Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DNA aptamer-based carrier for loading proteins and enhancing the enzymatic activity†
Jieun Kim,
Dajeong Kim and
Jong Bum Lee *
*
Department of Chemical Engineering, University of Seoul, 163 Seoulsiripdaero, Dongdaemun-gu, Seoul, 130-743, South Korea. E-mail: jblee@uos.ac.kr; Fax: +82 26490 2364; Tel: +82 26490 2372
First published on 14th December 2016
Abstract
Here, we synthesized DNA microparticles comprised of thrombin binding aptamers via rolling circle amplification (RCA). These DNA aptamer particles could successfully load a number of thrombins and the complexes have shown improved thrombin activity. This new platform offers a protein loading and delivery system for medical applications.
Functional DNA, especially DNA aptamers, have attracted much attention with their ability to bind to their targets with high affinity and specificity. Since their introduction, various aptamers have been reported in the past decades via in vitro selection or systematic evolution of ligands by exponential enrichment (SELEX).1–5 These DNA aptamers have been consistently studied with a variety of nanomaterials such as metals,6 silica,7 carbon8,9 and polymers.10 In addition, aptamers have been applied for biomedical applications such as cancer imaging,11,12 protein detection9,13 and targeted drug delivery.7,14 However, only a few studies that applied aptamers for loading and delivery of proteins have been reported.15 Moreover, there has been no attempt to utilize aptamers as building blocks forming self-assembled particles for high molecular loading.
In our previous study, RNA microparticles having numerous siRNA have been generated via a rolling circle replication technique16 and DNA hydrogel and DNA microspheres also have been generated similarly.17 Inspired by this enzymatic approach, here we synthesized DNA microparticles having numerous aptamer sites via rolling circle amplification (RCA) as illustrated schematically in Fig. 1. Highly concentrated multiple ssDNA chains are randomly crosslinked and form spherical DNA particles. We designed two kinds of particles comprised of aptamers binding to the fibrinogen-binding exosite of thrombin (F-TBA) and heparin-binding exosite of thrombin (H-TBA), which are the most commonly used thrombin binding aptamers. The synthesized particles could effectively bind to thrombin and the activity of thrombin was also confirmed. Moreover, by loading thrombin on the DNA aptamer particles, the thrombin activity was improved up to 1.7 times. Therefore, this new platform for carrying locally concentrated thrombin could be applied in medical applications to promote blood coagulation. In addition, this system could be applied for other proteins and drugs by modifying sequence and it offers the promise of potent loading and delivery system for improving the ability of proteins.
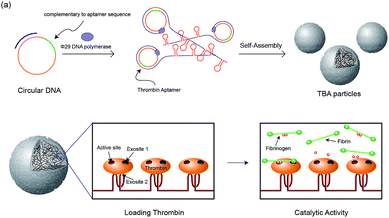 | ||
| Fig. 1 Schematic illustration of the construction of the TBA particles and the catalytic activity of thrombin with the particles. | ||
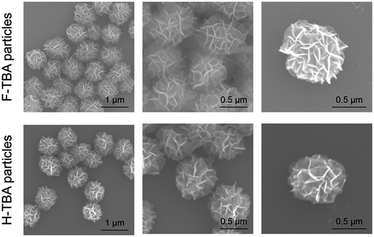
The two kinds of particles were generated from the designed circular templates as thrombin (TB) carriers. One circular DNA contains complementary sequence to F-TBA and the other one is comprised of complementary sequence to H-TBA. Each circular DNA was incubated for 20 h with phi29 DNA polymerase to synthesize F-TBA DNA particle (F-TBAP) and H-TBA DNA particle (H-TBAP). The sequences of circular DNAs and experimental details are given in the ESI.† The morphology and structure of the synthesized particles were analyzed by scanning electron microscopy (SEM). As shown in Fig. 2, both aptamer particles have similar porous spherical structure. This structure has advantages for carrying proteins because of their large surface area. In addition, numerous aptamer sites could be concentrated in a particle because the particles have large sized (∼0.9 μm).
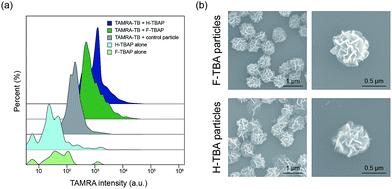
To confirm the selective binding of TB to TBAPs, fluorescent-labeled thrombin (TAMRA-TB) was utilized. The TAMRA-TB was incubated with the particles for 3 h at room temperature and the fluorescence intensity was measured after incubation. As shown in Fig. 3a and S1,† the fluorescence intensities of the F-TBAPs and H-TBAPs after incubation increased than non-labeled particles as negative control. Intensity of the DNA particles without aptamer was also measured after incubation with TAMRA-TB, however, they have shown distinctly lower fluorescence intensity than both TBAPs. These results mean that TB could selectively bind to the TBAPs, and thus the function of aptamer of TBAPs were successfully confirmed.
To observe the morphology of particles after incubation with TB, SEM was utilized. Interestingly, the morphology of the particles was slightly changed after binding with TB. As seen in the Fig. 3b, both particles became slightly smaller and shrank. The average size of the F-TBAPs decreased from 0.88 ± 0.07 μm to 0.71 ± 0.06 μm. The average size of the H-TBA particles also slightly decreased from 0.79 ± 0.08 μm to 0.75 ± 0.09 μm. These changes probably due to the formation of G-quadruplex structure when KCl and TB were added.18 The aptamer sites of the particles could be switched to G-quadruplex form by KCl or TB and this change could cause morphological change. To verify the effect of KCl, the sizes of KCl treated TBAP without TB were also confirmed by utilizing SEM (see Fig. S2†). As expected, the sizes of F-TBAP and H-TBAP decreased (0.77 ± 0.07 μm, 0.74 ± 0.06 μm each) when KCl was added. In spite of this alteration, the particles still maintain spherical and porous structures with large surface area. These results show that the giant aptamer particles could be effective protein carriers.
The activity of the TB conjugated particles were confirmed by thrombin activity assay. Various TBAP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TB volume ratios (from 1
TB volume ratios (from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 32
1 to 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
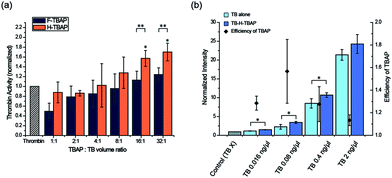
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were tested to optimize the binding condition. F-TBAPs and H-TBAPs were incubated with TB and washed after binding respectively. Then the TB activity of TB-loaded TBAPs (TB-TBAPs) were compared with same amount of TB assuming 100% binding. As seen in the Fig. 4a, the activities of the both TB-F-TBAPs and TB-H-TBAPs were gradually improved when the ratio of TBAP increased. It is probably due to the fact that TB was saturated at the small volumes of TBAPs. However, as ratio of TBAPs increased, the amount of particles would be sufficient to load TB, and thus the efficiency of particles could increase. To verify this idea, the loading efficiency of thrombin was measured by fluorescence. As shown in Fig. S3 and Table S2,† loading efficiency increased as the ratio of TBAP increased. Interestingly, TB-H-TBAPs have shown better activity than TB-F-TBAPs. Since the majority of thrombin binding to fibrinogen is mediated by exosite 1, this exosite 1 binding F-TBA could hinder fibrinogen cleavage.19 On the other hand, H-TBA doesn't blocking active site because it only binds on exosite 2 instead of exosite 1.20 This is consistent with the result, suggesting that H-TBAPs are better for blood coagulation than F-TBAPs. In the case of TB-H-TBAPs, they have shown better activities than TB control from the 8
1) were tested to optimize the binding condition. F-TBAPs and H-TBAPs were incubated with TB and washed after binding respectively. Then the TB activity of TB-loaded TBAPs (TB-TBAPs) were compared with same amount of TB assuming 100% binding. As seen in the Fig. 4a, the activities of the both TB-F-TBAPs and TB-H-TBAPs were gradually improved when the ratio of TBAP increased. It is probably due to the fact that TB was saturated at the small volumes of TBAPs. However, as ratio of TBAPs increased, the amount of particles would be sufficient to load TB, and thus the efficiency of particles could increase. To verify this idea, the loading efficiency of thrombin was measured by fluorescence. As shown in Fig. S3 and Table S2,† loading efficiency increased as the ratio of TBAP increased. Interestingly, TB-H-TBAPs have shown better activity than TB-F-TBAPs. Since the majority of thrombin binding to fibrinogen is mediated by exosite 1, this exosite 1 binding F-TBA could hinder fibrinogen cleavage.19 On the other hand, H-TBA doesn't blocking active site because it only binds on exosite 2 instead of exosite 1.20 This is consistent with the result, suggesting that H-TBAPs are better for blood coagulation than F-TBAPs. In the case of TB-H-TBAPs, they have shown better activities than TB control from the 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, and they are significantly effective (up to 1.7 times) than TB control at the 16
1 ratio, and they are significantly effective (up to 1.7 times) than TB control at the 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 32
1 and 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. Since the active site of TB could be well oriented on the particle surface, TB in the particles could effectively bind to fibrinogen. These results suggest a new platform for loading TB and enhancing their activity using DNA aptamer particles.
1 ratio. Since the active site of TB could be well oriented on the particle surface, TB in the particles could effectively bind to fibrinogen. These results suggest a new platform for loading TB and enhancing their activity using DNA aptamer particles.
Since the difference in activities between 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 32
1 and 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio was minor, we have used 16
1 ratio was minor, we have used 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio for further study considering cost effectiveness. To confirm the efficiency for the various concentrations, 0.016 ng μL−1 to 2 ng μL−1 of TB and TBA-TB particles were tested. From 0.016 ng μL−1 to 0.4 ng μL−1, H-TBA-TB particles were significantly effective than using TB alone and the particles have shown highest efficiency at 0.08 ng μL−1 as shown in Fig. 4b. These results confirmed that using TBA particles could help the enzymatic reaction of TB at various concentrations. Therefore, this TBAP system could be useful to save proteins especially in optimal condition because same performance could be achieved with the smaller amount of proteins. In addition, considering the loading efficiency of thrombin, the effectiveness could be improved more by re-incubating the residual thrombin with additional TBAP.
1 ratio for further study considering cost effectiveness. To confirm the efficiency for the various concentrations, 0.016 ng μL−1 to 2 ng μL−1 of TB and TBA-TB particles were tested. From 0.016 ng μL−1 to 0.4 ng μL−1, H-TBA-TB particles were significantly effective than using TB alone and the particles have shown highest efficiency at 0.08 ng μL−1 as shown in Fig. 4b. These results confirmed that using TBA particles could help the enzymatic reaction of TB at various concentrations. Therefore, this TBAP system could be useful to save proteins especially in optimal condition because same performance could be achieved with the smaller amount of proteins. In addition, considering the loading efficiency of thrombin, the effectiveness could be improved more by re-incubating the residual thrombin with additional TBAP.
In conclusion, we have proposed a general approach for loading a large amount of proteins using DNA aptamer particles. DNA particles having numerous aptamer sites were synthesized via RCA and the interaction between TBA particles and TB was successfully confirmed by increase of the fluorescence intensity using TAMRA-labeled TB. Moreover, at the optimal conditions, TB-F-TBAPs and TB-H-TBAPs have shown higher activity than that of TB alone. The improvement of protein activities was monitored at wide range of concentration of TB. These DNA aptamer particle–protein complexes could be applied to promote blood coagulation. Furthermore, this system can be easily applied in any aptamer-binding proteins to promote their activity for medical or biological applications.
Acknowledgements
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation of Korea (NRF) funded by the Korean Government (NRF-2016M3A9C6917402).Notes and references
- C. L. Hamula, J. W. Guthrie, H. Zhang, X.-F. Li and X. C. Le, TrAC, Trends Anal. Chem., 2006, 25, 681–691 CrossRef CAS.
- G. Mayer, Angew. Chem., Int. Ed., 2009, 48, 2672–2689 CrossRef CAS PubMed.
- R. Stoltenburg, C. Reinemann and B. Strehlitz, Biomol. Eng., 2007, 24, 381–403 CrossRef CAS PubMed.
- S. M. Shamah, J. M. Healy and S. T. Cload, Acc. Chem. Res., 2008, 41, 130–138 CrossRef CAS PubMed.
- X. Fang and W. Tan, Acc. Chem. Res., 2009, 43, 48–57 CrossRef PubMed.
- H.-Q. Chen, F. Yuan, S.-Z. Wang, J. Xu, Y.-Y. Zhang and L. Wang, Analyst, 2013, 138, 2392–2397 RSC.
- L. L. Li, Q. Yin, J. Cheng and Y. Lu, Adv. Healthcare Mater., 2012, 1, 567–572 CrossRef CAS PubMed.
- R. Yang, Z. Tang, J. Yan, H. Kang, Y. Kim, Z. Zhu and W. Tan, Anal. Chem., 2008, 80, 7408–7413 CrossRef CAS PubMed.
- Y. Pu, Z. Zhu, D. Han, H. Liu, J. Liu, J. Liao, K. Zhang and W. Tan, Analyst, 2011, 136, 4138–4140 RSC.
- X. He, B. Wei and Y. Mi, Chem. Commun., 2010, 46, 6308–6310 RSC.
- H. Shi, X. He, K. Wang, X. Wu, X. Ye, Q. Guo, W. Tan, Z. Qing, X. Yang and B. Zhou, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3900–3905 CrossRef CAS PubMed.
- J. Li, H. Zheng, P. J. Bates, T. Malik, X.-F. Li, J. O. Trent and C. K. Ng, Nucl. Med. Biol., 2014, 41, 179–185 CrossRef PubMed.
- N. Hamaguchi, A. Ellington and M. Stanton, Anal. Biochem., 2001, 294, 126–131 CrossRef CAS PubMed.
- Y. Park, H. Kim and J. B. Lee, ACS Biomater. Sci. Eng., 2016, 2, 616–624 CrossRef CAS.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831–834 CrossRef CAS PubMed.
- J. B. Lee, J. Hong, D. K. Bonner, Z. Poon and P. T. Hammond, Nat. Mater., 2012, 11, 316–322 CrossRef CAS PubMed.
- J. B. Lee, S. Peng, D. Yang, Y. H. Roh, H. Funabashi, N. Park, E. J. Rice, L. Chen, R. Long and M. Wu, Nat. Nanotechnol., 2012, 7, 816–820 CrossRef CAS PubMed.
- R. F. Macaya, P. Schultze, F. W. Smith, J. A. Roe and J. Feigon, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3745–3749 CrossRef CAS.
- S. M. Nimjee, S. Oney, Z. Volovyk, K. M. Bompiani, S. B. Long, M. Hoffman and B. A. Sullenger, RNA, 2009, 15, 2105–2111 CrossRef CAS PubMed.
- D. M. Tasset, M. F. Kubik and W. Steiner, J. Mol. Biol., 1997, 272, 688–698 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra25507h |
| This journal is © The Royal Society of Chemistry 2017 |