Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Aptamer functionalized silver clusters for STED microscopy
Lan Wanga,
Haisen Tab,
Chaitanya Ullalc,
Fu Wang *a,
Chuanyi Wang
*a,
Chuanyi Wang a and
Guohui Donga
a and
Guohui Donga
aLaboratory of Environmental Sciences and Technology, Key Laboratory of Functional Materials and Devices for Special Environments, Xinjiang Technical Institute of Physics & Chemistry, Chinese Academy of Sciences, Urumqi 830011, China. E-mail: wangfu@ms.xjb.ac.cn; Fax: +86-991-3838957; Tel: +86-991-3835879
bDepartment of NanoBiophotonics, Max Planck Institute for Biophysical Chemistry, Göttingen 37077, Germany
cDepartment of Materials Science and Engineering, Rensselaer Polytechnic Institute, Troy, New York 12180, USA
First published on 16th February 2017
Abstract
Alternative to traditional dye or fluorescent protein, a novel stable probe for stimulated emission depletion (STED) microscopy is critically needed. In this work, aptamer functionalized Ag clusters with high photostability were successfully prepared. Moreover, improved resolution has been observed both at the single particle level and in specific cell structures, suggesting that aptamer functionalized Ag clusters are promising candidates for STED microscopy.
Introduction
For more than a century, the spatial resolution of light microscopy was thought to be fundamentally limited, as described by Abbé's theory, which states that the resolution cannot be better than half the wavelength of the applied light.1–3 When imaging with visible light, the resolution is typically limited to ∼200 nm in the lateral direction and ∼500 nm in axial directions, which inhibits high-resolution imaging of sub-cellular structure and hinders a molecular-level understanding of cell structure and dynamics.4 Stimulated emission depletion (STED) microscopy was the first concept that defeated the diffraction barrier by shrinking the effective point spread function (PSF),5,6 fluorescence emission in the periphery of the diffraction limited focal spot is depleted by additional laser light, whose wavelength is tuned to the red edge of the emission spectrum of the fluorophore. The resolution of STED microscopy, depends primarily on the intensity of the depletion laser, and has no theoretical limitation.3 Recent studies unveiling cellular details with resolution better than 100 nm have been achieved in cells, living tissue and organisms.7–10 Locating targets of interest requires delivery of appropriate fluorophores in close proximity of the target structure. Typically, a pair of primary and dye labelled secondary antibodies is used for fluorescent staining. Since the size of a common antibody is on the order of 10 nm, therefore the actual distance between the protein of interest and the observed fluorescent spot can be as large as 20 nm.7,11 Moreover, due to spatial constraints, the antibody based labelling strategy cannot achieve the level of binding to every target molecule, resulting in spotty images.12–14 Alternatives to antibody staining, such as genetically fused fluorescent proteins and tag-based protein labelling strategies have been established for STED microscopy.15,16 However, the introduction and overexpression of foreign coding in a cell might lead to undesired protein aggregation, mis-localization, and loss or gain of function of the target.17,18Aptamer based labelling has been proposed as a complement to the use of existing probes as they possess many favourable characteristics in terms of low molecular weight, little or no immune response, low cost, and ease of preparation and modification.19–22 Aptamers are single-stranded DNA or RNA oligonucleotides with molecular weights of 10–15 kDa and lengths down to 2–3 nm, which can spontaneously fold into well-defined 3-dimensional structures and bind to a broad diversity of targets with high affinity and specificity, such as proteins, phospholipids, sugars, nucleic acids, and even whole bacteria and cells.20–24 In 2012, Opazo et al. demonstrated the potential of dye coupled aptamers for super-resolution imaging by imaging details of endosomal structures such as thin endosomal tubules.25 However, the photostability of traditional fluorescent dyes remains a concern for super resolution microscopy. In contrast, oligonucleotide stabilized fluorescent silver clusters have emerged as an alternative class of fluorescence contrast agents with distinct advantages.26–28 Fluorescent silver clusters, featured by their subnanometer size, good quantum yield and photostability, low toxicity, high extinction coefficients, large Stokes shift, tuneable emission spectrum, and unobtrusive blinking on all relevant time scales, fill a void between organic dye and much larger semiconductor-based probes.28–31
In this paper, we report on the preparation of aptamer functionalized silver clusters and their viability of using aptamer functionalized silver clusters as fluorophores for STED microscopy. Specifically, the aptamer functionalized silver clusters were constructed by linking the anti-proliferative G-rich aptamer AS1411 with a single stranded CCCTTAATCCC DNA oligomer. The aptamer AS1411 was used to specifically bind with the nucleolin protein in the plasma membranes of cancer cells,32,33 while the selected CCCTTAATCCC worked as the template for the formation of the silver cluster. The as prepared silver clusters have a fluorescence quantum yield of 35%, and show maximum absorption and emission at 590 nm and 640 nm, respectively. To STED microscopy, the benefit of the as-prepared silver clusters is to be effectively de-excited by 770 nm laser. Improved stability was observed compared to Alexa Fluor 594 and Atto 594. The specificity of aptamer functionalized silver clusters was evaluated using confocal microscopy by comparing the performance between a cancer cell line (Hela) and a normal cell line (Chinese hamster ovary, CHO), the results clearly showing that the AS1411 functionalized silver clusters can bind to the Hela cells. Nucleolin protein distribution in Hela cells was demonstrated in STED microscopy with resolved resolution, proving that the aptamer functionalized Ag cluster is a promising fluorescent probe for STED microscopy.
Experimental
Chemicals and materials
Silver nitrate (99.99%), sodium borohydride, NaBH4 (powder, 98%), disodium hydrogen phosphate, sodium dihydrogen phosphate, trisodium citrate, citric acid, paraformaldehyde, 4,6-diamidino-2-phenylindole (DAPI), rhodamine 6G, ATTO 594 and cell cultures were all purchased from SIGMA ALDRICH. Alexa Fluor 594 was purchased from Thermo Fisher Scientific. All single strand DNA were synthesized and purified by EBA Co. Ltd. (Germany). All other chemicals involved in this work were analytical-grade. All aqueous solutions were freshly prepared in ultrapure water (≥18 MΩ, Milli-Q, Millipore).Synthesis of fluorescent silver nanoclusters
DNA stabilized AgNCs were synthesized according to previous procedure with slight modification.32 In brief, 3 μM DNA template and 20 μM AgNO3 were sequentially added and mixed with various buffer by vigorous shaking for 1 min in an Eppendorf tube, and then kept at room temperature in the dark. After 20 min, 20 μM freshly prepared NaBH4 was added, thoroughly mixed, and then stored without further mixing at room temperature in the dark for 12 hours. The ratio of base to Ag+ and to BH4− was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
Apparatus and characterization
UV-vis spectra were recorded on a UV-3600 spectrophotometer, and fluorescence spectra were recorded using an F-4500 fluorescence spectrophotometer. TEM images were recorded on a JEM-2100F operated at 200 kV. The fluorescence lifetime and STED imaging were test on home build super-resolution microscopy. Other whole cell imaging and nuclear staining study were conducted with a commercial confocal microscope (TCS SP5, Leica, Germany).Quantum yield measurement
PL quantum yields (QYs) were determined by comparing the integrated emission of the samples in water with that of rhodamine 6G (QY = 94% in ethanol) or fluorescent Atto 594 (QY = 85% in water) according to the following equation:
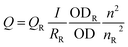 | (1) |
STED and confocal imaging
Single color STED microscopy was performed as described.34,35 STED and the corresponding confocal images were recorded by piezo stage scanning equipped with a pulsed excitation laser at 595 nm (PicoQuant, Berlin, Germany) and a Titanium-Sapphire STED laser emitting 770 nm pulses with reputation rate of 80 MHz. The STED beam passes a polymeric phase plate (Vortex pattern, RPC Photonics, NY, USA). In combination with a λ/4 waveplate, the phase plate is responsible for the doughnut shaped STED-focus. The excitation and the STED beams were co-aligned and coupled into a 1.4 numerical aperture oil immersion lens (NA 1.4 HCX PL APO, 100x, Leica Microsystems, Wetzlar, Germany). The fluorescence was collected by the same lens, spectrally separated from 620 to 680 nm. The fluorescence was detected by fiber-coupled single-photon-counting modules (SPCMAQRH13, Perkin Elmer), with their fiber cores acting as confocal pinholes.Preparation of samples for confocal and STED microscopy
Immobilized single-molecule experiments were performed by spun-coating diluted Alexa Fluor 594, Atto 594 and fluorescent Ag cluster solution on a poly-L-lysine coated glass coverslip at 1500 rpm for 1 min. Coverslips were cleaned by sequential sonication in acetone, 1 M NaOH, and Milli-Q water, each for 15 min. The coverslips were then rinsed with Milli-Q water, dried with compressed N2.The cultivation of cells, and immune labelling was based on established protocols.32,36 HeLa cells (1 × 105) were seeded onto 12 mm sterile coverslips in a 24-well plate. After 24 h, cells were incubated with AS1411-A5-AgNCs for 30 min at 4 °C after being washed three times with phosphate-buffered saline (PBS). To remove the unbound AgNCs, the cells were washed three times with PBS. Then cells were fixed for 20 min in 200 μL of 3.7% paraformaldehyde. After the fixation, cells were washed three times with PBS, followed by nucleus staining with DAPI. Ten minutes later, the cells were further washed three times with PBS buffer. The side of coverslip with fixed cells were mounted in Mowiol. In addition, HeLa cell with CCCTTAATCCC functionalized Ag cluster and CHO cell with AS1411-A5-AgNCs were also prepared as control samples.
Results and discussion
Ag clusters, termed as AgNCs, were prepared by simply mixing single strand CCCTTAATCCC DNA scaffold and silver nitrate in a phosphate buffer, followed by reduction with sodium borohydride. As shown in Fig. 1, the type of buffer, pH value and concentration of the buffer play important roles in the synthesis of luminescent Ag clusters, which might be due to the folding structure of aptamers being slightly different in various conditions.32,37 Four different buffers were compared in the synthesis of AgNCs, as shown in Fig. 1A, phosphate buffer gives the highest fluorescence with maximum emission around 560 nm when excited at 470 nm. The pH value and concentration of phosphate buffer (Fig. 1B and C) demonstrated that 20 mM phosphate buffer with pH at 7.0 present the strongest fluorescence emission. These optimized parameters were also used in the synthesis of aptamer functionalized AgNCs.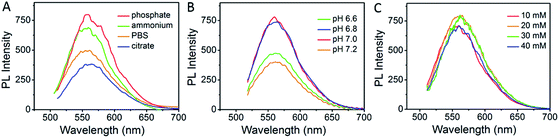 | ||
| Fig. 1 Optimizing the synthesis of AgNCs. (A) Buffer selection, (B) pH value and (C) concentration of phosphate buffer selection. | ||
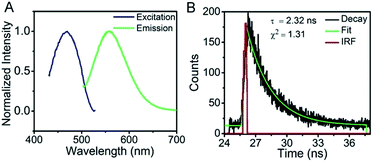
The resultant AgNCs have an excitation maximum of ∼470 nm and an emission maximum ∼560 nm (Fig. 2A). The corresponding fluorescence quantum yield determined by comparison with rhodamine 6G was ∼17%, and the AgNCs exhibit a fluorescence lifetime of 2.32 ns with a mono-exponential behavior (Fig. 2B), suggesting monodispersity of the AgNCs.
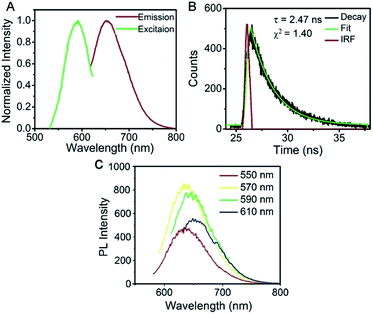
Aptamer functionalized AgNCs were prepared under the same conditions, with the exception of using AS1411 linked CCCTTAATCCC template. To avoid the incidence of steric hindrance,38 an AAAAA spacer was found more bright (termed as AS1411-A5-AgNCs) than the previously used TTTTT spacer.32 The resulted AS1411-A5-AgNCs show strong red emission with quantum yield of 35% using ATTO 594 as the reference standard. As shown in Fig. 3A, with a maximum excitation at around 570 nm, the emission peak is about 650 nm, the value is slightly red shift in excitation wavelength between 550 nm and 610 nm (Fig. 3C). The lifetime of AS1411-A5-AgNCs is 2.47 ns (Fig. 3B). It is well approximated by a single exponential decay, indicating that a single species dominates the emission. The red shift in the emission spectra and increased lifetime of the AgNCs suggest that the added DNA sequence influences the property of the final AgNCs. A similar effect was observed on aptamer functionalized single stranded DNA consisting of 12 cytosines.32
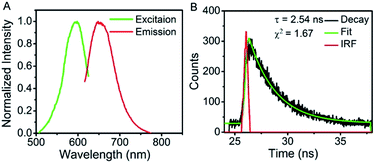
To confirm that CCCTTAATCCC template is critical to the formation of the AgNCs, aptamer mucin 1 was incorporated onto CCCTTAATCCC using an AAAAA spacer, termed as mucin-A5-AgNCs. The resulted excitation and emission peaks are located at 590 nm and 650 nm (Fig. 4A), respectively. The fluorescent decay profiles (Fig. 4B) once again display a mono-exponential fit with a lifetime of 2.54 ns. It should be mentioned that in the absence of CCCTTAATCCC, both AS1411 and mucin 1 do not produce significant fluorescence, suggesting that CCCTTAATCCC is crucial for the formation of fluorescent silver clusters, while the aptamer mainly serves the role of labelling the target structure and only slightly influences the spectra.
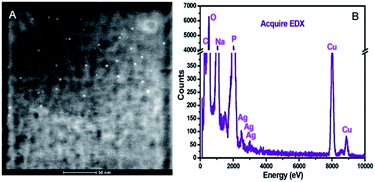
Size and elemental mapping of the AS1411-A5-AgNCs were performed using high-angle annular dark field (HAADF) imaging and EDX analysis. It can be seen in Fig. 5 that the AS1411-A5-AgNCs are monodispersed with a size distribution around 1.8 nm, while the EDX pattern confirms that the major constituent of clusters is Ag. The major elements like the Na, C, O, P, Cu should came from the buffer and the TEM substrate.
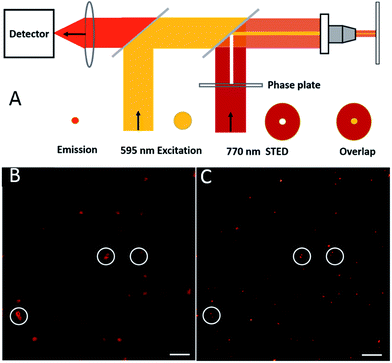
A stage scanning STED microscope was built to evaluate the performance of single AS1411-A5-AgNCs (Fig. 6A). A picosecond 595 nm laser with 80 MHz pulse train was used for excitation (yellow), while a 770 nm mode-locked Ti:sapphire laser whose output was engineered to produce a donut shaped focal spot (red) was used to switch off peripheral fluorescence, the shrunk fluorescence was collected by a detector (orange). As shown in Fig. 6B, isolated bright point-like fluorescent objects (around 220 nm), most likely due to individual AS1411-A5-AgNCs clusters, were observed in confocal image. In contrast, the STED counterpart (Fig. 6C) shows excellent resolution improvement with good signal-to noise ratio, especially the objects marked in white cycles, which are difficult to distinguish in confocal image, but well separated in STED image. For a power of 70 mW in the focal plane, an average optical resolution of approximately 52 nm was achieved. It should be noted that this resolution is attained in raw data, exclusively based on optically induced molecular transitions. Further increasing the power of STED laser should lead to an improvement in resolution. However, the high power depletion light also increases the probability to drive the AS1411-A5-AgNCs transiting into a recoverable long-lived dark state, which is similar to previous observation.39,40 For instance, upon increasing the STED power to 120 mW, almost one fourth of the clusters go to the dark state, before gradually turning fluorescent over the timescale of seconds. Previously research demonstrated that charge transfer from Ag to DNA mechanism possibly give rise to the photo induce dark states.41 In order to further improving the performance of the Ag clusters as what have been done to reduce the blinking of quantum dots,42 more extensive evaluation of the dark state need to be pursued in future studies.
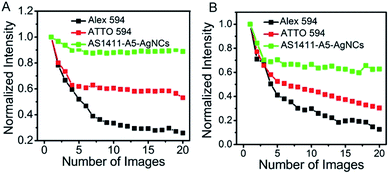
The photostability of AS1411-A5-AgNCs under confocal and STED conditions was evaluated, quantified and compared with two other conventional STED probes, Alexa flour 594 and ATTO 594. For this purpose, thin layers of AS1411-A5-AgNCs and dyes were spin-coated on poly-L-lysine coated glass coverslips. The normalized total fluorescence signal from a 2 × 2 μm size area (averaging from 3 different selected areas) in the samples over the course of 20 scans were then compared. Excitation and STED powers at the objective back aperture of 10 mW and 100 mW were used, respectively. As shown in Fig. 7, the AS1411-A5-AgNCs exhibit superior photostability compared with Alexa flour 594 and ATTO 594 under both confocal and STED conditions. The fluorescence of AS1411-A5-AgNCs persists after a relatively fast drop in the first few scans then to ∼90% and ∼60% after 20 scans in confocal and STED, respectively. In contrast, the fluorescence of Alexa flour 594 dramatically decreased to 26% and 13%, and ATTO 594 decreased to 53% and 30%, respectively.
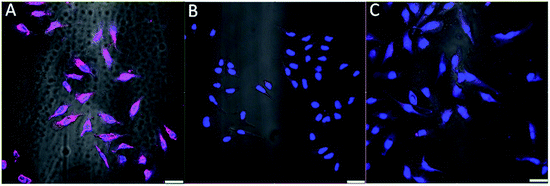
The aptamer AS1411 is reported to bind to nucleolin in the plasma membranes of cancer cells.32,33 To verify if the AS1411-A5-AgNCs could be used as a specific fluorescent probe for cancer cell imaging, the sample was incubated with HeLa and healthy CHO cells for 30 min at 4 °C and subsequently washed with PBS buffer. To increase phase-contrast, the nucleus of both cell lines were stained with DAPI (blue). As illustrated in Fig. 8, strong fluorescence (magenta) along the cellular membranes was observed in the HeLa cells (Fig. 8A), while no significant fluorescence signal was recorded for the CHO cells (Fig. 8B). The confocal microscopy images confirm that AS1411-A5-AgNCs bind specifically to nucleolin protein in the cellular membrane of cancer cells. Additional control experiment was performed by replacing AS1411-A5-AgNCs with AgNCs during incubation with HeLa cells; comparatively negligible fluorescence was observed from the plasma membranes in the corresponding image (Fig. 8C). Therefore, AS1411-A5-AgNCs preserve the specificity of the aptamer.
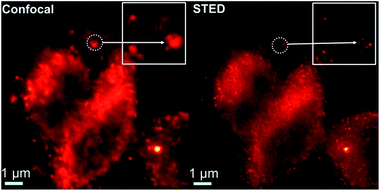
A closer observation of the distribution of AS1411-A5-AgNC labelled nucleolin in plasma membranes was further performed under confocal and STED conditions (Fig. 9). From the confocal image, it is hard to resolve the detail of the structure, while their STED counterpart consistently provide superior contrast and resolution. The specificity and reliability of the AS1411-A5-AgNC in compare with the conventional antibody labelling strategy is vital to promote its practical application and worthy to perform further.
Conclusions
In summary, aptamer functionalized fluorescent Ag clusters were successfully synthesized by integrating AS1411 to a new DNA scaffold via an AAAAA loop. The maximum absorption and emission of the prepared AS1411-A5-AgNCs are located at 590 nm and 640 nm, and the fluorescence quantum yield is 35%. The AS1411-A5-AgNCs demonstrate to have specific binding affinity to nucleolin in the plasma membranes of Hela cells. Moreover, AS1411-A5-AgNCs are proved to be an excellent probe for STED microscopy both at the single cluster level and in cell imaging. Considering the biological and chemical advantages of aptamers over antibodies and fluorescent proteins, including the small size, ease of production and chemical modification, and high affinity to targets, in combination with the photostability of silver clusters, the proposed aptamer functionalized silver clusters offer a promising labelling strategy for novel analytical and biomedical applications and will open up a new avenue for synthesizing small but stable fluorescent probes for super resolution microscopy.Acknowledgements
Financial support by the ‘1000 Talent Program’ (The Recruitment Program of Global Experts), and the National Natural Science Foundation of China (21503271, 21303258).References
- S. W. Hell, Angew. Chem., Int. Ed., 2015, 54, 8054–8066 CrossRef CAS PubMed.
- E. Abbe, Archiv für mikroskopische Anatomie, 1873, 9, 413–418 CrossRef.
- S. W. Hell, Nat. Methods, 2009, 6, 24–32 CrossRef CAS PubMed.
- P. Hunter, EMBO Rep., 2011, 12, 643–646 CrossRef CAS PubMed.
- S. W. Hell, Nat. Biotechnol., 2003, 21, 1347–1355 CrossRef CAS PubMed.
- T. A. Klar, S. Jakobs, M. Dyba, A. Egner and S. W. Hell, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8206–8210 CrossRef CAS.
- H. Blom and H. Brismar, J. Intern. Med., 2014, 276, 560–578 CrossRef CAS PubMed.
- R. Han, Z. Li, Y. Fan and Y. Jiang, J. Genet. Genomics, 2013, 40, 583–595 CrossRef CAS PubMed.
- R. Chéreau, J. Tønnesen and U. V. Nägerl, Methods, 2015, 88, 57–66 CrossRef PubMed.
- T. Müller, C. Schumann and A. Kraegeloh, ChemPhysChem, 2012, 13, 1986–2000 CrossRef PubMed.
- G. Patterson, M. Davidson, S. Manley and J. Lippincott-Schwartz, Annu. Rev. Phys. Chem., 2010, 61, 345–367 CrossRef CAS PubMed.
- T. Lang and S. O. Rizzoli, Physiology, 2010, 25, 116–124 CrossRef CAS PubMed.
- U. Agrawal, D. T. Reilly and C. M. Schroeder, Curr. Opin. Biotechnol., 2013, 24, 646–653 CrossRef CAS PubMed.
- F. Opazo, Super-Resolution Microscopy Techniques in the Neurosciences, 2014, vol. 86, pp. 369–386 Search PubMed.
- F. Stagge, G. Y. Mitronova, V. N. Belov, C. A. Wurm and S. Jakobs, PLoS One, 2013, 8, e78745 CAS.
- J. Chojnacki, T. Staudt, B. Glass, P. Bingen, J. Engelhardt, M. Anders, J. Schneider, B. Muller, S. W. Hell and H. G. Krausslich, Science, 2012, 338, 524–528 CrossRef CAS PubMed.
- D. M. Chudakov, M. V. Matz, S. Lukyanov and K. A. Lukyanov, Physiol. Rev., 2010, 90, 1103–1163 CrossRef CAS PubMed.
- M. W. Davidson and R. E. Campbell, Nat. Methods, 2009, 6, 713–717 CrossRef CAS PubMed.
- A. Aravind, Y. Yoshida, T. Maekawa and D. S. Kumar, Drug Delivery Transl. Res., 2012, 2, 418–436 CrossRef CAS PubMed.
- S. D. Jayasena, Clin. Chem., 1999, 45, 1628–1650 CAS.
- K. C. Liu, B. S. Lin and X. P. Lan, J. Cell. Biochem., 2013, 114, 250–255 CrossRef CAS PubMed.
- A. Z. Wang and O. C. Farokhzad, J. Nucl. Med., 2014, 55, 353–356 CrossRef CAS PubMed.
- C. L. A. Hamula, H. Q. Zhang, L. L. Guan, X. F. Li and X. C. Le, Anal. Chem., 2008, 80, 7812–7819 CrossRef CAS PubMed.
- C. X. Zhang, X. F. Lv, X. Han, Y. Man, Y. Saeed, H. Qing and Y. L. Deng, Anal. Methods, 2015, 7, 6339–6345 RSC.
- F. Opazo, M. Levy, M. Byrom, C. Schafer, C. Geisler, T. W. Groemer, A. D. Ellington and S. O. Rizzoli, Nat. Methods, 2012, 9, 938–939 CrossRef CAS PubMed.
- J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126, 5207–5212 CrossRef CAS PubMed.
- B. Y. Han and E. K. Wang, Anal. Bioanal. Chem., 2012, 402, 129–138 CrossRef CAS PubMed.
- S. Choi, R. M. Dickson and J. H. Yu, Chem. Soc. Rev., 2012, 41, 1867–1891 RSC.
- X. H. Tan and R. C. Jin, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 569–581 CrossRef CAS PubMed.
- L. Shang, S. J. Dong and G. U. Nienhaus, Nano Today, 2011, 6, 401–418 CrossRef CAS.
- Y. Z. Lu and W. Chen, Chem. Soc. Rev., 2012, 41, 3594–3623 RSC.
- J. L. Li, X. Q. Zhong, F. F. Cheng, J. R. Zhang, L. P. Jiang and J. J. Zhu, Anal. Chem., 2012, 84, 4140–4146 CrossRef CAS PubMed.
- H. G. Sun, X. Zhu, P. Y. Lu, R. R. Rosato, W. Tan and Y. L. Zu, Mol. Ther.--Nucleic Acids, 2014, 3, e182 CrossRef CAS PubMed.
- F. Göttfert, C. A. Wurm, V. Mueller, S. Berning, V. C. Cordes, A. Honigmann and S. W. Hell, Biophys. J., 2013, 105, L01–L03 CrossRef PubMed.
- C. A. Wurm, K. Kolmakov, F. Göttfert, H. Ta, M. Bossi, H. Schill, S. Berning, S. Jakobs, G. Donnert, V. N. Belov and S. W. Hell, Opt. Nanoscopy, 2012, 1, 1–7 CrossRef.
- J. B. Zhu, L. B. Zhang, Y. Teng, B. H. Lou, X. F. Jia, X. X. Gu and E. K. Wang, Nanoscale, 2015, 7, 13224–13229 RSC.
- C. I. Richards, S. Choi, J. C. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y. L. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038–5039 CrossRef CAS PubMed.
- A. Latorre, R. Lorca and A. Somoza, J. Chem., 2013, 631421 Search PubMed.
- J. Sharma, H. C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2010, 46, 3280–3282 RSC.
- T. Vosch, Y. Antoku, J. C. Hsiang, C. I. Richards, J. I. Gonzalez and R. M. Dickson, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12616–12621 CrossRef CAS PubMed.
- S. A. Patel, M. Cozzuol, J. M. Hales, C. I. Richards, M. Sartin, J. C. Hsiang, T. Vosch, J. W. Perry and R. M. Dickson, J. Phys. Chem. C, 2009, 113, 20264–20270 CAS.
- X. Y. Wang, X. F. Ren, K. Kahen, M. A. Hahn, M. Rajeswaran, S. Maccagnano-Zacher, J. Silcox, G. E. Cragg, A. L. Efros and T. D. Krauss, Nature, 2009, 459, 686–689 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |