Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
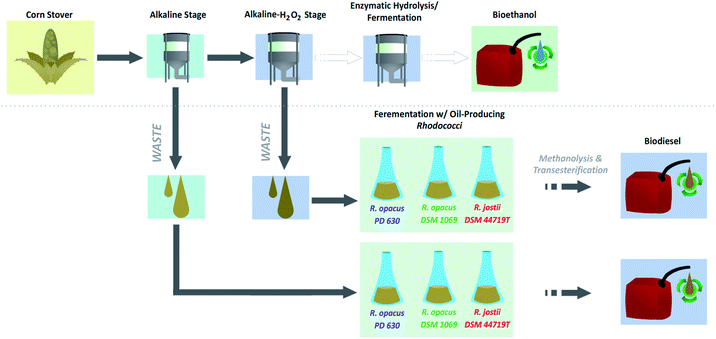
Conversion of corn stover alkaline pre-treatment waste streams into biodiesel via Rhodococci†
Rosemary K. Le‡ab,
Tyrone Wells Jr.‡
ab,
Parthapratim Dasab,
Xianzhi Mengab,
Ryan J. Stoklosacd,
Aditya Bhallade,
David B. Hodgecdf,
Joshua S. Yuang and
Arthur J. Ragauskas*abh
aDepartment of Chemical & Biomolecular Engineering, University of Tennessee Knoxville, Knoxville, Tennessee 37996, USA
bBioscience Division, Oak Ridge National Laboratory, Oakridge, Tennessee 37831, USA
cDepartment of Chemical Engineering & Materials Science, Michigan State University, East Lansing, Michigan 48824, USA
dGreat Lakes Bioenergy Research Center, Michigan State University, East Lansing, Michigan 48824, USA
eDepartment of Biochemistry, Michigan State University, East Lansing, Michigan 48824, USA
fDepartment of Biosystems & Agricultural Engineering, Michigan State University, East Lansing, Michigan 48824, USA
gSynthetic and Systems Biology Innovation Hub, Department of Plant Pathology and Microbiology, Texas A&M University, 21230 TAMU, College Station, Texas 77843, USA
hDepartment of Forestry, Wildlife, and Fisheries, Center for Renewable Carbon, University of Tennessee, Knoxville, Tennessee 37996, USA. E-mail: aragauskas@utk.edu
First published on 13th January 2017
Abstract
The bioconversion of second-generation cellulosic ethanol waste streams into biodiesel via oleaginous bacteria is a novel optimization strategy for biorefineries with substantial potential for rapid development. In this study, one- and two-stage alkali/alkali-peroxide pretreatment waste streams of corn stover were separately implemented as feedstocks in 96 h batch reactor fermentations with wild-type Rhodococcus opacus PD 630, R. opacus DSM 1069, and R. jostii DSM 44719T. Here we show using 31P-NMR, HPAEC-PAD, and SEC analyses, that the more rigorous and chemically-efficient two-stage chemical pretreatment effluent provided higher concentrations of solubilized glucose and lower molecular weight (∼70–300 g mol−1) lignin degradation products thereby enabling improved cellular density, viability, and oleaginicity in each respective strain. The most significant yields were by R. opacus PD 630, which converted 6.2% of organic content with a maximal total lipid production of 1.3 g L−1 and accumulated 42.1% in oils based on cell dry weight after 48 h.
1. Introduction
The need for sustainable and supplemental energy platforms will dramatically increase over the next decade as newly industrialized countries continue to modernize and place a greater demand on nonrenewable fossil fuel reserves.1–3 Studies conducted by the International Energy Agency (IEA) determined that the global demand for energy will increase 50% by 2030, with most of this increase originating by China and India. The strongest growing demand for energy comes from the global transportation sector, which accounts for ∼60% of the total oil demand and is projected to steadily grow by 1.8% per year towards 2035; approximately 80% of energy in this sector is dedicated strictly to road transport.4 Hence, the development of renewable transportation energy has been a growing and apt target for sustainability research.4 Accordingly, advanced transportation biofuels sourced from renewable lignocellulosic biomass will, in part, better leverage limited energy resources as well as address security and environmental issues that will arise as demands for fuel increase.5 Specifically, advanced biofuels sourced from inedible second-generation agricultural feedstocks and biorefinery waste streams will be key to further optimizing plant-to-fuel processes while uniquely circumventing public concerns regarding the food vs. fuel debate.6–8Advanced transportation biofuels are composed of two distinct categories: liquid alcohol-based fuels that include butanol and ethanol, and diesel-like hydrocarbon-based fuels such as biodiesel.9 Together, bioethanol and biodiesel are the most commonly blended liquid biofuels used in modern global transportation markets at roughly 150 billion liters per annum, an output that is predicted to nearly double by 2021 and address roughly 25% of worldwide transport fuel demand by 2050.10,11 Bioethanol is commonly produced from the fermentation of sugar or starch largely sourced from plant biomass (e.g. energy crops, agricultural residues, wood materials, etc.).12 Alternatively, biodiesel is a product of the transesterification of triacylglycerides (TAG) typically sourced from animal fats and plant oils into long chain fatty acid methyl esters (FAME).4,10,11 Although the use of bioethanol as a supplemental liquid fuel additive is noteworthy, the comparative energy density (26.8 MJ kg−1) is roughly 60% of pure gasoline (46 MJ kg−1), and the extent of blending is limited in some countries due to an incompatible dated transportation infrastructure.13,14 Comparatively, the energy density of biodiesel (38 MJ kg−1) is ∼84% compared to diesel (45 MJ kg−1) and can be used as a compatible fungible fuel for existing diesel engines.15 Despite this, sourcing biodiesel at commercial scale is often inhibited by the cost of limited production capacity.16,17 Interestingly, the conversion of biorefinery waste streams into biodiesel can provide a strategic avenue to better optimize biofuel production.18,19
The use of microbial autotrophs (e.g. algae) and heterotrophs (e.g. fungi and bacteria) that can accumulate intracellular lipids for the sake of biodiesel production is a rapidly growing area of study towards biorefinery optimization.20,21 While algae- and yeast-to-biofuel processes are prominent and are continuously being refined, oil-producing bacteria that are well-adapted for the degradation of plant biomass, also boasting a broader threshold of viable nutrients and tolerable growth conditions, represent an area of research with considerable potential for advancement.17,18,22,23 The genera of bacteria used in this work commonly include Acinetobacter, Mycobacterium, Streptomyces, and more recently Rhodococcus.24 R. opacus and R. jostii have extensively been studied in the last few years and are well noted for their ability to accumulate oleaginous extents of intracellular single cell oils based on cell dry weight (CDW) >20%. For example, R. opacus PD 630 has exhibited high rates of oleaginicity (lipids per cell dry weight), above 80% CDW, when utilizing glucose as a carbon source under nitrogen-limited conditions.24,25 Beyond glucose, these wild-type species have also demonstrated the effective degradation of aromatic material common in lignocellulosic biomass, specifically, substituted aryl species found in lignin. The biochemical degradation of aryl units into lipids is made feasible via enzyme-mediated sequences such as the β-ketoadipate pathway (β-KAP).26 For example, R. jostii was previously shown to accumulate over 55% CDW in TAG utilizing benzoate as a carbon source, which is a comparable analog of lignin depolymerization products.27 Furthermore, R. opacus strains PD 630 and DSM 1069 have been shown to exercise the β-KAP pathway to utilize highly purified kraft lignin as a sole source of carbon and energy, albeit with limited extents of oleaginicity (<5.0% CDW), which was attributed to the recalcitrant nature of lignin.
Pretreatment methodologies for cellulosic ethanol are being implemented in order to degrade, solubilize, and reduce the integrity of the lignin–hemicellulose matrix, thereby providing greater access to cellulose fibers for subsequent bioethanol processing.28,29 For example, standalone alkali pretreatment strategies have been designed to degrade and solubilize lignin and hemicellulose with corn stover.30–33 These alkali processes are being further optimized for industrial application33–39 and have been reviewed by Kim et al.40 The efficacy of a multi-stage pretreatment processes involving alkali pre-extraction with alkaline-oxidative post-treatments with corn stover has been investigated by Liu et al. and demonstrated that mild NaOH pre-extraction (<100 °C) followed by alkali-hydrogen peroxide post-extraction (ALK-AHP) significantly reduced the amount of required active chemical agents needed to extract lignin.33 ALK-AHP pretreated corn stover also exhibited improved enzyme digestibility and fermentability towards second generation bioethanol production over standalone alkali processing (ALK).33
Notably, both the standalone ALK and two-stage ALK-AHP pretreatments generate an aqueous waste effluent fraction that contains low molecular weight lignin oligomers and carbohydrate content that could be further suited as a substrate for oleaginous Rhodococci towards biodiesel generation in a method that could substantially optimize biorefinery waste streams (Fig. 1).34,35 Several research groups have demonstrated the advantages in using alkaline pretreatment of biomass to yield effluents that can be utilized by microorganism to generate a variety of useful chemicals, including lipids in yeast41,42 and plastic building block, polyhydroxyalkanoate in Pseudomonas putitda.43 Furthermore, work by Linger et al. and Katahira et al. have demonstrated that using alkaline or base-catalyzed pretreatments, which depolmerize lignin, produce low molecular weight aromatics, and yield a polysaccharide rich effluent. These low molecular weight molecules can then be biologically funneled by bacterial aromatic catabolic pathways to produce useful fuels and chemicals and overcome the intrinsically heterogeneous nature of lignin.43,44
Due to the chemical-efficient and rigorous two-stage ALK-AHP treatment producing a superior feedstock effluent with enhanced concentrations of degraded lignin and carbohydrate products, we hypothesize this substrate will be more apt for microbial upgrading into biodiesel. We expect to see increased lipid yield compared to traditional standalone ALK pretreatment.
To investigate this matter, the efficacy of ALK and ALK-AHP pretreatment effluents (prepared as previously detailed)33 with corn stover (Zea mays L. Pioneer hybrid 36H56) as a feedstock for wild-type R. opacus PD 630, R. opacus DSM 1069, and R. jostii DSM 44719T was studied.
2. Experimental
2.1. Corn stover feedstock
Corn stover effluents were sourced from the Great Lakes Bioenergy Research Center (GLBRC) and harvested from the Arlington Research Facility in Wisconsin, USA. Untreated corn stover had a composition of 30.2% glucan, 18.5% xylan, 1.1% arabinan, 24.1% Klason lignin, 6% ASL, and 17.7% ash, with 2.3% attributable to acetate groups. Alkali loading was 8% NaOH and solid–liquid separation was performed utilizing a 200 mesh stainless steel screen and centrifugation at 8000 rpm for 5 min to remove fine particulates. Pretreatments were performed as previously detailed.33 Approximately 20.8% of total dry solids from the ALK effluent wash and 52.1% ALK-AHP effluent was non-ash content.2.2. Cell cultivation and monitoring
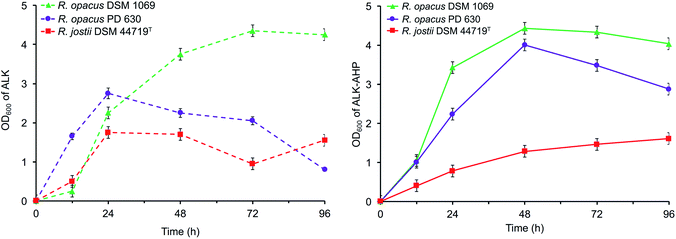
Cell growth, assaying, monitoring, and methanolysis were performed based on previous literature.45,46 Wild-type R. opacus PD 630, R. opacus DSM 1069, and R. jostii DSM 44719T were sourced from DSMZ and cultured following conventional methods.46 After an initial adaptation period of 48 h in the ALK and ALK-AHP effluents at pH 7.2, 40.0 mg mL−1 w/v solids loading, amended with minimal salt medium,39 and 0.1% w/v nitrogen content, the effluents were implemented as a sole source of energy and carbon under nitrogen-limited conditions (pH 7.2, 40.0 mg mL−1, 0.05 w/v% nitrogen content) in a separate and parallel series of 96 h batch reactor fermentations (150 mL) to analyse the composition of intracellular lipids produced during the exponential and stationary phases of cell growth; the duration of these phases were resolved by tracking the cell density via optical density measurements taken at 600 nm (OD600) and were determined to occur within 0–24 h and 24–72 h, respectively (Fig. 2).2.3. Instrumental analyses
Sugar release of monosaccharides was quantified using high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD). Specifically, a Dionex ICS-3000 ion chromatography with CarboPac PA-1 column was implemented. The temperature of the column was set to 23 °C and the eluents A and B were 100% DI water (18 MΩ cm) and 200 mM NaOH, respectively. The flow rate was set to 0.3 mL min−1. Triplicate samples of the fermentation broths were analyzed and each analyte was quantified using linear regression based on calibration curves of an external standard of glucose, xylose, mannose, galactose, and arabinose based on a 1 × 10−3 to 1 × 10−1 mg mL−1 concentrations were used with an R2 values calculated above 0.995 (following NREL/TP-510-42618). Errors associated with Dionex monosaccharide concentration determinations were below ±1%.4631P NMR analysis of the lignin samples was resolved according to previous studies.47 In brief, endo-N-hydroxyl-5-norbornene-2,3-dicarboximide, Cr(III) acetylacetonate and 2-chloro-4,4,5,5-tetramethyl-1,3,2-dioxaphospholane were used as the internal standard, relaxation and phosphitylating agent, respectively. Spectra were collected on a Bruker Avance III 400 MHz NMR spectrometer (fitted with a 5 mm Broadband Observe probe) that utilized an inverse-gated decoupling pulse sequence with a 90° pulse. The spectra were run for 64 scans with 1 s acquisition time, a delay time of 25 s, and referenced with respect to the residual water peak in the NMR solvent at 132.2 ppm.
GC-MS procedures were followed as described elsewhere.48 Likewise, SEC (size exclusion chromatography) analysis was performed as described previously.49 In detail, molecular mass distributions of the acetylated lignin samples were then analysed with an Agilent GPC SECurity 1200 system equipped with four Waters Styragel columns (HR1, HR2, HR4, HR6), and an Agilent UV detector set to 270 nm, using tetrahydrofuran (THF) as the mobile phase (1.0 mL min−1), and an injection volume of 20.0 μL. A standard polystyrene sample was used for calibration.
3. Results and discussion
3.1. Tracking cellular growth
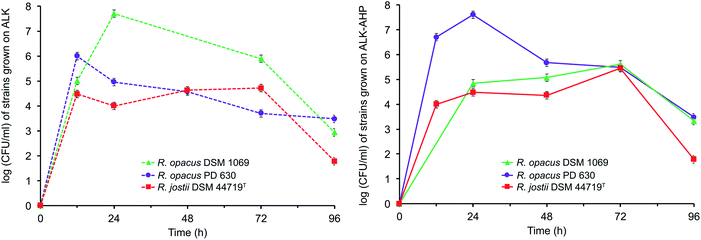
The data showed that the approximate optical cell densities of R. jostii DSM 44719T were largely consistent in either ALK or ALK-AHP effluent, but significantly lower than the two R. opacus strains as shown in Fig. 2. In ALK media, the absorbance of PD 630 and DSM 1069 were 1.2 and 2.2-fold greater than DSM 44719T at 48 h, the median point of incubation, respectively. Moreover, when using ALK-AHP, the approximate cell densities of PD 630 and DSM 1069 were more improved at roughly 3.5-fold higher than DSM 44719T at the same time period. In summary, the cell densities were improved with ALK-AHP as the feedstock and particularly for PD 630 and DSM 1069. Additionally, fluctuations of cell dry weight (CDW) as an estimate of total cell biomass per volume (mg mL−1) were measured, which complimented OD600 data, in that the optimal cell density and cell dry weight occurred at approximately 48 h, and is illustrated and detailed in ESI Fig. 1.† The CDW was significantly higher for ALK-AHP, which reached maxima of 2.13, 2.02 and 1.10 mg mL−1 for PD 630, DSM 1069, and DSM 44719T at 48 h. In contrast, dry cells per volume for strains grown in ALK media remained below 0.30 mg mL−1 throughout 96 h of incubation, thereby supporting that ALK-AHP effluent was the more optimal substrate for the propagation of cellular biomass.To assess cell viability, optical density measurements (OD600) and the concentration of colony forming units per mL (CFU mL−1) was monitored via serial dilute plating (SDP), illustrated in Fig. 2 and 3, respectively. As shown by OD600 and SDP, the viability of DSM 44719T was the lowest of the three strains and particularly consistent in either media while viability of cells grown in ALK-AHP media were distinctly improved, particularly for PD 630. When utilizing ALK effluent as a substrate, both PD 630 and DSM 1069 reached CFU mL−1 concentrations of a magnitude of 105 after 12 h of incubation, compared to DSM 44719T, which exhibited a magnitude of 104. At 24 h, near the end of the exponential phase, DSM 1069 displayed the highest viability with ALK effluents (7.7 × 107 CFU mL−1) while, at the same period, PD 630 exhibited the highest viability for ALK-AHP effluents (7.6 × 107 CFU mL−1) as shown in Fig. 3. In contrast, CFU mL−1 for DSM 44719T cells principally remained within a magnitude of 104 for 12–72 h with both feedstocks. This result suggests that both effluent feedstocks were capable of supporting the proliferation of new Rhodococci cells. Notably, while both R. opacus strains exhibited substantial increases in cell density when utilizing ALK-AHP as an effluent from 0–48 h, the viability of PD 630 was markedly improved over DSM 1069 (and consequently DSM 44719T) during the same interval, which may support that PD 630 experienced the most optimal growth, overall, of the three strains based on cellular density and viability.
3.2. Tracking of lipid yields
Using gas chromatography-mass spectrometry (GC-MS), the total yield of FAME (mg L−1) and the resultant oleaginicity (% CDW, lipids per cell dry weight) of each strain were calculated. Overall, the yield of FAME and oleaginicity were substantially improved with ALK-AHP effluent as a feedstock compared to ALK effluent as summarized in Fig. 4. In detail, when ALK-AHP effluent was implemented as a substrate, PD 630 exhibited the highest total FAME and oleaginicity (1.28 g mL−1, 42.1%) at 48 h. Interestingly, at this same period, DSM 44719T exhibited the second highest yield of FAME and oleaginicity (1.05 g mL−1; 23.3%) despite exhibiting lower cell density and viability compared to DSM 1069 (358.5 mg L−1; 12.6%). These results indicate that under the given conditions, DSM 1069 was more adept at generating cells while DSM 44719T more effectively accumulated lipids. Meanwhile, R. opacus PD 630 showcased the comparatively higher concentrations of viable cells and oleaginicity yields. In contrast, when ALK was the feedstock, lipid yields were 100-, 175-, and 180-fold lower for DSM 1069, PD 630, and DSM 44179T, respectively. Overall, total FAME production and oleaginicity on the ALK effluent were universally <16 mg L−1 and <0.5%, respectively with the most substantial outcome exhibited by DSM 1069 (15.7 mg L−1, 0.4%) after 96 h of incubation. This finding is consistent to the extent that DSM 1069 displayed the most optimal growth in ALK media based on OD600, CDW, and CFU mL−1 data. In all cases, the resulting FAME were determined to be enriched with palmitic (hexadecanoic), stearic (octadecanoic), and oleic acids, which is consistent with previous reports involving wild-type Rhodococci.45 A detailed distribution of the amount and variety of FAME produced during cell proliferation on the ALK and ALK-AHP effluents are shown in ESI Tables 1–3.†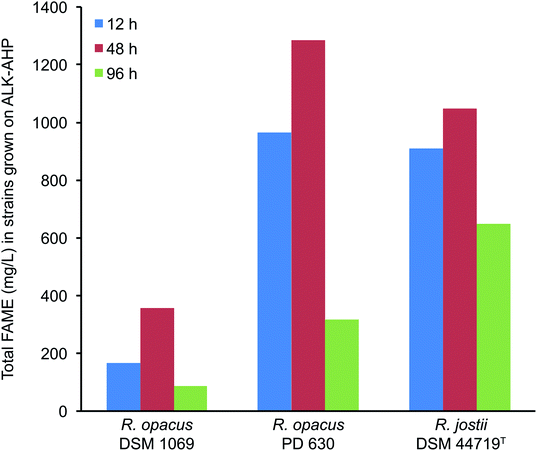 | ||
| Fig. 4 Total FAME production by Rhodococci strains using ALK-AHP effluent. Largest overall yields were exhibited by R. opacus PD 630, followed by R. jostii DSM 44719T and R. opacus DSM 1069. | ||
3.3. Tracking changes in monomeric glucose and molecular weight distribution
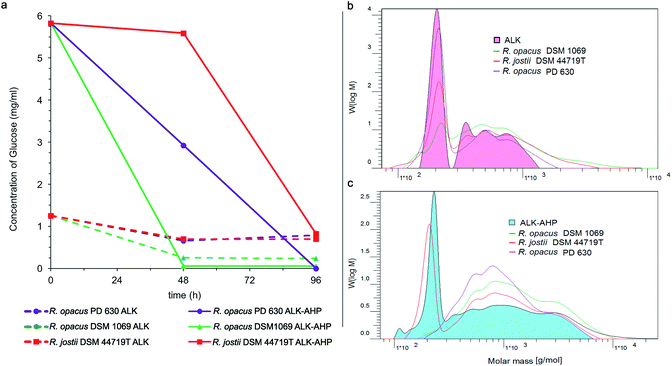
A comprehensive characterization of the feedstock broths was performed to determine changes of the degradation materials present during pretreatment. For example, alkaline corn stover pretreatments at moderate conditions (<100 °C) can release oligomeric fragments of lignin as well as hemicelluloses such as glucan and xylan along with monomeric glucose.33 As previously detailed, glucose can be targeted as a key metabolic unit by oleaginous Rhodococci.24,46 Hence, high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PED, Dionex, Sunnyvale, CA, USA) was used to track the concentrations of glucose monomers during the initial, median, and final hour of incubation. Ultimately, it was determined that the initial concentrations of glucose were 4.5-fold higher in the multistage ALK-AHP effluent (5.82 mg mL−1) compared to the standalone ALK effluent (1.25 mg mL−1). These concentrations universally declined from 0–96 h to values below 1.00 mg mL−1 for both effluents as illustrated in Fig. 5(a). The tracking of other sugars are detailed in ESI Fig. 2(a)–(c).† For example, in ALK-AHP, DSM 1069 nearly depleted solubilized glucose to a threshold below 1.00 mg mL−1 within 0–48 h and PD 630 and DSM 44719T reached this threshold during the 48–96 h interval; this result concurs with the respective order at which viability was observed to significantly depreciate for each strain as cells likely transitioned to alternative constituents.Size exclusion chromatography (SEC) was preformed to further assess changes in the distribution of molecular mass of the effluent components before and after incubation with Rhodococci. As shown in Fig. 5(b) and (c), the initial molecular weight distribution profile of both effluents generally exhibited two distinct regions: a mono-modal low molecular weight peak (100–300 g mol−1) and a multimodal shelf spanning 300–2500 g mol−1 for ALK and 300–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 for ALK-AHP. As expected, the more rigorous multistage pretreatment process yielded a substantially broader assortment of molecular weight components in the effluent. Overall, the most significant changes were observed in the <300 g mol−1 regions. For example, DSM 1069 in ALK effluent displayed the most substantial depletion of low molecular weight constituents followed by DSM 44719T and PD 630, in that order. More thorough depletion of components smaller than 300 g mol−1 was observed when ALK-AHP effluent was used as a feedstock, especially by R. opacus PD 630 and DSM 1069.
000 g mol−1 for ALK-AHP. As expected, the more rigorous multistage pretreatment process yielded a substantially broader assortment of molecular weight components in the effluent. Overall, the most significant changes were observed in the <300 g mol−1 regions. For example, DSM 1069 in ALK effluent displayed the most substantial depletion of low molecular weight constituents followed by DSM 44719T and PD 630, in that order. More thorough depletion of components smaller than 300 g mol−1 was observed when ALK-AHP effluent was used as a feedstock, especially by R. opacus PD 630 and DSM 1069.
3.4. 31P-NMR analysis
31P-NMR analysis was performed on the ALK and ALK-AHP feedstock effluents before and after 96 h of incubation with Rhodococci to assess changes to solubilized lignin oligomers, illustrated in Fig. 6. Both effluents provided signals that were evident of grass-type lignin (GSH-lignin). In detail, syringyl- (142.7 ppm), guaiacyl- (139.0–140.2 ppm), and para-hydroxyphenyl-based (137.8 ppm) monolignol units were clearly resolved.47,50 The ALK-AHP feedstock had an initial ∼30% and ∼80% higher concentration of total phenolic and aliphatic content compared to ALK, respectively. Moreover, carboxylic hydroxyl groups were higher in ALK-AHP compared to ALK, which is expected of a more rigorous delignification process.36,51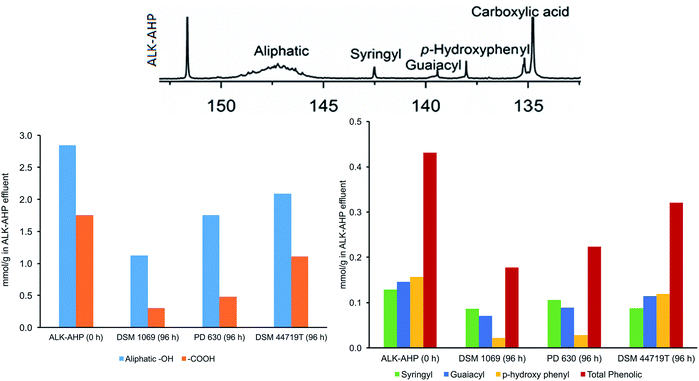 | ||
| Fig. 6 Quantitative 31P-NMR results. The 31P-NMR spectra of ALK-AHP at 0 h is shown above. Below are the results of integrated regions respective to aliphatic and carboxylic –OH groups (left) as well as syringyl, guaiacyl, p-hydroxyphenyl, and total phenolic –OH groups (right). Results for the standalone ALK effluent are provided in ESI.† | ||
After 96 h of incubation, all strains exhibited significant depletion of phenolic, aliphatic, and carboxylic regions, particularly by PD 630 and DSM 1069 when utilizing ALK-AHP as a substrate. The total phenolic content decreased by 58, 48, and 26% by DSM 1069, PD 630, and DSM 44719T, respectively. Likewise, aliphatic OH content decreased by 60, 38, and 26% by DSM 1069, PD 630, and DSM 44719T, in that same order; furthermore, losses in carboxylic hydroxyl content were by 83, 73, 37% following the same succession. These results support that simultaneous degradation of lignin along with carbohydrates during incubation was evident in all strains of Rhodococci utilizing ALK-AHP as a substrate, particularly by the R. opacus strains. Using model compounds, it has been reported that DSM 1069 and PD 630 exhibit preferences towards para-hydroxylphenyl, guaiacyl, and syringyl monolignol units, in that order.52 This may be due to the amount of energy required for demethoxylation in order to yield protocatechuate or catechol as initial metabolites for the β-KAP.52,53 Therefore, it is intriguing that the R. opacus strains, which both gained relatively substantial cell density in ALK-AHP media, exhibited a clear preference toward para-hydroxylphenyl units, followed by guaiacyl, and syringyl units. This observation is consistent with previous studies.46,52 This feature was not observed for DSM 44719T, which depleted approximately 25–30% of each phenolic type with no observable preference, and could be due to the lower degree of cell proliferation that occurred over the course of the experiment. In a similar manner, phenolic preference was generally irregular for Rhodococci grown on ALK substrates. The most noteworthy modifications were caused by DSM 1069 in ALK effluent, which exhibited the highest cell proliferation, and depleted a mere 15, 20, and 16% of total phenolic, aliphatic, and carboxylic content, respectively (ESI Fig. 4†).
4. Conclusions
Multistage ALK-AHP, which couples alkaline pre-extraction with an alkaline hydrogen peroxide post-treatment, is a more chemical-efficient pretreatment strategy compared to standalone alkali pre-extraction that serves to enhance enzymatic hydrolysis and fermentability towards bioethanol production. Moreover, when compared to ALK, ALK-AHP produces an effluent stream that contains more solubilized components and serves as a more practical feedstock for microbial biodiesel production. In this work, it was reported that oleaginous wild-type Rhodococci can simultaneously catabolize glucose and low molecular weight (<300 g mol−1) compounds in ALK-AHP effluent and accumulate substantial yields of intracellular oils. Wild-type R. opacus PD 630 was the most optimal strain based on lipid production for a total FAME yield of 1.3 g L−1 and oleaginicity of 42.1%, based on cell dry weight. Total organic content in ALK-AHP was determined to be 20.8 g L−1, of which 6.2% was converted into lipids by PD 630 after 48 h (ESI Table 3†). The considerable amount of inorganic content was due to processing conditions of the primary feedstock and could be a concern with regard to cellular inhibition. Despite this, and even lacking metabolic engineering, the titer yields reported here are largely comparable to current benchmark of effective pathways established for biodiesel production in bacteria, which is approximately ∼1.5 g L−1 in fatty acid ethyl esters.54 Furthermore, the lipid titer in this work is a promising starting point for non-engineered Rhodococcus strains. In contrast with the work by Kurosowa et al. that showed that the R. opacus PD 630 could yield an optimal value of 25.2 g L−1 of fatty acids when grown on media containing 240 g L−1 glucose,55 the ALK and ALK-AHP are suitable effluents that can balance growth and lipid accumulation on significantly less glucose, approximately 40 times less at, 1.2 g L−1 and 5.8 g L−1 glucose, respectively, initially, and the lignin was utilized, as seen by the transformation of the phenolic, aliphatic, and carboxylic groups present via NMR. These findings demonstrate that utilizing industrial effluents that contain a fraction of sugars and lignin to support cellular biomass generation could be an advantageous pathway for biofuel production that warrants further investigation into industrially-derived effluents containing low-molecular weight carbon sources, which could also be optimized if coupled with engineered oleaginous bacterial strains to target TAG production or other value-added targets.Acknowledgements
The research team acknowledges the Department of Energy (DE—EE0006112) for funding the microbial conversion results reported in this manuscript.References
- Y. He, Y. Xu, Y. Pang, H. Tian and R. Wu, Renewable Energy, 2016, 89, 695–705 CrossRef.
- S. Ahmed, A. Mahmood, A. Hasan, G. A. S. Sidhu and M. F. U. Butt, Renewable Sustainable Energy Rev., 2016, 57, 216–225 CrossRef.
- B. D. Solomon, A. Banerjee, A. Acevedo, K. E. Halvorsen and A. Eastmond, Environ. Manage., 2015, 56, 1276–1294 CrossRef PubMed.
- A. E. Atabani, A. S. Silitonga, I. A. Badruddin, T. Mahlia, H. Masjuki and S. Mekhilef, Renewable Sustainable Energy Rev., 2012, 16, 2070–2093 CrossRef.
- D. Tilman, R. Socolow, J. A. Foley, J. Hill, E. Larson, L. Lynd, S. Pacala, J. Reilly, T. Searchinger and C. Somerville, Science, 2009, 325, 270–271 CrossRef CAS PubMed.
- R. Singh, M. Srivastava and A. Shukla, Renewable Sustainable Energy Rev., 2016, 54, 202–216 CrossRef CAS.
- J. C. Liao, L. Mi, S. Pontrelli and S. Luo, Nat. Rev. Microbiol., 2016, 14, 288–304 CrossRef CAS PubMed.
- G. R. Timilsina and S. Mevel, Environ. Resour. Econ., 2013, 55, 1–19 CrossRef.
- J. M. Bergthorson and M. J. Thomson, Renewable Sustainable Energy Rev., 2015, 42, 1393–1417 CrossRef CAS.
- I. Gelfand, R. Sahajpal, X. Zhang, R. C. Izaurralde, K. L. Gross and G. P. Robertson, Nature, 2013, 493, 514–517 CrossRef CAS PubMed.
- G. R. Timilsina, Philos. Trans. R. Soc., A, 2014, 372, 20120323 CrossRef PubMed.
- A. Gupta and J. P. Verma, Renewable Sustainable Energy Rev., 2015, 41, 550–567 CrossRef CAS.
- L. Caspeta, N. A. Buijs and J. Nielsen, Energy Environ. Sci., 2013, 6, 1077–1082 CAS.
- I. Hore-Lacy, Nuclear Energy in the 21st Century: World Nuclear University Press, Academic press, 2010 Search PubMed.
- Y. Chisti, Biotechnol. Adv., 2007, 25, 294–306 CrossRef CAS PubMed.
- M. K. Lam and K. T. Lee, Appl. Energy, 2012, 94, 303–308 CrossRef CAS.
- D. Pimentel and T. W. Patzek, Nat. Resour. Res., 2005, 14, 65–76 CrossRef CAS.
- G. T. Beckham, C. W. Johnson, E. M. Karp, D. Salvachúa and D. R. Vardon, Curr. Opin. Biotechnol., 2016, 42, 40–53 CrossRef CAS PubMed.
- A. J. Ragauskas, C. K. Williams, B. H. Davison, G. Britovsek, J. Cairney, C. A. Eckert, W. J. Frederick, J. P. Hallett, D. J. Leak and C. L. Liotta, Science, 2006, 311, 484–489 CrossRef CAS PubMed.
- X. Miao and Q. Wu, Bioresour. Technol., 2006, 97, 841–846 CrossRef CAS PubMed.
- F. Deeba, V. Pruthi and Y. S. Negi, Bioresour. Technol., 2016, 213, 96–102 CrossRef CAS PubMed.
- J. A. Gomez, K. Höffner and P. I. Barton, Green Chem., 2016, 18, 461–475 RSC.
- X. Meng, J. Yang, X. Xu, L. Zhang, Q. Nie and M. Xian, Renewable Energy, 2009, 34, 1–5 CrossRef CAS.
- H. M. Alvarez, R. Kalscheuer and A. Steinbuchel, Appl. Microbiol. Biotechnol., 2000, 54, 218–223 CrossRef CAS PubMed.
- H. M. Alvarez and A. Steinbuechel, Appl. Microbiol. Biotechnol., 2002, 60, 367–376 CrossRef CAS PubMed.
- T. Wells and A. J. Ragauskas, Trends Biotechnol., 2012, 30, 627–637 CrossRef CAS PubMed.
- S. Amara, N. Seghezzi, H. Otani, C. Diaz-Salazar, J. Liu and L. D. Eltis, Sci. Rep., 2016, 6, 24985 CrossRef CAS PubMed.
- A. J. Ragauskas and F. Huang, in Pretreatment Techniques for Biofuels and Biorefineries, Springer, 2013, pp. 151–179 Search PubMed.
- A. J. Ragauskas, G. T. Beckham, M. J. Biddy, R. Chandra, F. Chen, M. F. Davis, B. H. Davison, R. A. Dixon, P. Gilna, M. Keller, P. Langan, A. K. Naskar, J. N. Saddler, T. J. Tschaplinski, G. A. Tuskan and C. E. Wyman, Science, 2014, 344, 709 CrossRef CAS PubMed.
- K. L. Kadam and J. D. McMillan, Bioresour. Technol., 2003, 88, 17–25 CrossRef CAS PubMed.
- D. M. Alonso, S. G. Wettstein, J. Q. Bond, T. W. Root and J. A. Dumesic, ChemSusChem, 2011, 4, 1078–1081 CrossRef CAS PubMed.
- R. E. Sims, W. Mabee, J. N. Saddler and M. Taylor, Bioresour. Technol., 2010, 101, 1570–1580 CrossRef CAS PubMed.
- T. Liu, D. L. Williams, S. Pattathil, M. Li, M. G. Hahn and D. B. Hodge, Biotechnol. Biofuels, 2014, 7, 1 CrossRef PubMed.
- Y. Yamashita, M. Shono, C. Sasaki and Y. Nakamura, Carbohydr. Polym., 2010, 79, 914–920 CrossRef CAS.
- B. C. Saha and M. A. Cotta, Biotechnol. Prog., 2006, 22, 449–453 CrossRef CAS PubMed.
- M. Li, C. Foster, S. Kelkar, Y. Pu, D. Holmes, A. Ragauskas, C. M. Saffron and D. B. Hodge, Biotechnol. Biofuels, 2012, 5, 38 CrossRef CAS PubMed.
- D. Donald and G. Göran, in Lignin and Lignans, CRC Press, 2010, pp. 349–391, DOI:10.1201/EBK1574444865-c1010.1201/EBK1574444865-c10.
- E. M. Karp, M. G. Resch, B. S. Donohoe, P. N. Ciesielski, M. H. O'Brien, J. E. Nill, A. Mittal, M. J. Biddy and G. T. Beckham, ACS Sustainable Chem. Eng., 2015, 3, 1479–1491 CrossRef CAS.
- B. Wang, Y. H. Rezenom, K.-C. Cho, J. L. Tran, D. G. Lee, D. H. Russell, J. J. Gill, R. Young and K.-H. Chu, Bioresour. Technol., 2014, 161, 162–170 CrossRef CAS PubMed.
- J. S. Kim, Y. Y. Lee and T. H. Kim, Bioresour. Technol., 2016, 199, 42–48 CrossRef CAS PubMed.
- Z. Gong, H. Shen, X. Yang, Q. Wang, H. Xie and Z. K. Zhao, Biotechnol. Biofuels, 2014, 7, 158 CrossRef PubMed.
- M.-H. Joe, J.-Y. Kim, S. Lim, D.-H. Kim, S. Bai, H. Park, S. G. Lee, S. J. Han and J.-I. Choi, Biotechnol. Biofuels, 2015, 8, 125 CrossRef PubMed.
- J. G. Linger, D. R. Vardon, M. T. Guarnieri, E. M. Karp, G. B. Hunsinger, M. A. Franden, C. W. Johnson, G. Chupka, T. J. Strathmann, P. T. Pienkos and G. T. Beckham, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12013–12018 CrossRef CAS PubMed.
- R. Katahira, A. Mittal, K. McKinney, X. Chen, M. P. Tucker, D. K. Johnson and G. T. Beckham, ACS Sustainable Chem. Eng., 2016, 4, 1474–1486 CrossRef CAS.
- Z. Wei, G. Zeng, M. Kosa, D. Huang and A. J. Ragauskas, Appl. Biochem. Biotechnol., 2015, 175, 1234–1246 CrossRef CAS PubMed.
- T. Wells, Z. Wei and A. Ragauskas, Biomass Bioenergy, 2015, 72, 200–205 CrossRef CAS.
- Y. Pu, S. Cao and A. J. Ragauskas, Energy Environ. Sci., 2011, 4, 3154–3166 CAS.
- M. Kosa and A. J. Ragauskas, Green Chem., 2013, 15, 2070–2074 RSC.
- T. Wells, M. Kosa and A. J. Ragauskas, Ultrason. Sonochem., 2013, 20, 1463–1469 CrossRef CAS PubMed.
- R. John and L. L. Larry, in Lignin and Lignans, CRC Press, 2010, pp. 137–243, DOI:10.1201/EBK1574444865-c510.1201/EBK1574444865-c5.
- J.-D. Mao, K. Holtman and D. Franqui-Villanueva, J. Agric. Food Chem., 2010, 58, 11680–11687 CrossRef CAS PubMed.
- M. Kosa and A. J. Ragauskas, Appl. Microbiol. Biotechnol., 2012, 93, 891–900 CrossRef CAS PubMed.
- T. Wells Jr and A. J. Ragauskas, Trends Biotechnol., 2012, 30, 627–637 CrossRef PubMed.
- S. Sherkhanov, T. P. Korman, S. G. Clarke and J. U. Bowie, Sci. Rep., 2016, 6, 24239 CrossRef CAS PubMed.
- K. Kurosawa, P. Boccazzi, N. M. de Almeida and A. J. Sinskey, J. Biotechnol., 2010, 147, 212–218 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra28033a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |