study. A series of overlapped signals was observed because of their highly similar chemical environments. Thus, the signals of these protons were generally assigned and did not affect the judgment on the binding of CBS and CBS-X to HSA. Fig. 2 shows the similar characteristic signals between the reference 1H NMR and 1H STD-NMR spectra in CBS–HSA and CBS-X–HSA systems, indicating that CBS and CBS-X binds to HSA. The values of relative STD (%) were calculated and the results are shown in Table 1. The obtained relative STD values indicated that a slight binding preference exists through the H-a7, b7, a10, b10 protons, given that this resonance is the highest saturation-receiving proton. Moreover, the hydrogen in benzene had intense signal. Thus, these findings indicated that CBS and CBS-X exhibit strong affinity to HSA.
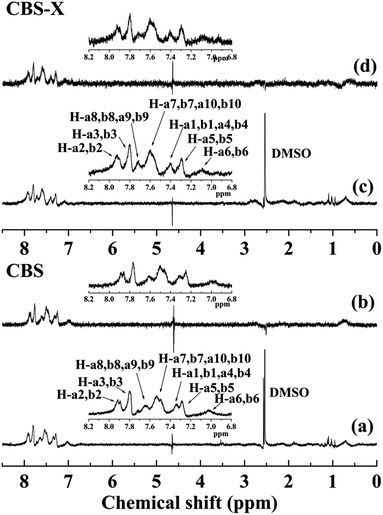 |
| | Fig. 2 The 1H NMR (a) and 1H STD-NMR spectra (b) of CBS in the presence of HSA in 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio; 1H NMR (c) and 1H STD-NMR (d) spectra of CBS-X in the presence of HSA in 40 1 ratio; 1H NMR (c) and 1H STD-NMR (d) spectra of CBS-X in the presence of HSA in 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. Watergate scheme was used for solvent suppression [CBS] = 400 μM, [CBS-X] = 400 μM, [HSA] = 10 μM, pH = 7.4, T = 298 K. 1 ratio. Watergate scheme was used for solvent suppression [CBS] = 400 μM, [CBS-X] = 400 μM, [HSA] = 10 μM, pH = 7.4, T = 298 K. | |
Table 1 Relative degrees of saturation of individual protons in CBS and CBS-X 1H STD-NMR spectra. The protons of H-a7, b7, a10, b10 have the strongest STD effect, which is assigned with a value of 100%
| |
H-a6, b6 |
H-a5, b5 |
H-a1, b1, a4, b4 |
H-a7, b7, a10, b10 |
H-a8, b8, a9, b9 |
H-a3, b3 |
H-a2, b2 |
| CBS relative STD (%) |
20 |
24 |
36 |
100 |
60 |
56 |
76 |
| CBS-X relative STD (%) |
17.24 |
34.48 |
27.59 |
100 |
34.48 |
55.17 |
65.52 |
3.1.2. Recognition of binding site. Subdomains IIA and IIIA in HSA are two major binding regions for endogenous and exogenous small molecules named Sudlow's sites I and II, respectively.14,15 To investigate the possible interactive sites of CBS and CBS-X, competitive STD-NMR experiments were performed with warfarin and ibuprofen at two different concentrations. Warfarin and ibuprofen are widely used as references that preferably bind to sites I and II of HSA, respectively.29,34 Direct competition and interaction at specific sites in 1H STD-NMR can be reflected by changes in the resonance of STD signals.35 Fig. 3 shows the 1H NMR and 1H STD-NMR spectra after the addition of site markers into the systems of CBS–HSA and CBS-X–HSA. The relative STD (%) values of the samples with the site marker added are shown in Table 2. The relative STD (%) values of H-a3, b3, a6, b6, are not presented in Table 2 because their chemical shifts are the same as those of warfarin and ibuprofen. In the case of CBS–HSA, the STD intensity was decreased when the warfarin and ibuprofen concentrations increased, indicating that both warfarin and ibuprofen competed with CBS in binding toward the protein. Moreover, increased remarkable STD signal changes were observed in the presence of ibuprofen. This observation revealed that CBS was more likely bound to the site II of HSA. Similarly, the STD intensity was decreased as the warfarin and ibuprofen concentrations increased in the CBS-X–HSA system. Furthermore, the STD signals of the H-a8, b8, a9, b9 of CBS-X disappeared as the warfarin and ibuprofen concentrations increased. This finding revealed that competition occurred with CBS-X from both sites I and II of HSA. However, the STD intensity changes of CBS-X were highly visible in the presence of varying concentrations warfarin, and the H-a1, b1, a4, b4 signals for warfarin did not appear. Thus, CBS-X preferentially binds to HSA at site I. The primary binding site of CBS and CBS-X are different due to the different CBS and CBS-X structures.
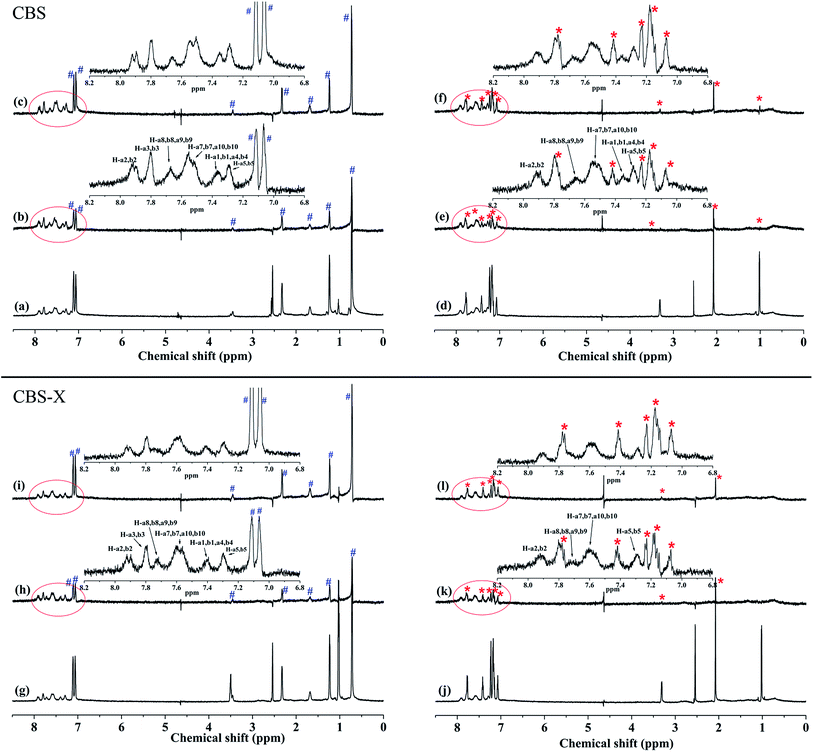 |
| | Fig. 3 1H NMR spectrum of the mixture of CBS and HSA after adding 600 μM ibuprofen (a) and 600 μM warfarin (d). 1H STD-NMR spectrum of the mixture of CBS and HSA after adding 600 μM ibuprofen (b), 1200 μM ibuprofen (c), 600 μM warfarin (e), and 1200 μM warfarin (f). 1H NMR spectrum of the mixture of CBS-X and HSA after adding 600 μM ibuprofen (g) and 600 μM warfarin (j). 1H STD-NMR spectrum of the mixture of CBS-X and HSA after adding 600 μM ibuprofen (h), 1200 μM ibuprofen (i), 600 μM warfarin (k), and 1200 μM warfarin (l) [CBS] = 400 μM, [CBS-X] = 400 μM, [HSA] = 10 μM. The signals of competitors are indicated by # (ibuprofen) and * (warfarin). | |
Table 2 The relative STD (%) values of CBS and CBS-X competitive with different concentrations ibuprofen and warfarin
| |
CBS |
CBS-X |
| Ibfa |
Ibfb |
Wrfa |
Wrfb |
Ibfa |
Ibfb |
Wrfa |
Wrfb |
| The concentration of ibuprofen and warfarin are 600 μM. The concentration of ibuprofen and warfarin are 1200 μM. |
| H-a5, b5 |
33.33 |
33.33 |
36.84 |
38.46 |
36.36 |
50 |
47.06 |
35.29 |
| H-a1, b1, a4, b4 |
33.33 |
25 |
31.58 |
30.77 |
36.36 |
33.33 |
— |
— |
| H-a7, b7, a10, b10 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
| H-a8, b8, a9, b9 |
46.67 |
25 |
42.11 |
38.46 |
27.27 |
— |
41.18 |
— |
| H-a3, b3 |
46.67 |
33.33 |
— |
— |
45.45 |
33.33 |
— |
— |
| H-a2, b2 |
66.67 |
25 |
57.89 |
53.85 |
45.45 |
33.33 |
64.71 |
29.41 |
3.2. Analysis of binding mechanisms and models
3.2.1. Binding mechanisms. Fluorescence technique is a common method for studying the binding properties of small molecules to a protein.36 Generally, the intrinsic fluorescence of HSA is caused by tryptophan (Trp), tyrosine (Tyr), and phenylalanine (Phe) residues, with the last residue contributing only slightly.37 The microenvironment of a fluorophore and the intrinsic fluorescence intensity of HSA may change when the small molecules interact with HSA. In this study, the interactions between CBS and CBS-X and HSA were investigated by measuring the intrinsic fluorescence intensities of HSA in the absence and presence of CBS and CBS-X. The fluorescence spectral changes of HSA with different amounts of CBS and CBS-X at 298 K are illustrated in Fig. 4(a) and (b). When HSA was mixed with varying amounts of CBS and CBS-X, the fluorescence signal decreased. Moreover, the quenching was stronger in the case of CBS, whereas the extent of quenching lessened with the addition of CBS-X. Moreover, remarkable blue shifts at maximum λem (from 335 to 327 nm) indicated that the microenvironment around the fluorophore changed, and the Trp residues might transfer to a more hydrophobicity environment. The observed decrease in the fluorescence intensity can be ascribed to the interactions of CBS and CBS-X with HSA, which are consistent with the results obtained in the 1H STD-NMR.
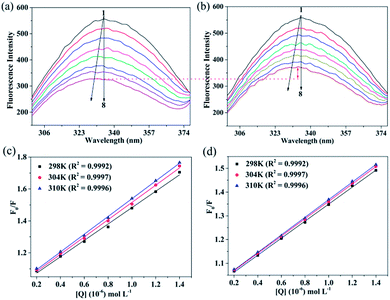 |
| | Fig. 4 Fluorescence emission spectra of HSA with different amounts of CBS (a) and CBS-X (b) additions. HSA is constant at 2 μM and CBS and CBS-X vary from 0–1.4 μM with a gradient growth of 0.2 μM (peaks from top to bottom). Stern–Volmer plots of the CBS–HSA (c) and CBS-X–HSA (d) systems at different temperatures. | |
The fluorescence quenching of protein could be attributed to various mechanisms, including excited-state reactions, molecular rearrangements, ground-state complex formations, and collisional quenching.38 Any observed fluorescence quenching is likely related to ground-state complex formation or collisional quenching when the inner-filter effect is removed.39 Collisional quenching, also named as dynamic quenching, which can be distinguished from static quenching by varying the temperature; in this case, the Stern–Volmer equation is used:
where
F0 and
F are the fluorescence intensities in the absence and presence of a quencher, respectively;
KSV is the Stern–Volmer quenching constant; [Q] is the quencher concentration. The
KSV increases during dynamic quenching because high temperatures results in fast diffusion, whereas it decreases during static quenching because high temperatures typically result in the dissociation of weakly bound complexes.
40 The calculated
KSV values are summarized in
Table 3 (Stern–Volmer plots are shown in
Fig. 4(c) and (d)). In both CBS–HSA and CBS-X–HSA interactions, the values of
KSV gradually increased as the temperature increased, which indicated that HSA fluorescence quenching by CBS and CBS-X may follow a dynamic quenching rather than a static mechanism.
41,42
Table 3 Stern–Volmer, effective quenching constants and thermodynamic parameters for the two ligands binds to HSA at different temperatures
| Compounds |
T (K) |
KSV × 105 (L mol−1) |
Ka × 105 (L mol−1) |
n |
K × 105 (L mol−1) |
ΔG (kJ mol−1) |
ΔH (kJ mol−1) |
ΔS (J mol−1 K−1) |
| CBS |
298 |
5.134 |
3.158 |
1.360 |
3.100 |
−31.318 |
21.967 |
178.811 |
| 304 |
5.411 |
3.563 |
1.261 |
3.651 |
−32.391 |
| 310 |
5.565 |
4.418 |
1.145 |
4.367 |
−33.465 |
| CBS-X |
298 |
3.583 |
2.830 |
1.193 |
2.752 |
−31.082 |
17.998 |
164.701 |
| 304 |
3.656 |
3.611 |
1.046 |
3.385 |
−32.070 |
| 310 |
3.706 |
3.876 |
1.007 |
3.637 |
−33.058 |
To further substantiate the quenching mechanism, fluorescence lifetime measurements were conducted.43 Static quenching failed to decrease the fluorescence decay time of the uncombined fluorophores, whereas dynamic quenching apparently altered the mean decay time of the entire excited-state population. The data obtained from the time-resolved fluorescence spectra were analyzed using the tail-fitting method. The qualities of the fits were assessed using χ2 values and residuals. The average fluorescence lifetime (〈τ〉) for tail-fitting was calculated using the amplitude-averaged lifetime method as follows:44
| | |
〈τ〉 = τ1α1 + τ2α2 + τ3α3
| (3) |
where
τ1,
τ2, and
τ3 are the decay times; and
α1,
α2, and
α3 are the pre-exponential factors. The fluorescence decay of free HSA and the protein with ligands (CBS and CBS-X) additions are illustrated in
Fig. 5(a) and (b), and the fitted results are summarized in
Table 4. The average lifetime of HSA showed a significant reduction with the additions of CBS and CBS-X, indicating that the quenching follows a dynamic mechanism. This finding further verified the conclusion obtained from the
KSV analysis. Furthermore, the extent of the average lifetime reduction of HSA in the case of CBS was stronger than that of CBS-X, and this is consistent with the fluorescence quenching analytical results.
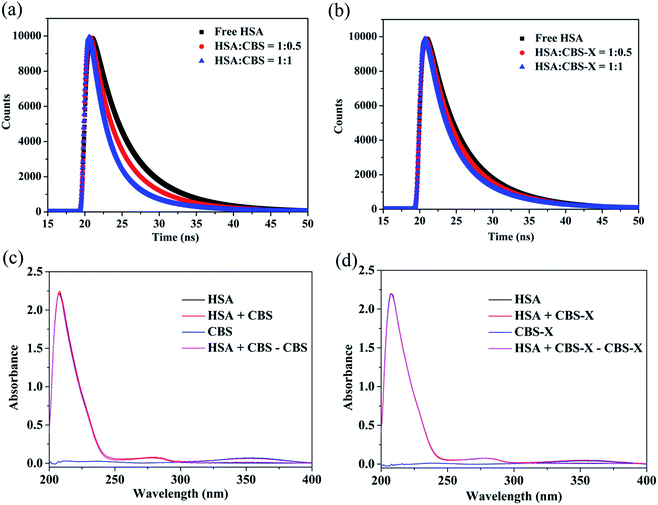 |
| | Fig. 5 The time-resolved fluorescence decay of the CBS–HSA (a) and CBS-X–HSA (b) systems, [HSA] = 2 μM, [CBS] = 1 and 2 μM, [CBS-X] = 1 and 2 μM. UV-vis absorption spectra of HSA, ligands, and ligands–HSA system, (c) for the CBS–HSA system, (d) for the CBS-X–HSA system, [HSA] = 2 μM, [CBS] = 1.4 μM, [CBS-X] = 1.4 μM. | |
Table 4 Lifetimes of fluorescence decay of HSA with different concentrations of ligands
| System |
Cligand × 10−6 (mol L−1) |
τ1 (ns) |
τ2 (ns) |
τ3 (ns) |
α1 |
α2 |
α3 |
〈τ〉 (ns) |
χ2 |
| HSA |
0 |
3.118 |
0.527 |
6.892 |
0.334 |
0.038 |
0.628 |
5.389 |
1.016 |
| HSA–CBS |
1 |
2.559 |
0.443 |
6.551 |
0.371 |
0.079 |
0.550 |
4.588 |
1.119 |
| 2 |
2.218 |
0.445 |
6.029 |
0.454 |
0.127 |
0.419 |
3.588 |
1.107 |
| HSA–CBS-X |
1 |
2.959 |
0.539 |
6.740 |
0.335 |
0.057 |
0.608 |
5.121 |
1.115 |
| 2 |
2.747 |
0.519 |
6.642 |
0.366 |
0.076 |
0.557 |
4.747 |
1.103 |
One additional method to distinguish static from dynamic quenching is by careful examination of the absorption spectra of the fluorophore. Dynamic quenching only affects the excited states of the fluorophores; thus, no changes in the absorption spectra are expected. In contrast, the ground-state complex formation will frequently result in the perturbation of the absorption spectrum of the fluorophore.40 The UV-vis absorption spectra of HSA (Fig. 5(c) and (d)) and the corresponding difference absorption spectra can be superposed with each other in the experimental error. Herein, this result further confirmed that the proposed quenching mechanism by CBS and CBS-X are dynamic quenching procedures.
3.2.2. Binding parameters. In the case of dynamic quenching, the quencher must diffuse to the fluorophore during the lifetime of the excited state. Upon contact, the fluorophore returns to the ground state without photon emission.40 The nature of the dynamic quenching can be attributed to an exciplex formation and/or energy transfer.45 The effective binding constant (Ka) of the accessible fluorophores was determined from the steady state fluorescence data with variations of the ligand (CBS and CBS-X) concentrations. The Ka for the association of a small molecule to a protein was determined according to the following equation:41,46| | |
F0/(F0 − F) = 1/fa + 1/Kafa[Q]
| (4) |
In the present case, (F0 − F) is the difference in fluorescence in the absence and presence of the quencher at concentration [Q], and fa is the fraction of the accessible fluorescence. The plots of F0/(F0 − F) versus 1/[Q] for CBS–HSA and CBS-X–HSA systems are shown in Fig. 6(a) and (b). The value of Ka can be obtained by the ratio of the intercept and slope, and the corresponding results at the three investigated temperatures are listed in Table 3. The Ka values in both CBS–HSA and CBS-X–HSA interactions increases gradually as temperature increases, and this is consistent with a dynamic quenching mechanism. The Ka values of the two compounds at the same temperature decreased in the order CBS > CBS-X, demonstrating that the compound with the sodium sulfonate group showed a stronger interaction with HSA than that with the sulfonic acid group.
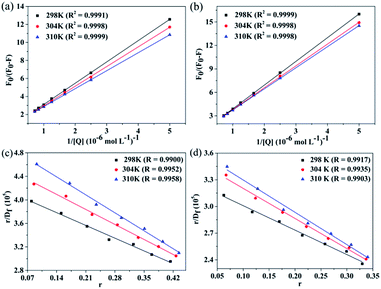 |
| | Fig. 6 Plot of F0/(F0 − F) versus 1/[Q] for the CBS–HSA (a) and CBS-X–HSA systems (b) at different temperatures. The Scatchard plots of the CBS–HSA (c) and CBS-X–HSA systems (d) at different temperatures. | |
The binding constant and number of binding sites can be deduced from the Scatchard equation:20,47
where
r (
r = Δ
F/
F0) represents the number of moles of bound small molecules per mole of protein,
Df is the molar concentration of the free ligand, and
n and
K are the number of binding sites and constants, respectively.
Fig. 6(c) and (d) show the plots of
eqn (5) and
Table 3 lists the
n and
K values at different temperatures. All values of
n were close to 1, and we may infer that one mainly independent class of binding sites exists in HSA for CBS and CBS-X.
3.2.3. Interactive model and distance. The interactive forces between small molecules and biomacromolecules include hydrogen bonds, hydrophobic interaction, electrostatic interaction, and van der Waals forces.48 The intermolecular forces can be estimated using thermodynamic parameters, such as Gibbs free-energy change (ΔG), enthalpy change (ΔH), and entropy change (ΔS). Ignoring the effects of temperature on ΔH, these parameters, namely, ΔG, ΔH, and ΔS, can be calculated using the van't Hoff equations:| |
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = −ΔH/RT + ΔS/R K = −ΔH/RT + ΔS/R
| (6) |
| |
ΔG = ΔH − TΔS = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K
| (7) |
where K corresponds to Ka at specific temperatures, R is the gas constant, and T is the absolute temperature. The results obtained from eqn (6) and (7) are presented in Table 3. The negative ΔG values indicate that the binding processes of CBS and CBS-X to the protein are spontaneous at the corresponding temperatures. The ΔH and ΔS values are positive for the CBS–HSA and CBS-X–HSA systems, demonstrating that hydrophobic forces play a major role in the interaction of CBS and CBS-X with HSA.49Förster's energy transfer theory (FRET) is often applied to calculate the energy transfer efficiency and the distance between the donor and acceptor.50,51 In this study, the fluorescence of CBS and CBS-X were increased when they interacted with HSA (Fig. S1†). Moreover, a nearly complete overlap was found between the fluorescence emission spectrum of HSA and the absorption spectra of CBS (Fig. 7(a)) and CBS-X (Fig. 7(b)), and this overlap is the primary requirement for the occurrence of FRET. The molecular distance (r) between the acceptor (CBS and CBS-X) and donor (HSA) was estimated using FRET. Furthermore, the efficiency of energy transfer (E) between a donor and an acceptor can be calculated using the following equation:
| | |
E = (F0 − F)/F0 = R06/(R06 + r6)
| (8) |
where
R0 is the critical distance at a 50% transfer efficiency, which can be obtained according to the following equation:
| | |
R06 = 8.79 × 10−25K2n−4ΦJ
| (9) |
where
K2 is the spatial orientation factor involving the geometry of the donor–acceptor dipole;
n is the average refractive index of the medium;
Φ represents the fluorescence quantum yield of the donor; and
J expresses the integral of the spectral overlap between the fluorescence emission of the donor and the absorption of the acceptor.
| |
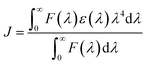 | (10) |
where
F(
λ) is the fluorescence intensity of the donor at a wavelength range
λ, and
ε(
λ) is the molar absorption coefficient of the acceptor at a certain wavelength
λ. Using
K2 = 2/3,
n = 1.336, and
Φ = 0.118, we obtained
J = 1.231 × 10
−14 cm
3 L mol
−1,
R0 = 2.37 nm,
E = 50.5%, and
r = 2.36 nm for the CBS–HSA system; and
J = 1.256 × 10
−14 cm
3 L mol
−1,
R0 = 2.38 nm,
E = 40.6%, and
r = 2.54 nm for the CBS-X–HSA system.
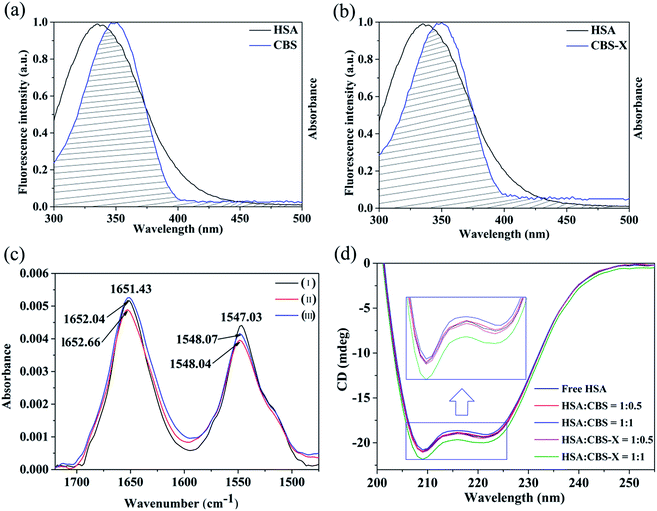 |
| | Fig. 7 (a) Spectral overlap between the UV-vis absorption spectrum of CBS and the fluorescence spectrum of HSA, (b) spectral overlap between the UV-vis absorption spectrum of CBS-X, and the fluorescence spectrum of HSA. (c) FT-IR spectra of free HSA (I) and spectra of the CBS–HSA system (II) and CBS-X–HSA system (III), with a molar concentration ratio of HSA to CBS (or CBS-X) of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.7 in pH 7.4 PBS, [HSA] = 0.2 mM. (d) Circular dichroism spectra of 2 μM HSA in the absence or presence of CBS and CBS-X. 0.7 in pH 7.4 PBS, [HSA] = 0.2 mM. (d) Circular dichroism spectra of 2 μM HSA in the absence or presence of CBS and CBS-X. | |
In case of average distance r < 8 nm that this follows the condition, we obtained 0.5R0 < r < 1.5R0.47 Thus, the energy transfer occurred during the CBS–HSA and CBS-X–HSA interaction.
3.3. Investigation of the conformational changes of HSA
3.3.1. FT-IR spectroscopy. The distinctive HSA fluorescence quenching and the blue shift of the maximum emission wavelength (Fig. 4(a) and (b)) suggested that the Trp residue microenvironment was changed after CBS and CBS-X interacted with HSA. The secondary structure changes of a protein can be conveniently measured using the FT-IR spectrum. In the IR spectra of proteins, amides I and II are the most widely used vibrational bands for studying the conformation of protein secondary structures.52 Amide I (1600–1700 cm−1) is primarily attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration, whereas amide II (1500–1600 cm−1) is attributed to the coupling of the N–H in plane bending and C–N stretching modes.53 As shown in Fig. 7(c), the characteristic amide I and II absorption peak positions of free HSA are at 1652.04 and 1547.03 cm−1, respectively. After CBS addition, the two amide bands apparently decreased with a peak shift (amide I: from 1652.04 cm−1 to 1652.66 cm−1; amide II: from 1547.03 cm−1 to 1548.04 cm−1) in absorbance intensity; whereas after CBS-X addition, the amide I increased with a peak shift (from 1652.04 cm−1 to 1651.43 cm−1) and the amide II decreased with a peak shift (from 1547.03 cm−1 to 1548.07 cm−1) in absorbance intensity. The peak position shift of amide I, along with the changes in peak intensity, implied that the α-helical content of HSA has been changed after additions of CBS and CBS-X. All changes of these peak positions and intensities suggested that CBS and CBS-X interacted with C
O stretching vibration, whereas amide II (1500–1600 cm−1) is attributed to the coupling of the N–H in plane bending and C–N stretching modes.53 As shown in Fig. 7(c), the characteristic amide I and II absorption peak positions of free HSA are at 1652.04 and 1547.03 cm−1, respectively. After CBS addition, the two amide bands apparently decreased with a peak shift (amide I: from 1652.04 cm−1 to 1652.66 cm−1; amide II: from 1547.03 cm−1 to 1548.04 cm−1) in absorbance intensity; whereas after CBS-X addition, the amide I increased with a peak shift (from 1652.04 cm−1 to 1651.43 cm−1) and the amide II decreased with a peak shift (from 1547.03 cm−1 to 1548.07 cm−1) in absorbance intensity. The peak position shift of amide I, along with the changes in peak intensity, implied that the α-helical content of HSA has been changed after additions of CBS and CBS-X. All changes of these peak positions and intensities suggested that CBS and CBS-X interacted with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–N groups in the protein polypeptides, thereby resulting in the rearrangement of polypeptide carbonyl hydrogen bonding networks and, ultimately, in the change in the secondary structure of HSA.54
O and C–N groups in the protein polypeptides, thereby resulting in the rearrangement of polypeptide carbonyl hydrogen bonding networks and, ultimately, in the change in the secondary structure of HSA.54
3.3.2. CD spectroscopy. CD spectroscopy is a powerful and sophisticated tool to distinguish conformational changes of a protein in protein–ligand interactions, which are conducted to evaluate further the structural changes of the secondary structures of HSA after the addition of ligands (CBS and CBS-X). The obtained CD spectra of HSA in the absence and presence of CBS and CBS-X are displayed in Fig. 7(d). The two typical negative bands of native HSA at ∼208 (π–π*) and ∼222 nm (n–π*) represent the α-helix structures of the protein. Generally, the MRE208 is the mean residue ellipticity (deg cm2 dmol−1) value at 208 nm and can be used to calculate the α-helix content of free or combined protein using the following equations:55| |
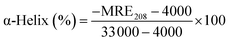 | (11) |
| |
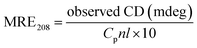 | (12) |
where Cp is the molar concentration of the protein; n is the number of amino acid residues in the protein, and l is the path length. The calculated results exhibited a decrease in α-helix content from 54.09% in the free HSA to 52.70% at a CBS to HSA molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, whereas α-helix structures increased to 57.10% at a CBS-X to HSA molar ratio of 1
1, whereas α-helix structures increased to 57.10% at a CBS-X to HSA molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. These changes in α-helix content obtained from CD data are consistent with the results of FT-IR spectroscopy, indicating that the secondary structure of HSA has been altered during the reaction processes of HSA with CBS and CBS-X.
1. These changes in α-helix content obtained from CD data are consistent with the results of FT-IR spectroscopy, indicating that the secondary structure of HSA has been altered during the reaction processes of HSA with CBS and CBS-X.
3.4. Molecular docking studies
The flexible molecular docking was complemented in this study to describe the difference between CBS–HSA and CBS-X–HSA interactions. The optimal docked results are shown in Fig. 8. CBS and CBS-X molecules interacted with HSA at the active sites II and I, respectively. The binding position is consistent with the previous results of 1H STD-NMR. The binding energy values for CBS–HSA and CBS-X–HSA interactions were −9.76 and −9.48 kcal mol−1, indicating that the protein–ligand complex stability followed the order CBS > CBS-X, which is consistent with the previous fluorescence spectra. Fig. 8 shows the binding microenvironments of CBS and CBS-X. In the CBS–HSA interaction, the CBS molecule was surrounded by some hydrophobic residues (Met 548, Val 409, Ala 406), polar residues (Glu 492, Lys 414, Lys 402, Arg 410, Lys 545), and a hydrophilic residue (Asn 405), indicating that hydrophobic forces combined with certain electrostatic interactions promoted the binding process of CBS to HSA. Some hydrophobic residues (Ala 291, Leu 238, Trp 214, Val 455), hydrophilic residues (Tyr 150, Cys 448, Tyr 452), and polar residues (Arg 257, His 242, Lys 199, Lys 195, Arg 222, Asp 451, Lys 436) around CBS-X molecule demonstrated the same binding model (hydrophobic forces combined with certain electrostatic interactions) in the CBS-X–HSA interaction. Moreover, CBS formed three hydrogen bonds with the Lys 545, Lys 414, and Arg 410 residues, whereas CBS-X formed three hydrogen bonds with Tyr 150, His 242, Arg 257, and Lys 436 residues. These docking results further proved that the major hydrophobic forces that are coupled with electrostatic interactions and hydrogen bonds promote the interactions between CBS and CBS-X and HSA.
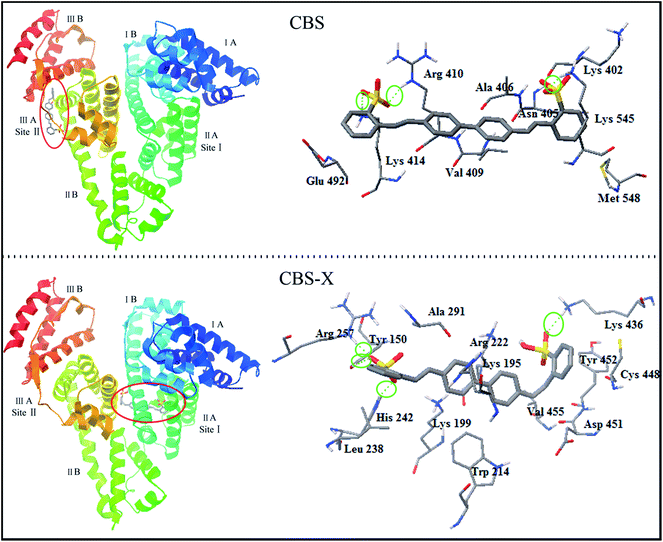 |
| | Fig. 8 Three-dimensional docking representations for CBS–HSA and CBS-X–HSA interactions. The sites of the interaction of CBS and CBS-X are magnified on the right panel along with the labeling of the HSA residues near the ligands at the interaction site. The H-bonds (green dotted line) are surrounded by a green coil. | |
4. Conclusions
In this work, the molecular interaction between fluorescent whitening agents (CBS and CBS-X) and the important transport protein HSA was first studied using 1H STD-NMR combined with multi-spectroscopy. The 1H STD-NMR data indicated that CBS–HSA and CBS-X–HSA interactions could be efficiently conformed, and CBS and CBS-X exhibit a strong affinity to HSA. The fluorescence studies revealed that CBS and CBS-X can interact with HSA through dynamic quenching, and the binding affinity follows the order: CBS > CBS-X. 1H STD-NMR successfully identified the binding sites that CBS and CBS-X can bind to HSA at sites II and I, respectively, which was further verified by molecular docking. Different degrees of microenvironmental and conformational changes occurred to HSA affected by CBS and CBS-X, which was proven by FT-IR and CD spectra. The results reported in this paper are helpful to improve the understanding of the biological effects and toxicity of CBS and CBS-X in biological processes, and imply a potentially theoretical foundation for studying the harm of DSBP-FWAs to the human body.
Acknowledgements
This work was financially supported by Ningbo Municipal Science and Technology Bureau (15H0640) and Chengdu Science and Technology Bureau (2015-HM01-00380-SF). Thanks to School of Life Sciences, University of Science and Technology of China for NMR technical assistance.
References
- W. C. Shu and W. H. Ding, J. Chromatogr. A, 2005, 1088, 218–223 CrossRef CAS PubMed.
- H. C. Chen, S. P. Wang and W. H. Ding, J. Chromatogr. A, 2006, 1102, 135–142 CrossRef CAS PubMed.
- Z. Wu, Y. Xu, M. Li, X. Guo, Y. Xian and H. Dong, RSC Adv., 2016, 6, 17941–17946 RSC.
- Q. Wang, X. Chen, B. Qiu, L. Zhou, H. Zhang, J. Xie, Y. Luo and B. Wang, J. Sep. Sci., 2016, 39, 1242–1248 CrossRef CAS PubMed.
- E. T. Iamazaki and T. D. Z. Atvars, Langmuir, 2007, 23, 12886–12892 CrossRef CAS PubMed.
- S. Toffanin, S. Kim, S. Cavallini, M. Natali, V. Benfenati, J. J. Amsden, D. L. Kaplan, R. Zamboni, M. Muccini and F. G. Omenetto, Appl. Phys. Lett., 2012, 101, 091110 CrossRef.
- S. Cavallini, S. Toffanin, C. Chieco, A. Sagnella, F. Formaggio, A. Pistone, T. Posati, M. Natali, M. Caprini, V. Benfenati, N. Di Virgilio, G. Ruani, M. Muccini, R. Zamboni and F. Rossi, Composites, Part B, 2015, 71, 152–158 CrossRef CAS.
- K. J. Smit and K. P. Ghiccino, J. Polym. Sci., Part B: Polym. Phys., 1991, 29, 1397–1405 CrossRef CAS.
- X. Guo, Y. Xian, H. Luo, Y. Wu, D. Luo, Y. Chen, Y. Lu and D. Xu, Anal. Methods, 2013, 5, 6086–6093 RSC.
- C. H. Gloxhuber and H. Blochmg, Clin. Toxicol., 1978, 13, 171–203 CrossRef CAS PubMed.
- H. Liao, J. Sep. Sci., 2016, 39, 3683–3689 CrossRef CAS PubMed.
- C. D. Kanakis, P. A. Tarantilis, M. G. Polissiou, S. Diamantoglou and H. A. Tajmir-Riahi, J. Mol. Struct., 2006, 798, 69–74 CrossRef CAS.
- X. Li and S. Wang, New J. Chem., 2015, 39, 386–395 RSC.
- T. G. Gantchev, R. Ouellet and J. E. van Lier, Arch. Biochem. Biophys., 1999, 366, 21–30 CrossRef CAS PubMed.
- N. V. Rakotoarivelo, P. Perio, E. Najahi and F. Nepveu, J. Phys. Chem. B, 2014, 118, 13477–13485 CrossRef CAS PubMed.
- Q. Wang, X. Ma, J. He, Y. Li and H. Li, RSC Adv., 2015, 5, 44696–44704 RSC.
- D. Wu, Y. Zhai, J. Yan, K. Xu, Q. Wang, Y. Li and H. Li, RSC Adv., 2015, 5, 11036–11042 RSC.
- D. Wu, J. Yan, J. Wang, Q. Wang and H. Li, Food Chem., 2015, 170, 423–429 CrossRef CAS PubMed.
- A. Basu and G. S. Kumar, J. Hazard. Mater., 2014, 273, 200–206 CrossRef CAS PubMed.
- X. Zhou, X. Li and X. Chen, Dyes Pigm., 2013, 98, 212–220 CrossRef CAS.
- C. Ràfols, S. Zarza and E. Bosch, Talanta, 2014, 130, 241–250 CrossRef PubMed.
- L. Trnková, I. Boušová, V. Staňková and J. Dršata, J. Mol. Struct., 2011, 985, 243–250 CrossRef.
- J. Yan, Y. Cai, Y. Wang, X. Lin and H. Li, Food Chem., 2014, 143, 82–89 CrossRef CAS PubMed.
- M. Šimšíková, Arch. Biochem. Biophys., 2016, 593, 69–79 CrossRef PubMed.
- J. Yan, Q. Wang, Q. Pan, Z. Rao, Y. Su and H. Li, J. Lumin., 2013, 137, 180–185 CrossRef CAS.
- C. F. Martinez-Farina, N. McCormick, A. W. Robertson, H. Clement, A. Jee, A. Ampaw, N. L. Chan, R. T. Syvitski and D. L. Jakeman, Org. Biomol. Chem., 2015, 13, 10324–10327 CAS.
- A. Antanasijevic, B. Ramirez and M. Caffrey, J. Biomol. NMR, 2014, 60, 37–44 CrossRef CAS PubMed.
- B. Tang, Y. Huang, H. Yang, P. Tang and H. Li, J. Photochem. Photobiol., B, 2016, 165, 24–33 CrossRef CAS PubMed.
- H. Yang, Y. Huang, D. Wu, J. Yan, J. He and H. Li, New J. Chem., 2016, 40, 2530–2540 RSC.
- M. Mayer and B. Meyer, J. Am. Chem. Soc., 2001, 123, 6108–6117 CrossRef CAS PubMed.
- S. A. K. Tanoli, N. U. Tanoli, T. M. Bondancia, S. Usmani, Z. Ul-Haq, J. B. Fernandes, S. S. Thomasi and A. G. Ferreira, RSC Adv., 2015, 5, 23431–23442 RSC.
- S. Mudgal, I. Keresztes, G. W. Feigenson and S. S. H. Rizvi, Food Chem., 2016, 197, 84–91 CrossRef CAS PubMed.
- X. L. Jin, X. Wei, F. M. Qi, S. S. Yu, B. Zhou and S. Bai, Org. Biomol. Chem., 2012, 10, 3424–3431 CAS.
- Z. Chi and R. Liu, Biomacromolecules, 2011, 12, 203–209 CrossRef CAS PubMed.
- D. M. Dias, J. P. G. L. M. Rodrigues, N. S. Domingues, A. M. J. J. Bonvin and M. M. C. A. Castro, Eur. J. Inorg. Chem., 2013, 2013, 4619–4627 CrossRef CAS.
- P. B. Kandagal, J. Seetharamappa, S. Ashoka, S. M. Shaikh and D. H. Manjunatha, Int. J. Biol. Macromol., 2006, 39, 234–239 CrossRef CAS PubMed.
- A. S. Sharma, S. Anandakumar and M. Ilanchelian, RSC Adv., 2014, 4, 36267–36281 RSC.
- S. Li, J. He, Y. Huang, Q. Wang, H. Yang, K. Xu and H. Li, RSC Adv., 2016, 6, 85811–85819 RSC.
- M. van de Weert and L. Stella, J. Mol. Struct., 2011, 998, 144–150 CrossRef CAS.
- J. R. Lakowicz, Principles of fluorescence spectroscopy, Springer, Singapore, 3rd edn, 2009 Search PubMed.
- Z. Q. Xu, Q. Q. Yang, J. Y. Lan, J. Q. Zhang, W. Peng, J. C. Jin, F. L. Jiang and Y. Liu, J. Hazard. Mater., 2016, 301, 242–249 CrossRef CAS PubMed.
- S. Huang, F. Zhu, Q. Qian, Q. Xiao and W. Su, Biol. Trace Elem. Res., 2015, 164, 150–161 CrossRef CAS PubMed.
- S. Li, K. Huang, M. Zhong, J. Guo, W. Z. Wang and R. Zhu, Spectrochim. Acta, Part A, 2010, 77, 680–686 CrossRef PubMed.
- K. Ray, R. Badugu and J. R. Lakowicz, RSC Adv., 2015, 5, 54403–54411 RSC.
- I. Vayá, V. Lhiaubet-Vallet, M. C. Jiménez and M. A. Miranda, Chem. Soc. Rev., 2014, 43, 4102–4122 RSC.
- M. D. Meti, S. T. Nandibewoor, S. D. Joshi, U. A. More and S. A. Chimatadar, J. Iran. Chem. Soc., 2016, 13, 1325–1338 CrossRef CAS.
- P. Mitra, U. Pal, N. Chandra Maiti, A. Ghosh, A. Bhunia and S. Basu, RSC Adv., 2016, 6, 53454–53468 RSC.
- X. Li, D. Chen, G. Wang and Y. Lu, Eur. J. Med. Chem., 2013, 70, 22–36 CrossRef CAS PubMed.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef CAS PubMed.
- S. Huang, H. Qiu, Y. Liu, C. Huang, J. Sheng, W. Su and Q. Xiao, Colloids Surf., B, 2015, 136, 955–962 CrossRef CAS PubMed.
- O. K. Abou-Zied and S. A. J. Sulaiman, Dyes Pigm., 2014, 110, 89–96 CrossRef CAS.
- M. Shahlaei, B. Rahimi, A. Nowroozi, M. R. Ashrafi-Kooshk, K. Sadrjavadi and R. Khodarahmi, Chem.-Biol. Interact., 2015, 242, 235–246 CrossRef CAS PubMed.
- H. Chen, H. Rao, J. Yang, Y. Qiao, F. Wang and J. Yao, J. Environ. Sci. Health, Part B, 2016, 51, 154–159 CrossRef CAS PubMed.
- J. He, H. Yang, S. Li, K. Xu, Q. Wang, Y. Huang and H. Li, RSC Adv., 2016, 6, 61119–61128 RSC.
- N. Shahabadi and S. M. Fili, Spectrochim. Acta, Part A, 2014, 118, 422–429 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra04008c |
|
| This journal is © The Royal Society of Chemistry 2017 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article c
c
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) at room temperature. Competition studies were performed with ibuprofen and warfarin, respectively. Competitive NMR samples, including identical HSA and ligand concentrations containing two different concentrations of warfarin (600 and 1200 μM) or ibuprofen (600 and 1200 μM), were analyzed. In the STD experiments, 50 ms Gaussian pulses were applied at an on-resonance saturation of −0.5 ppm and an off-resonance saturation of 34 ppm. All spectra were acquired with a sweep width of 8389.26 Hz, 256 transients, and acquisition time of 1 s. The total scan number was 1024 and typically used 16 ppm spectral widths for the 1H STD spectra. The subtraction was performed after every STD scan by phase cycling. Each sample was recorded with Watergate solvent suppression to obtain a standard 1H spectrum. All the spectra were processed and analyzed using the ACD/CNMR software (Advanced Chemistry Development, Inc., version 11.0).
40) at room temperature. Competition studies were performed with ibuprofen and warfarin, respectively. Competitive NMR samples, including identical HSA and ligand concentrations containing two different concentrations of warfarin (600 and 1200 μM) or ibuprofen (600 and 1200 μM), were analyzed. In the STD experiments, 50 ms Gaussian pulses were applied at an on-resonance saturation of −0.5 ppm and an off-resonance saturation of 34 ppm. All spectra were acquired with a sweep width of 8389.26 Hz, 256 transients, and acquisition time of 1 s. The total scan number was 1024 and typically used 16 ppm spectral widths for the 1H STD spectra. The subtraction was performed after every STD scan by phase cycling. Each sample was recorded with Watergate solvent suppression to obtain a standard 1H spectrum. All the spectra were processed and analyzed using the ACD/CNMR software (Advanced Chemistry Development, Inc., version 11.0).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratios) were observed in sequence. Experimental data were analyzed using the tail-fitting method, and the qualities of the fits were assessed using χ2 values and residuals.
1 molar ratios) were observed in sequence. Experimental data were analyzed using the tail-fitting method, and the qualities of the fits were assessed using χ2 values and residuals.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratios).
1 molar ratios).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 generations, as well as a medium number of 2
000 generations, as well as a medium number of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 energy evaluations. Other parameters were set as the default protocol settings. Subsequently, the lowest-energy docked conformation was applied to further locate the most suitable binding mode.
000 energy evaluations. Other parameters were set as the default protocol settings. Subsequently, the lowest-energy docked conformation was applied to further locate the most suitable binding mode.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = −ΔH/RT + ΔS/R
K = −ΔH/RT + ΔS/R
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K
K
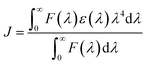
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration, whereas amide II (1500–1600 cm−1) is attributed to the coupling of the N–H in plane bending and C–N stretching modes.53 As shown in Fig. 7(c), the characteristic amide I and II absorption peak positions of free HSA are at 1652.04 and 1547.03 cm−1, respectively. After CBS addition, the two amide bands apparently decreased with a peak shift (amide I: from 1652.04 cm−1 to 1652.66 cm−1; amide II: from 1547.03 cm−1 to 1548.04 cm−1) in absorbance intensity; whereas after CBS-X addition, the amide I increased with a peak shift (from 1652.04 cm−1 to 1651.43 cm−1) and the amide II decreased with a peak shift (from 1547.03 cm−1 to 1548.07 cm−1) in absorbance intensity. The peak position shift of amide I, along with the changes in peak intensity, implied that the α-helical content of HSA has been changed after additions of CBS and CBS-X. All changes of these peak positions and intensities suggested that CBS and CBS-X interacted with C
O stretching vibration, whereas amide II (1500–1600 cm−1) is attributed to the coupling of the N–H in plane bending and C–N stretching modes.53 As shown in Fig. 7(c), the characteristic amide I and II absorption peak positions of free HSA are at 1652.04 and 1547.03 cm−1, respectively. After CBS addition, the two amide bands apparently decreased with a peak shift (amide I: from 1652.04 cm−1 to 1652.66 cm−1; amide II: from 1547.03 cm−1 to 1548.04 cm−1) in absorbance intensity; whereas after CBS-X addition, the amide I increased with a peak shift (from 1652.04 cm−1 to 1651.43 cm−1) and the amide II decreased with a peak shift (from 1547.03 cm−1 to 1548.07 cm−1) in absorbance intensity. The peak position shift of amide I, along with the changes in peak intensity, implied that the α-helical content of HSA has been changed after additions of CBS and CBS-X. All changes of these peak positions and intensities suggested that CBS and CBS-X interacted with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–N groups in the protein polypeptides, thereby resulting in the rearrangement of polypeptide carbonyl hydrogen bonding networks and, ultimately, in the change in the secondary structure of HSA.54
O and C–N groups in the protein polypeptides, thereby resulting in the rearrangement of polypeptide carbonyl hydrogen bonding networks and, ultimately, in the change in the secondary structure of HSA.54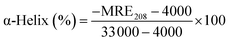
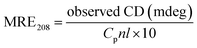
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, whereas α-helix structures increased to 57.10% at a CBS-X to HSA molar ratio of 1
1, whereas α-helix structures increased to 57.10% at a CBS-X to HSA molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. These changes in α-helix content obtained from CD data are consistent with the results of FT-IR spectroscopy, indicating that the secondary structure of HSA has been altered during the reaction processes of HSA with CBS and CBS-X.
1. These changes in α-helix content obtained from CD data are consistent with the results of FT-IR spectroscopy, indicating that the secondary structure of HSA has been altered during the reaction processes of HSA with CBS and CBS-X.







