Open Access Article
Open Access ArticleGuidance for engineering of synthetic methylotrophy based on methanol metabolism in methylotrophy
Wenming Zhang,
Ting Zhang,
Sihua Wu,
Mingke Wu,
Fengxue Xin,
Weiliang Dong,
Jiangfeng Ma,
Min Zhang and
Min Jiang *
*
State Key Laboratory of Materials-Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing Tech University, No. 30 Puzhu Road, Pukou District, 211816, Nanjing, China. E-mail: bioengine@njtech.edu.cn; Tel: +86-25-58139927
First published on 16th January 2017
Abstract
Methanol is increasingly becoming an attractive substrate for production of different metabolites, such as commodity chemicals, and biofuels via biological conversion, due to the increment of annual production capacity and decrement of prices. In recent years, genetic engineering towards native menthol utilizing organisms – methylotrophy has developed rapidly and attracted widespread attention. Therefore, it is vital to elucidate the distinct pathways that involve methanol oxidation, formaldehyde assimilation and disassimilation in the different methylotrophies for future synthetic work. In addition, this will also help to genetically construct some new and non-native methylotrophies. This review summarizes the current knowledge about the methanol metabolism pathways in methylotrophy, discusses and compares different pathways on methanol utilization, and finally presents the strategies to integrate the methanol metabolism with other chemicals, biofuels or other high value-added product formation pathways.
1. Introduction
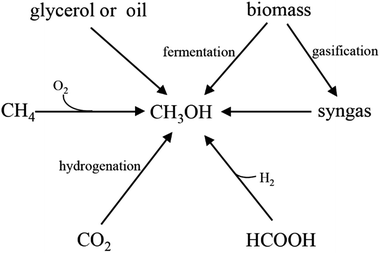
With the rapid growth of the world population and development of industry and society, energy demand is dramatically increasing annually. However, due to the depletion of fossil fuels and the concerns about environmental pollution, it is urgent to develop renewable and environmentally friendly fuels or chemicals. One of the alternative strategies is to generate these greener fuels or chemicals via microbial fermentation, in which monosaccharides, such as pentose and hexose are usually used as the major raw materials. However, due to the high cost of these sugars, it is not economically feasible when using these monosaccharides as substrates especially for large-scale applications. Although lignocellulose has been developed as a raw material, many bottlenecks still occur, such as the low yield of fermentable sugars from lignocellulose by acidolysis or enzymolysis, time-consuming, high content of inhibitors, and high cost of enzyme. Therefore, finding appropriate and cheap raw materials is crucial for the sustainable development of biotechnology.Methanol has been considered as an attractive C1 compound both from a biotechnological and a bulk chemical point of view.1,2 With a worldwide production of approximately 53 million tons per year and an expected annual growth rate in the range of 10–20%, a methanol-based bioeconomy has been proposed.3 Especially recently, with the rise of the methanol production process, the price of methanol steadily declined. Therefore, it is an important breach that reduces the cost of biotransformation process by using methanol as fermentation raw materials. Compared with existing raw materials, methanol has the following advantages: (1) abundant sources (Fig. 1). Methanol can be produced not only from abundant natural gas but also from biomass and factory waste gases; (2) low prices. The price of methanol is lower than sugars in the market;4 (3) easily handle. Methanol is non-corrosion liquid under room temperature, which is compatible with current transport systems;5 (4) pure substrate. Unlike some low-cost materials (such as molasses), methanol is a pure substrate which can be completely utilized in the fermentation process.6
Methylotrophs, which possess the capability of using C1 compounds such as methane, methanol and methylamine as carbon and energy sources, show great potential to convert C1 compounds into various applicable chemicals and materials.7 In nature, methylotrophs include a diverse group of microorganisms, mainly methylotrophic bacteria and yeasts. In all of methylotrophs, methanol will be enzymatically converted into toxic formaldehyde in vivo first, which is then further oxidized to CO2 by dissimilation or enter the carbon metabolic pathways for methylotrophic growth via certain assimilation pathways.8 Different methylotrophs possess different assimilation and dissimilation pathways due to biological diversity among them. Methylotrophs could directly use sole methanol as carbon sources, however, methanol could only be used for the production of single cell protein at an industrial scale so far, which significantly reduce the value of methanol.4 Currently, only limited understanding of the metabolism in methylotrophic organisms is available. Additionally, their genetic-transfer systems remain largely inefficient. So in this review, we will comprehensively discuss currently reported methanol metabolic pathways found in methylotrophs, which will provide a feasible scheme to help genetically construct some non-native methylotrophs.
2. Methanol oxidation to formaldehyde
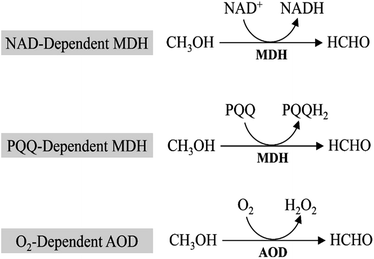
In all methylotrophs, oxidation of methanol to formaldehyde is the first step in methanol metabolism, which is conducted by the oxidoreductase, mainly methanol dehydrogenases (MDHs). MDHs are classified into 3 distinct groups based on their electron acceptor (Fig. 2): PQQ (pyrroloquinoline quinone)-dependent methanol dehydrogenase, NAD-dependent methanol dehydrogenase, and O2-dependent alcohol oxidase (AOD).2.1. PQQ-dependent MDHs
MDHs in Gram-negative methylotrophs contain a pyrroloquinoline quinone (PQQ) prosthetic group that captures electrons from methanol oxidation and passes them to cytochrome c.9,10 The PQQ prosthetic group connects redox reactions to the respiratory chain.11 In Methylobacterium extorquens AM1, 15 genes participate in methanol oxidation, and 14 of them are co-transcribed,12 including large and small subunits of MDH (mxaF and mxaI), electron acceptor cytochrome c (mxaG) and accessory proteins involved in transport and assembly (mxaJRSACKLD).13 However, PQQ biosynthesis requires molecular oxygen,14 which will restrict the applications of PQQ-dependent MDHs as some of metabolites must be produced under anaerobic conditions.2.2. NAD-dependent MDHs
NAD-dependent MDHs, which utilize nicotinamide adenine dinucleotide (NAD) as the cofactor, exist in thermophilic Gram-positive methylotrophs. Currently, most studies focused on the identification and characterization of MDHs from B. stearothermophilus and B. methanolicus.15,16 Unlike PQQ-dependent MDHs mediated methanol oxidation, NAD-dependent MDHs require only one gene for realization of its function.17 B. methanolicus MGA3, a well-known methylotroph that contains three NAD-dependent MDHs genes (mdh, mdh2, and mdh3) and the MDH activator protein (ACT) modulates the activity of all three MDHs.18 In vitro studies suggest that ACT increases methanol affinity, oxidation rates and activity of NAD-dependent MDHs,16,19 while it shows no apparent effect in vivo.20 Compared with PQQ-dependent MDHs, NAD-dependent MDHs utilize a ubiquitous cofactor (NAD) that can be used to provide electron for metabolite production and generate reducing equivalents in the form of NADH. Moreover, it can implement function under both aerobic and anaerobic conditions, whereas the PQQ-dependent MDHs would be limited to aerobic conditions. In view of this, NAD-dependent MDHs may be better candidates for synthetic methylotrophy.2.3. O2-Dependent AODs
In eukaryotic methylotrophs, O2-dependent AODs were used to oxidize methanol.21 Unlike periplasmic PQQ-dependent MDHs or cytoplasmic NAD-dependent MDHs, O2-dependent AODs locate in the peroxisome of yeasts. Methanol will be oxidized by O2-dependent AODs and converted to formaldehyde and hydrogen peroxide first, which are highly toxic compounds to cells. Then, dihydroxyacetone synthase (DAS) and catalase (CTA) located in peroxisome will transform them into non-toxic compounds. And this is one of the most important steps in methanol utilization. Similar to PQQ-dependent MDHs, O2-dependent AODs only implement function under aerobic conditions, however, conservation of electrons of O2-dependent AODs is inferior to that of PQQ-dependent MDHs. While compared to NAD-dependent MDHs (ΔrG′ = 34.2 kJ mol−1) and PQQ-dependent MDHs (ΔrG′ = −24.8 kJ mol−1), O2-dependent AODs (ΔrG′ = −99.2 kJ mol−1) have higher efficiency in methanol oxidation.103. Formaldehyde assimilated into biomass
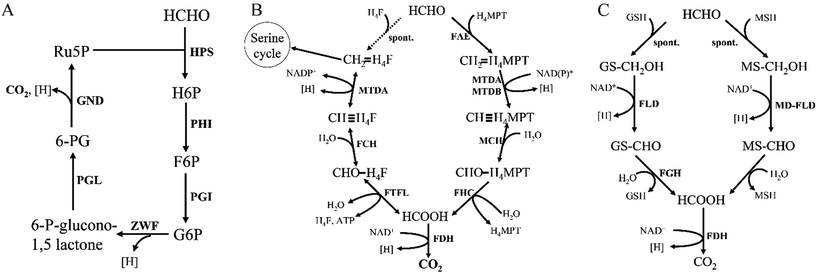
Formaldehyde (HCHO), the first intermediate in methanol oxidation, will get assimilated into biomass once converted. Although HCHO is an important intermediate in methylotrophic metabolism, it is highly toxic. The toxicity arises from the high level of nonspecific reactivity of HCHO with proteins and nucleic acids.8 Thus, HCHO must be rapidly assimilated into biomass or dissimilated to CO2. Three different assimilation cyclic pathways have been discovered and biochemically described, which are: the ribulose monophosphate pathway (RuMP), the serine pathway, and the xylulose monophosphate pathway (XuMP).22 The first two pathways are mainly found in prokaryotes, while the last pathway occurred in yeast. Despite differences in pathway, the end-product in these three pathways is a three-carbon compound synthesized from three C1 compounds.3.1. Formaldehyde assimilation via the RuMP pathway
The RuMP pathway involves synthesis of a three-carbon compound from three molecules of HCHO,22,23 which can be divided into three parts: the fixation part, the cleavage part and the rearrangement part (Fig. 3). In the fixation part, HCHO reacted with ribulose-5-phosphate (Ru5P) to eventually form fructose 6-phosphate (F6P) catalyzed by two key enzymes, 3-hexulose-6-phosphate synthase (HPS) and 6-phospho-3-hexuloisomerase (PHI).24 F6P is then metabolized via the main metabolic cellular pathways: glycolysis (the EMP pathway), the Entner–Doudoroff (ED) pathway, or Pentose Phosphate Pathway (PPP).10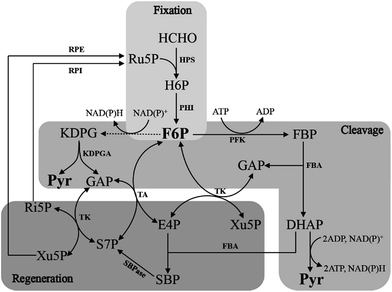 | ||
| Fig. 3 Outline of the three RuMP pathways. Solid arrows represent one step reactions, while dashed arrows show multi-step reactions. | ||
The cleavage part mainly contains F6P which is converted by a three-carbon compound and glyceraldehyde 3-phosphate (GAP). Two variants are found in this part, during which, one is FBP aldolase (FBPA) variant, and the other is KDPG aldolase (KDPGA) variant.25 In the FBPA variant, F6P is converted to FBP and subsequently cleaved by FBPA to GAP and dihydroxyacetone phosphate (DHAP), which can be further converted into pyruvate, generating one NAD(P)H and two ATP. However, in the KDPGA variant, F6P is converted to 2-keto-3-deoxy-6-phosphogluconate (KDPG) by several enzymes, and then cleaved by KDPGA to GAP and pyruvate. However, only one NAD(P)H can be generated in this variant (Fig. 3).
The regeneration of Ru5P is vital to keep the cycle running. There are two variants existing in the rearrangement part: the transaldolase (TA) one and the sedoheptulose 1,7-bisphosphatase (SBPase) one (Fig. 3). However, the last few steps are the same in two variants, which are, GAP and sedoheptulose-7-phosphate (S7P) rearranged by transketolase (TK) to two 5-carbon sugar phosphates, xylulose 5-phosphate (Xu5P) and ribose 5-phosphate (Ri5P), which is further converted to Ru5P and completed the regeneration pathway. While the transaldolase variant exhibits a more efficient stoichiometry: 3HCHO + NAD+ + ADP = pyruvate + NADH + ATP.26 And the effect of temperature is very important, because both of NAD-dependent MDHs and FBA have optimal activity at 50 °C, which is also the optimal growth temperature of B. methanolicus.27
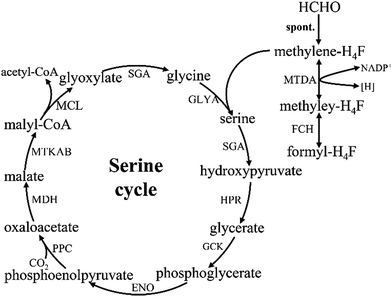
3.2. Formaldehyde assimilation via the serine pathway
The serine cycle is another HCHO assimilation pathway in methylotrophic bacteria. M. extorquens AM1 is the most well-studied methylotrophic bacterium for serine cycle pathway.28 There are two routes existing for generating methylene-H4F from HCHO in methylotrophy (Fig. 4). One is HCHO and H4F spontaneously converted into methylene-H4F.29,30 In the other route, formaldehyde is firstly converted to formate via tetrahydromethanopterin (H4MPT)-dependent enzymes, subsequently formate is further converted to methylene-H4F via H4F-dependent enzymes. Although in the former route, less enzymes are involved and ATP is consumed, it had suggested that methylene-H4F is mainly formed by the route involving the H4MPT and H4F pathways.31 It is possible that the spontaneous condensation reaction serves as an overflow valve for transient episodes of formaldehyde overproduction.32 The carbon balance of the pathway as shown here is: CO2 + 2HCHO + 2NADH + 2ATP = pyruvate + 2NAD+ + 2ADP + FPH2.103.3. Formaldehyde assimilation via the XuMP pathway
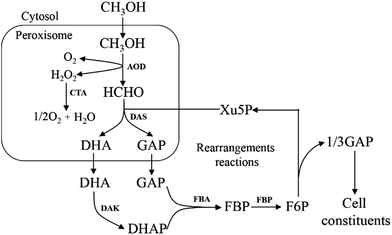
All methylotrophic yeasts share the same HCHO assimilation pathway, which is XuMP pathway (Fig. 5). In this pathway, HCHO is firstly combined with xylulose 5-phosphate (Xu5P) by peroxisomal enzyme DAS, forming dihydroxyacetone (DHA) and GAP. Then DHA and GAP will be further assimilated in the cytosol.33 DHA is phosphorylated to form dihydroxyacetone phosphate (DHAP), which can be condensed with GAP to form fructose-1,6-bisphosphate (FBP). FBP will be subsequently dephosphorylated to form F6P, and a portion of them is then utilized for regeneration of Xu5P, while the left parts will be used for biosynthesis of cell constituents (Fig. 5). Two key enzymes for HCHO assimilation, AOD and DAS, will be highly expressed in vivo when grown on methanol, but their enzymatic activities are not detectable in cells when grown on other carbon sources, such as glucose, glycerol, or ethanol,34 indicating that they are substrate inducible. The overall carbon balance of this cycle is then: 3HCHO + NAD+ + 2ADP = pyruvate + NADH + 2ATP.4. Formaldehyde dissimilation to CO2
HCHO dissimilation is ubiquitous in living organisms, as HCHO is commonly produced from various demethylation reactions in cells.8 Microorganisms possess a variety of pathways to fulfill formaldehyde-oxidizing reactions. One way to oxidize HCHO in some methylotrophs is through the dissimilatory RuMP cycle (Fig. 6A). Research shows that key enzyme in the dissimilatory RuMP pathway was upregulated when cells grown on methanol.35 However, the main routes of HCHO dissimilation takes place by HCHO oxidation to formate and subsequently to CO2, involving four cofactors, tetrahydrofolate (H4F), tetrahydromethanopterin (H4MPT), glutathione (GSH) and mycothiol (MSH) (Fig. 6). These cofactor-dependent pathways have some common characteristics, for instance: HCHO and the respective C1 carrier can condense spontaneously or enzyme catalyzed into cofactor-bound C1 unit, then converted by several enzymes' oxidation to formate, which further oxidized to CO2.364.1. H4F/H4MPT-dependent pathway
Reactions related with tetrahydrofolate (H4F)/tetrahydromethanopterin (H4MPT) are commonly found in methylotrophic and nonmethylotrophic organisms.37 Formaldehyde oxidation is initiated by the condensation of formaldehyde and H4F/H4MPT to methylene-H4F/H4MPT, which is spontaneous reaction. However, the H4MPT-dependent reaction can be accelerated by a specific enzyme, formaldehyde-activating enzyme (Fae).29 Then methylene-H4F/H4MPT is oxidized to formate through a series of reactions, which can ultimately be dissimilated to CO2 via FDH (Fig. 6B).There are two distinct pathways existing in methylotrophy, which show that both the H4MPT and H4F pathways are necessary to sustain cell growth.38 Interestingly, the H4F-dependent reactions are all completely reversible, therefore, an alternative hypothesis suggests that there are two routes for generating the key intermediate methylene-H4F from HCHO: one is the direct condensation step via H4F pathway and the other is indirect condensation step via both H4MPT and H4F pathways. In the latter one, formaldehyde is firstly catalyzed to methylene-H4MPT, which is further converted to formate under H4MPT pathway, and finally generated to methylene-H4F via reductive H4F pathways direction. If this hypothesis is correct, the H4MPT pathway would play a role in both dissimilatory and assimilatory metabolism, in much the same way that the tricarboxylic acid cycle plays a dual role in growth on multicarbon compounds.38
4.2. Thiol (glutathione/mycothiol)-dependent pathways
Thiol-dependent formaldehyde oxidation is also a vital pathway for formaldehyde detoxification. Just like H4MPT-dependent pathways, HCHO spontaneously reacts with glutathione (GSH) to form S-hydroxymethylglutathione (Fig. 6C). And this spontaneous reaction can be accelerated by glutathione-dependent formaldehyde-activating enzyme (Gfa) in Paracoccus denitrificans.39 Currently, this pathway is the most commonly found one for formaldehyde conversion in many nonmethylotrophic organisms, including plants and mammals, as well as methylotrophic bacteria and yeasts.40Several Gram-positive bacteria do not contain glutathione, instead, they possess a “sugar thiol”, mycothiol (MSH).41 The methylotrophic Gram-positive bacterium Amycolatopsis methanolica contains a mycothiol-dependent formaldehyde dehydrogenase (MD-FalDH), which is similar to the zinc-containing medium chain alcohol dehydrogenases.42,43 It is hypothesized that, MSH also spontaneously reacts with HCHO to form an S-hydroxymethylmycothiol just as GSH, and then it was converted by MD-FalDH to S-formylmycothiol, which might be further hydrolyzed to formate and MSH via a hydrolase44 (Fig. 6C). FadH, an NAD-linked mycothiol-dependent formaldehyde dehydrogenase from Corynebacterium glutamicum, was also reported and shown to be active as a homotetramer. Mycothiol-dependent formaldehyde oxidation revealed Km values of 0.6 mM for mycothiol and 4.3 mM for formaldehyde and a Vmax of 7.7 U mg−1.45 Bacillithiol, a thiol cofactor from B. methanolicus, was found in formaldehyde oxidation by LC-MS (theoretical oxidized monoisotopic mass: 794.1833 amu). The results also showed that bacillithiol may be used for redox homeostasis and bacillithiol-dependent formaldehyde oxidation will be activated when the cells were exposed to a sudden increase in methanol concentration.46
5. Engineering of synthetic methylotrophy in non methylotrops
Methylotrophs have been used for the biotechnological production of various chemicals from methanol. The most typical products described in biotechnological processes by using methylotrophic microorganisms are poly-β-hydroxybutyrate (PHB),47 amino acids,48 alcohols,49,50 organic acids,51 cadaverine,6 γ-aminobutyric acid (GABA)52 as well as proteins.53 Although native methylotrophs grow quite fast on C1 substrates and produce various chemicals, several problems still exist, such as lackage of efficient genetic tools for strain modification, non-production of metabolites at high levels.10,54 Moreover, most native methylotrophs are obligate aerobes, which would limit the efficient production of metabolites, because more reducing power are consumed by electron transport chain rather than the accumulation of desirable metabolites.55 Based on these considerations, the concept of synthetic methylotrophy is attracting more and more attention.55,56As mentioned, many distinct methanol metabolism pathways exist in methylotrophic organisms. However, the formation of end metabolic products depends on the environmental conditions. For example, fatty acid and amino acid are produced in aerobic condition, while organic acid and biofuels are usually produced through anaerobic metabolism. Therefore, it is critical to choose suitable methanol metabolism pathway to achieve the goal of enabling aerobic, microaerobic, and anaerobic growth based on methanol as a carbon source in synthetic methylotrophy.
Three distinct methanol dehydrogenases have been described above. Compared to PQQ-dependent MDHs and O2-dependent AODs, NAD-dependent MDHs require only one enzyme for its functional assembly in both aerobic and anaerobic conditions. Therefore, it can be speculated that NAD-dependent MDHs could be the best candidates for synthetic methylotrophy, because it can implement function under both aerobic and anaerobic conditions and generate reducing equivalents (NADH), which can help promote strain growth. However, it is vitally important to choose an appropriate MDH in synthetic methylotrophy. Six MDHs from two Bacillus methanolicus (MDH1, MDH2, MDH3 from B. methanolicus MGA3 and MDH, MDH1, MDH2 from B. methanolicus PB1) were produced recombinantly in Escherichia coli. Among those, only MDH2 and MDH3 from B. methanolicus MGA3 possessed significant activity in E. coli.16 Müller et al. (2015) have chosen E. coli as the host to implement metabolic construction for methanol conversion.20 They screened a series of enzymes from different microorganism donors and analyzed the matching of these enzymes in E. coli. The results showed that MDH2, HPS and PHI from B. methanolicus MGA3 were found to be the most effective in E. coli. Labeling experiments using 13C methanol demonstrated that approximately 40% of the methanol entered into central metabolites. However, when three MDHs from B. methanolicus MGA3 were expressed in Corynebacterium glutamicum, MDH and MDH3 showed the highest activity, whereas MDH2 showed the lowest catalytic activity. Interestingly, the expression of HPS and PHI from B. subtilis could also complete the RuMP pathway in C. glutamicum.19 Whitaker et al. (2016) reported that MDH from B. stearothermophilus has a lower Km and higher enzyme activity than MDH2 from B. methanolicus under normal physiological conditions for E. coli.57 Wu et al. (2016) characterized and engineered a group III NAD-dependent alcohol dehydrogenase (Mdh2) from a Gram-negative, mesophilic, non-methylotrophic organism Cupriavidus necator N-1. This enzyme exhibited higher or comparable activity and affinity toward methanol relative to the B. methanolicus Mdh with or without activator protein in a wide range of temperatures.58 I think it's a boon to the development of synthetic methylotrophy. Although these papers provide evidence that MDHs and the RuMP pathway can be successfully assembled to non-native methylotrophs, the rate of methanol consumption is significantly lower than those reported for native methylotrophs. One of the key reasons is the high activity of reverse formaldehyde reductase for Mdh, which is at least 1000-fold higher than methanol oxidation according to the kinetics analyses.16 To solve this problem, three strategies have been adopted to promote carbon flux in a desirable direction. One is modulating the expression levels of HPS and PHI (through manipulating promoter strengths or ribosome binding site strengths) or improving activities of them by directed evolution, which will reduce the accumulation of formaldehyde and then decrease the reversible methanol oxidation. Another strategy is constructing synthetic protein scaffolds that spatially recruit MDH, HPS and PHI in a designable manner. This will increase the overall pathway flux while preventing metabolic intermediates accumulation. Moreover, the production levels of different enzymes can be regulated through change the number of its specific ligands.59 Recently, Price et al. (2016) reported a scaffoldless, self-assembly approach to cluster MDH, HPS, and PHI using an SH3–ligand interaction pair. The use of engineered supramolecular enzyme complex yielded a 50-fold improvement in methanol conversion.60 The third strategy is reducing the concentration of NADH to preventing reversible formaldehyde reduction. This strategy can easily be achieved if the desired products is reducing metabolites, such as succinate acid or butanol.
To accelerate consumption of methanol in sugar-based fermentation processes, the synthetic formaldehyde assimilation and the endogenous dissimilation should be balanced. For example, in case of C. glutamicum, the pathway for methanol dissimilation was recently discovered.61 Deletion of the responsible genes improved the assimilation, as 13C-feeding experiments showed that more methanol-derived carbon could be assimilated, and also more toxic formaldehyde accumulated, which in turn inhibited the overall cell growth.19 Moreover, the regeneration of Ru5P is vital to keep the RuMP pathway running in the facultative methylotroph B. methanolicus MGA3. If the relative genes were deficient, the strains will not able to grow on methanol.62 B. methanolicus has a dynamic control system, the transcription levels of the genes in RuMP pathway were significantly upregulated when formaldehyde accumulated, which will benefit formaldehyde detoxification.63 However, no reports were presented as to whether non-native methylotrophs have this system when growing on methanol. Thus, it is crucial to sustain the proper concentration of Ru5P for formaldehyde assimilation in synthetic methylotrophy. Two strategies may be useful for strengthening the replenishment of Ru5P. One is overexpressing key genes for Ru5P replenishment, such as glpX (fructose-1,6-bisphosphatase), rpe (ribulose-5-phosphate 3-epimerase), tkt (transketolase). The other is finding appropriate promoters, which are sensitive to the concentration of formaldehyde, to substitute the native ones. If the regeneration of Ru5P can be solved in synthetic methylotrophy, using methanol as fermentation raw materials for fuel and chemical production will be applied widely in industrial biotechnology. One thing to note is that synthetic methylotrophy cannot growth on methanol solely.
So far, no metabolic product production has been reported in genetically modified methylotrophy. If methanol was used as the substrate instead of traditional raw materials, the production costs will be reduced. However, how to integrate the methanol metabolism with other chemicals, biofuels or high value-added products formation pathway in vivo is still the biggest obstacle. The following strategies are proposed based on literatures. (1) NAD-dependent MDHs and RuMP pathway, which can implement function under anaerobic conditions, could be recruited in the dicarboxylic acid production strains. One of the main reasons of low production and yield in these strains is the shortage of reducing equivalents (NADH) supply. While methanol oxidation by NAD-dependent MDHs generates NADH, which is beneficial to cell growth and products synthesis (Fig. 7). Moreover, because of successfully eliminated NADH accumulation, formaldehyde reduction will be slow down, which will lead to further enhancement in methanol utilization. (2) XuMP pathway is only applied to some yeast genera, as three key enzymes AOD, DAS and CTA are located in peroxisomes. So these genes can be introduced into Saccharomyces cerevisiae for production of amino acids or biodiesel. (3) Methylene-H4F, the intermediate metabolite in serine pathway, can also be synthesized from syngas. If it can be coupled with Wood–Ljungdahl pathway to synthesize acetyl-CoA which can further convert into various biofuels, the whole pathway will be shorter and less ATP and reducing equivalents (NADH) will be consumed. This strategy will promote syngas utilization efficiency and increased biofuels production.
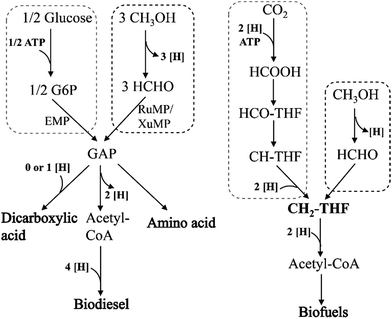 | ||
| Fig. 7 Comparison of different raw materials, glucose, methanol and syngas for producing series of bio-based chemicals. | ||
6. Conclusions
Methanol represents an attractive alternative non-food raw material in biotechnological processes from an economic, ecologic, and process point of view. Methylotrophs are widely used to produce varies products, for example single-cell protein, PHB and some amino acids. In particular, with the development of the genome editing tools, such as CRISPR-Cas9 system, it is possible to construct robust methylotrophs more efficiently. Therefore, the potential of using these species for the development of economically competitive bioprocesses based on methanol as an alternative carbon source will be more realistic. Moreover, metabolic engineering has recently become more powerful with the advanced in synthetic biology, which is allowing us to create novel methanol metabolism in non-native methylotrophs, so that they can utilize methanol as co-substrates with carbohydrates to produce metabolites as biofuels and chemicals. Based on these, novel technologies will be designed, and methanol will have a more prospective application in the near future.Abbreviations of intermediates
| Ru5P | Ribulose 5-phosphate |
| H6P | Hexulose 6-phosphate |
| F6P | Fructose 6-phosphate |
| FBP | Fructose-1,6-bisphosphate |
| KDPG | 2-Keto-3-deoxy-6-phosphogluconate |
| GAP | Glyceraldehyde phosphate |
| DHAP | Dihydroxyacetone phosphate |
| Pyr | Pyruvate |
| X5P | Xylulose 5-phosphate |
| E4P | Erythrose-4-phosphate |
| SBP | Sedoheptulose-1,7-bisphosphate |
| S7P | Sedoheptulose-7-phosphate |
| Ri5P | Ribose 5-phosphate |
| DHA | Dihydroxyacetone |
| G6P | Glucose 6-phosphate |
| 6-PG | 6-Phosphogluconate |
| GS-CH2OH | S-Hydroxymethylglutathione |
| GS-CHO | S-Formylglutathione |
| MS-CH2OH | S-Hydroxymethylmycothiol |
| MS-CHO | S-Formyl-mycothiol |
Abbreviations of enzymes
| HPS | 3-Hexulose-6-phosphate synthase |
| PHI | 6-Phospho-3-hexuloisomerase |
| PFK | Phosphofructokinase |
| KDPGA | 2-Keto-3-deoxy-6-phosphogluconate aldolase |
| FBA | Fructose-1,6-bisphosphate aldolase |
| TK | Transketolase |
| TA | Transaldolase |
| SBPase | Sedoheptulose-1,7-bisphosphatase |
| RPE | Ribulose-5-phosphate 3-epimerase |
| RPI | Ribose-5-phosphate isomerase |
| MTDA | Methylene-H4F dehydrogenase |
| FCH | Methenyl-H4F cyclohydrolase |
| GLYA | Serine hydroxymethyltransferase |
| HPR | Hydroxypyruvate reductase |
| GCK | Glycerate kinase |
| ENO | Enolase |
| PPC | Phosphoenolpyruvate carboxylase |
| MDH | Malate dehydrogenase |
| MTKAB | Malate thiokinase |
| MCL | Malyl-CoA lyase |
| SGA | Serine-glyoxylate aminotransferase |
| AOD | Alcohol oxidase |
| CTA | Catalase |
| DAS | Dihydroxyacetone synthase |
| DAK | Dihydroxyacetone kinase |
| FBP | Fructose 1,6-bisphosphatase |
| PGL | 6-Phosphogluconolactonase |
| GND | 6-Phosphogluconate dehydrogenase |
| FTFL | Formyl H4F ligase |
| FDH | Formate dehydrogenase |
| FAE | Formaldehyde activating enzyme |
| MTDB | Methylene H4MPT dehydrogenase |
| MCH | Methenyl H4MPT cyclohydrolase |
| FHC | Formyltransferase/hydrolase complex |
| FLD | Formaldehyde dehydrogenase |
| FGH | S-Formylglutathione hydrolase |
| MD-FLD | MSH-dependent formaldehyde dehydrogenase |
Acknowledgements
This work was supported by the “973” Program of China (Grant No. 2013CB733902), the National Natural Science Foundation of China (Grant No. 21306087), the Specialized Research Fund for the Doctoral Program of Higher Education (Grant No. 20123221110008), Program for New Century Excellent Talents in University, A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Program for Changjiang Scholars and Innovative Research Team in University.Notes and references
- J. D. Linton and H. G. D. Niekus, Antonie van Leeuwenhoek, 1987, 53, 55–63 CrossRef CAS PubMed.
- G. A. Olah, Angew. Chem., Int. Ed., 2005, 44, 2636–2639 CrossRef CAS PubMed.
- A. M. Ochsner, F. Sonntag, M. Buchhaupt, J. Schrader and J. A. Vorholt, Appl. Microbiol. Biotechnol., 2015, 99, 517–534 CrossRef CAS PubMed.
- J. Schrader, M. Schilling, D. Holtmann, D. Sell, M. Villela, A. Marx and J. A. Vorholt, Trends Biotechnol., 2009, 27, 107–115 CrossRef CAS PubMed.
- I. Orita, K. Nishikawa, S. Nakamura and T. Fukui, Appl. Microbiol. Biotechnol., 2014, 98, 3715–3725 CrossRef CAS PubMed.
- I. Naerdal, J. Pfeifenschneider, T. Brautaset and V. F. Wendisch, Microb. Biotechnol., 2015, 8, 342–350 CrossRef CAS PubMed.
- J. R. Quayle and T. Ferenci, Microbiol. Rev., 1978, 42, 251–273 CAS.
- H. Yurimoto, N. Kato and Y. Sakai, Chem. Rec., 2005, 5, 367–375 CrossRef CAS PubMed.
- C. Anthony, Arch. Biochem. Biophys., 2004, 428, 2–9 CrossRef CAS PubMed.
- W. B. Whitaker, N. R. Sandoval, R. K. Bennett, A. G. Fast and E. T. Papoutsakis, Curr. Opin. Biotechnol., 2015, 33, 165–175 CrossRef CAS PubMed.
- K. Matsushita, J. C. Arents, R. Bader, M. Yamada, O. Adachi and P. W. Postma, Microbiology, 1997, 143, 3149–3156 CrossRef CAS PubMed.
- M. Zhang and M. E. Lidstrom, Microbiology, 2003, 149, 1033–1040 CrossRef CAS PubMed.
- L. Chistoserdova, S. W. Chen, A. Lapidus and M. E. Lidstrom, J. Bacteriol., 2003, 185, 2980–2987 CrossRef CAS PubMed.
- J. S. Velterop, E. Sellink, J. J. M. Meulenberg, S. David, I. Bulder and P. W. Postma, J. Bacteriol., 1995, 177, 5088–5098 CrossRef CAS PubMed.
- M. C. Sheehan, C. J. Bailey, B. C. Dowds and D. J. McConnell, Biochem. J., 1988, 252, 661–666 CrossRef CAS PubMed.
- A. Krog, T. M. Heggeset, J. E. Muller, C. E. Kupper, O. Schneider, J. A. Vorholt, T. E. Ellingsen and T. Brautaset, PLoS One, 2013, 8, e59188 CAS.
- A. M. Ochsner, J. E. N. Muller, C. A. Mora and J. A. Vorholt, FEBS Lett., 2014, 588, 2993–2999 CrossRef CAS PubMed.
- T. M. Heggeset, A. Krog, S. Balzer, A. Wentzel, T. E. Ellingsen and T. Brautaset, Appl. Environ. Microbiol., 2012, 78, 5170–5181 CrossRef CAS PubMed.
- S. Witthoff, K. Schmitz, S. Niedenfuhr, K. Noh, S. Noack, M. Bott and J. Marienhagen, Appl. Environ. Microbiol., 2015, 81, 2215–2225 CrossRef CAS PubMed.
- J. E. N. Müller, F. Meyer, B. Litsanov, P. Kiefer, E. Potthoff, S. Heux, W. J. Quax, V. F. Wendisch, T. Brautaset, J. C. Portais and J. A. Vorholt, Metab. Eng., 2015, 28, 190–201 CrossRef PubMed.
- S. Limtong, N. Srisuk, W. Yongmanitchai, H. Yurimoto and T. Nakase, Int. J. Syst. Evol. Microbiol., 2008, 58, 302–307 CrossRef CAS PubMed.
- L. Dijkhuizen, P. Levering and G. De Vries, in Methane and methanol utilizers, Springer, 1992, pp. 149–181 Search PubMed.
- C. Anthony, Biotechnology, 1991, 18, 79–109 CAS.
- N. Kato, H. Yurimoto and R. K. Thauer, Biosci., Biotechnol., Biochem., 2006, 70, 10–21 CrossRef CAS PubMed.
- Ø. M. Jakobsen, Doctoral Theses, NTNU, 2008, 255, pp. 10–15.
- C. Anthony, Biochemistry of methylotrophs, Academic Press, 1982 Search PubMed.
- J. Stolzenberger, S. N. Lindner and V. F. Wendisch, Microbiology, 2013, 159, 1770–1781 CrossRef CAS PubMed.
- S. Vuilleumier, L. Chistoserdova, M. C. Lee, F. Bringel, A. Lajus, Y. Zhou, B. Gourion, V. Barbe, J. Chang, S. Cruveiller, C. Dossat, W. Gillett, C. Gruffaz, E. Haugen, E. Hourcade, R. Levy, S. Mangenot, E. Muller, T. Nadalig, M. Pagni, C. Penny, R. Peyraud, D. G. Robinson, D. Roche, Z. Rouy, C. Saenampechek, G. Salvignol, D. Vallenet, Z. N. Wu, C. J. Marx, J. A. Vorholt, M. V. Olson, R. Kaul, J. Weissenbach, C. Medigue and M. E. Lidstrom, PLoS One, 2009, 4, e5584 Search PubMed.
- J. A. Vorholt, C. J. Marx, M. E. Lidstrom and R. K. Thauer, J. Bacteriol., 2000, 182, 6645–6650 CrossRef CAS PubMed.
- R. G. Kallen and W. P. Jencks, J. Biol. Chem., 1966, 241, 5851–5863 CAS.
- G. J. Crowther, G. Kosaly and M. E. Lidstrom, J. Bacteriol., 2008, 190, 5057–5062 CrossRef CAS PubMed.
- X. Guo and M. E. Lidstrom, Arch. Microbiol., 2006, 186, 139–149 CrossRef CAS PubMed.
- H. Yurimoto, M. Oku and Y. Sakai, Int. J. Microbiol., 2011, 2011, 101298 Search PubMed.
- J. L. Cereghino and J. M. Cregg, FEMS Microbiol. Rev., 2000, 24, 45–66 CrossRef CAS PubMed.
- J. E. N. Muller, B. Litsanov, M. Bortfeld-Miller, C. Trachsel, J. Grossmann, T. Brautaset and J. A. Vorholt, Proteomics, 2014, 14, 725–737 CrossRef PubMed.
- J. A. Vorholt, Arch. Microbiol., 2002, 178, 239–249 CrossRef CAS PubMed.
- B. Maden, Biochem. J., 2000, 350, 609–629 CrossRef CAS PubMed.
- C. J. Marx, S. J. Van Dien and M. E. Lidstrom, PLoS Biol., 2005, 3, 244–253 CAS.
- M. Goenrich, S. Bartoschek, C. H. Hagemeier, C. Griesinger and J. A. Vorholt, J. Biol. Chem., 2002, 277, 3069–3072 CrossRef CAS PubMed.
- B. Lee, H. Yurimoto, Y. Sakai and N. Kato, Microbiology, 2002, 148, 2697–2704 CrossRef CAS PubMed.
- J. A. Duine, BioFactors, 1999, 10, 201–206 CrossRef CAS PubMed.
- M. Misset-Smits, P. W. van Ophem, S. Sakuda and J. A. Duine, FEBS Lett., 1997, 409, 221–222 CrossRef CAS PubMed.
- A. Norin, P. W. Ophem, S. R. Piersma, B. Persson, J. A. Duine and H. Jörnvall, Eur. J. Biochem., 1997, 248, 282–289 CAS.
- G. L. Newton and R. C. Fahey, Arch. Microbiol., 2002, 178, 388–394 CrossRef CAS PubMed.
- L. Lessmeier, M. Hoefener and V. F. Wendisch, Microbiology, 2013, 159, 2651–2662 CrossRef CAS PubMed.
- J. E. Muller, F. Meyer, B. Litsanov, P. Kiefer and J. A. Vorholt, Mol. Microbiol., 2015, 98, 1089–1100 CrossRef PubMed.
- D. Bourque, Y. Pomerleau and D. Groleau, Appl. Microbiol. Biotechnol., 1995, 44, 367–376 CrossRef CAS.
- T. Brautaset, M. D. Williams, R. D. Dillingham, C. Kaufmann, A. Bennaars, E. Crabbe and M. C. Flickinger, Appl. Environ. Microbiol., 2003, 69, 3986–3995 CrossRef CAS PubMed.
- B. Hu and M. E. Lidstrom, Biotechnol. Biofuels, 2014, 7, 1–10 CrossRef PubMed.
- B. Hu, Y. M. Yang, D. A. C. Beck, Q. W. Wang, W. J. Chen, J. Yang, M. E. Lidstrom and S. Yang, Biotechnol. Biofuels, 2016, 9, 84 CrossRef PubMed.
- W. L. Zhu, J. Y. Cui, L. Y. Cui, W. F. Liang, S. Yang, C. Zhang and X. H. Xing, Appl. Microbiol. Biotechnol., 2016, 100, 2171–2182 CrossRef CAS PubMed.
- M. Irla, I. Nærdal, T. Brautaset and V. F. Wendisch, Ind. Crops Prod., 2016 DOI:10.1016/j.indcrop.2016.11.050.
- J. Gutierrez, D. Bourque, R. Criado, Y. J. Choi, L. M. Cintas, P. E. Hernandez and C. B. Miguez, FEMS Microbiol. Lett., 2005, 248, 125–131 CrossRef CAS PubMed.
- Q. Fei, M. T. Guarnieri, L. Tao, L. M. L. Laurens, N. Dowe and P. T. Pienkos, Biotechnol. Adv., 2014, 32, 596–614 CrossRef CAS PubMed.
- C. A. Haynes and R. Gonzalez, Nat. Chem. Biol., 2014, 10, 331–339 CrossRef CAS PubMed.
- Q. Fei, M. T. Guarnieri, L. Tao, L. M. Laurens, N. Dowe and P. T. Pienkos, Biotechnol. Adv., 2014, 32, 596–614 CrossRef CAS PubMed.
- W. B. Whitaker, J. A. Jones, R. K. Bennett, J. E. Gonzalez, V. R. Vernacchio, S. M. Collins, M. A. Palmer, S. Schmidt, M. R. Antoniewicz and M. A. Koffas, Metab. Eng., 2016 DOI:10.1016/j.ymben.2016.10.015.
- T.-Y. Wu, C.-T. Chen, J. T.-J. Liu, I. W. Bogorad, R. Damoiseaux and J. C. Liao, Appl. Environ. Microbiol., 2016, 100, 4969–4983 CAS.
- J. E. Dueber, G. C. Wu, G. R. Malmirchegini, T. S. Moon, C. J. Petzold, A. V. Ullal, K. L. Prather and J. D. Keasling, Nat. Biotechnol., 2009, 27, 753–759 CrossRef CAS PubMed.
- J. V. Price, L. Chen, W. B. Whitaker, E. Papoutsakis and W. Chen, Proc. Natl. Acad. Sci. U. S. A., 12691–12696 Search PubMed.
- S. Witthoff, A. Muhlroth, J. Marienhagen and M. Bott, Appl. Environ. Microbiol., 2013, 79, 6974–6983 CrossRef CAS PubMed.
- T. Brautaset, O. M. Jakobsen, M. C. Flickinger, S. Valla and T. E. Ellingsen, J. Bacteriol., 2004, 186, 1229–1238 CrossRef CAS PubMed.
- O. M. Jakobsen, A. Benichou, M. C. Flickinger, S. Valla, T. E. Ellingsen and T. Brautaset, J. Bacteriol., 2006, 188, 3063–3072 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |