Self assembly of a γ-amino butyric acid based derivative into tunable nano/micro structures†
Surinder K.
Brar
a,
Prabhjot
Singh
b,
Manish
Bajaj
a,
Akash
Deep
c,
Nishima
Wangoo
*bd and
Rohit K.
Sharma
*a
aDepartment of Chemistry & Centre for Advanced Studies in Chemistry, Panjab University, Sector-14, Chandigarh-160014, India. E-mail: rohitksg@pu.ac.in; Fax: +91-172-2545074; Tel: +91-172-2534409
bCentre for Nanoscience & Nanotechnology (U.I.E.A.S.T.), Panjab University, Sector-14, Chandigarh 160014, India
cCentral Scientific Instruments Organisation (CSIR-CSIO), Sector 30 C, Chandigarh 160030, India
dDepartment of Applied Sciences, University Institute of Engineering & Technology (U.I.E.T.), Panjab University, Sector-25, Chandigarh 160014, India
First published on 4th August 2016
Abstract
The self assembly and structural modification of a γ-amino butyric acid based derivative in different solvents were studied and the structural variations were characterized using FE-SEM, confocal microscopy and computational methods. In addition, concentration and temperature dependence studies were carried out exhibiting the importance of experimental conditions in the process of self assembly and structural modification. Such an approach is not only important for gaining significant new insights into the fabrication of variable architectures from simple self assembling organic molecules but can also pave the way for a better understanding of the self assembly process at the molecular level.
Introduction
The engineering of distinct three dimensional patterns at the supramolecular nano/micro scale by manipulating specific chemical moieties of an organic molecule has become an area of growing interest and demand in recent years.1,2 The fabrication of nanomaterials generally requires self assembly which is an attractive strategy for constructing advanced functional materials with control over nanoscale dimensions.3 Self-assembled constructs mainly employ non-covalent interactions which include electrostatic forces, hydrogen bonds, van der Waals interactions and π–π stacking interactions in a more or less ordered manner.4 Along with the covalent framework, these non-covalent interactions allow the creation of novel functional materials through a simple and versatile approach of molecular self assembly.5 The examples of natural systems have demonstrated that even these simple architectural arrangements have the potential to create novel materials leading researchers to understand their conformational arrangements. The self assembly of lipids in cell membranes, protein folding in enzymes, formation of the DNA double helix, and formation of viruses are some of the examples which have evolved in biological systems over the centuries.6The self assembly of organic building blocks such as peptides, nucleic acids, and phospholipids has led to the development of a new generation of biomaterials in recent years.7,8 This has ushered in a completely new and appealing area of materials research in which the formation and applications of ordered structures using simple organic molecules can be studied.7 The applicability of these ordered structures generally depends upon the ability of the chosen molecule for specific molecular recognition, biocompatibility, chemical modification and ease of availability.9 In particular, intense research efforts have been witnessed in the development of peptide based self assembling scaffolds in recent years.10 The origin of such an area highlights the fundamental importance in understanding the self assembling of the natural as well as synthetic peptide based structural and functional building blocks which include cyclic peptides,11 dendritic peptides,12 amphiphilic peptides,13 oligopeptides,14 aromatic dipeptides,15etc. These building blocks have been reported to spontaneously associate for forming different structural morphologies like nanotubes, nanospheres, nanofibrils, nanorods, nanotapes, and organogels under different conditions.16 Similarly, other organic materials such as p-distyrylbenzene (DSP),17trans-1,2-diamidocyclohexane,18 phthalimide end capped thiophene based organic molecules,19 and monosubstituted perylenediimide (PDI) derivatives20 get self assembled into a number of structures depending upon the functionality involved. Therefore, there is a need for understanding the mechanism behind self assembly of organic molecules and exploration for the generation of new age materials with much needed applications.
Recently, reports on the protected/unprotected amino acid derived self assembly have gained attention as the self assembly of amino acid based molecules in combination with other building blocks such as poly(N-methacryloyl-L-valine), poly(N-methacryloyl-L-phenylalanine), and poly(N-methacryloylglycyl-L-leucine) has been reported.21 Even the self assembly of single amino acids like phenylalanine and tyrosine has been recently reported in the literature.22 Similarly, unnatural amino acids like β-alanine, with an extra –CH2 unit compared to the α-amino acids, have been investigated for the formation of self-assembled structures.23 It was observed that the presence of an extra carbon atom providing a large degree of conformational variability leads to a greater diversity of backbone structures in peptides and polypeptides.24 Gamma amino butyric acid (GABA), a γ-amino acid with two extra carbon atoms compared to the α-amino acid, is an important inhibitory neurotransmitter present in the vertebrate central nervous system.25 Such amino acid neurotransmitters that produce responses within a few milliseconds play an important role in brain functions and neurological diseases and are critical for the function of the central nervous system (CNS).26
As proteins are combinations of several amino acid residues, a morphological study of amino acids under different conditions may help to understand the morphological pattern of proteins/peptides or amino acid based bio-molecules in different biological environments within bio-systems. Although the morphological study of single α-amino acid and ω-amino acid based molecules has been reported to the best of our knowledge, the study of γ-amino acids has not been carried out yet. Understanding the fact that the self assembly behaviour of molecules largely depends on molecular chemistry and the assembling environment such as solvents, co-assembling molecules and temperature, we focused our study on the self assembly of a GABA derivative under various experimental conditions.27 As the choice of the protecting group may play an important role in the study of the morphological behaviour of γ-amino butyric acid, the N-terminal of GABA was protected using the Boc (t-butyloxycarbonyl) group as it is known to play a key role in promoting various nanostructures due to its assembling selectivity. N,N′-Dicyclohexylurea was chosen as a protecting group for the carboxylic group of GABA due to its biological importance and easy availability.23 Thus, herein we report the synthesis of Boc-GABA-N,N′-dicyclohexylurea followed by the exploration of its self assembly under various experimental conditions. It was observed that the reported GABA derivative was not only able to self assemble into various structures but also demonstrated structural variations depending upon the solvent, temperature and concentration modulations.
Results and discussions
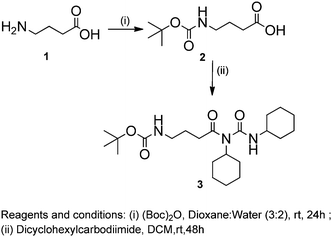
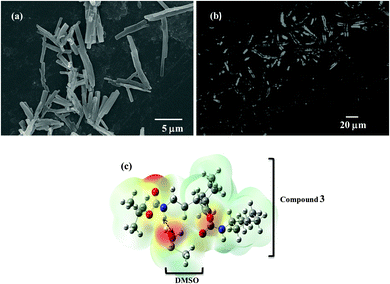
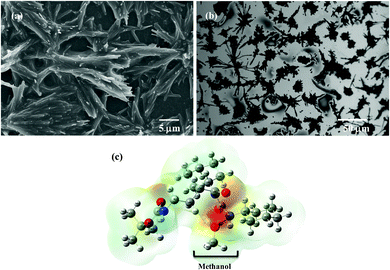
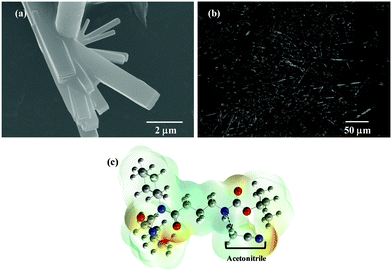
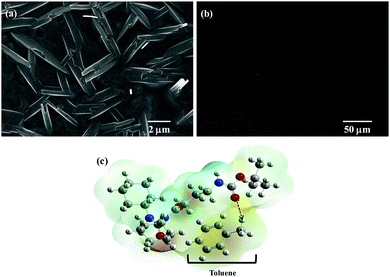
The chemical synthesis of the desired Boc-GABA-N,N′-dicyclohexylurea was carried out using a two-step synthetic route (Scheme 1). Firstly, the amino terminus of γ-amino butyric acid (1) was protected using di-tert-butyl dicarbonate to form the Boc-protected γ-amino butyric acid (2) followed by reaction with dicyclohexylcarbodiimide to form Boc-GABA-N,N′-dicyclohexylurea (3) which was characterized using NMR and mass spectrometry (ESI,† Fig. S1 and S2). The morphological diversity of the self assembled structures of synthesized γ-amino acid derivative Boc-GABA-N,N′-dicyclohexylurea 3 was examined using different spectroscopic and microscopic based techniques. It is quite well known that the self assembly behaviour can be influenced by factors such as nature and topography of the substrate, pH, temperature, concentration and the medium.28 Keeping this in mind, our initial aim was to study the formation of self assembled motifs of the GABA derivative (3) in different solvents such as dimethyl sulfoxide (DMSO), methanol, acetonitrile and toluene. The main objective behind such a study is based on the fact that the properties of the solvent influence the self assembly of an organic molecule through interactions such as hydrogen bond formation, π–π stacking and other electrostatic affinities.Initially, the self assembly of compound 3 was tried in dimethylsulfoxide (DMSO) and the morphology of the self assembled structures thereby formed was characterized using microscopic and computational methods. The field emission scanning electron microscopy (FE-SEM) results indicate the formation of dense and well-structured rod shaped clusters with an almost uniform size distribution and length of 5–10 µm [Fig. 1(a)]. The evidence of the formation of these rod clusters was further provided by the confocal microscopy images [Fig. 1(b)]. To understand the mechanism of the self assembly process, a computational model was generated and the optimisation of the structures was done using the Hartree–Fock method by employing the 3-21G basis set which indicated a hydrogen bond-like interaction between the hydrogen of the amine and oxygen of DMSO. These interactions were also confirmed by the generation of the molecular electrostatic potential (MEP) plot which depicts enhanced electron density at the DMSO oxygen (red color) and the corresponding less electron density at the amine hydrogen (blue color), thereby authenticating the interaction between these two atoms [Fig. 1(c)]. The calculated distance for this bond of 1.825 Å falls in the range of hydrogen bond interactions and the bond distance between the DMSO oxygen and amine nitrogen was found to be 2.780 Å which is in agreement for hydrogen bond formation. Indeed, this hydrogen bond seems likely to be responsible for the overall rod cluster shape. Further, powder X-ray diffraction studies were also carried out in order to check the emergence of new planes (ESI,† Fig. S3). This confirmed the self assembly of the compound. In addition to DMSO, methanol was also tested as a solvent for the self assembly of GABA derivative 3. In the case of methanol as the solvent, the well-defined rod structure obtained in DMSO appeared to have aggregated and resulted in overlapping motifs as observed in FE-SEM [Fig. 2(a)] and confocal microscopic images [Fig. 2(b)]. This observation may be explained on the basis of the difference in polarity between methanol and DMSO (methanol is more polar than DMSO). The observation of aggregated structures may be explained on the basis that the interaction of GABA derivative 3 is different with more polar methanol as compared to DMSO. Therefore, the GABA derivative may interact with methanol through the intermolecular hydrogen bonding, which may be the driving force for the aggregation of rods. The MEP plot generated for methanol as solvent for compound 3 indicated the possibility of the formation of intermolecular hydrogen bond interaction between the nitrogen atom of the cyclohexyl ring of the GABA derivative and the hydroxyl hydrogen atom of methanol as the bond distance between these two atoms which was observed to be 1.989 Å, suitably lies in the hydrogen bond interaction range [Fig. 2(c)]. Further, we observed the formation of well-defined nanorods of GABA derivative 3 in FE-SEM [Fig. 3(a)] and confocal microscopic images [Fig. 3(b)] when acetonitrile was used as the solvent system. In the MEP plot, the electrostatic interaction between the amino nitrogen of GABA derivative 3 and one of the methyl hydrogens of acetonitrile was observed to be 2.349 Å [Fig. 3(c)]. As a representative of aromatic solvents, toluene was used as a solvent to further understand the self assembly of compound 3 which resulted in the formation of sheet-like structures as depicted by FE-SEM [Fig. 4(a)] and confocal microscopic images [Fig. 4(b)]. The MEP plot confirmed interaction between the t-Boc carbonyl oxygen of the GABA derivative and methyl hydrogen of toluene with a bond distance of 2.574 Å which indicates a van der Waals type of interaction [Fig. 4(c)]. To further confirm the secondary structure of GABA derivative 3, Fourier-transform infra-red (FT-IR) spectroscopy was used. The FT-IR spectra obtained for compound 3 in different solvents generally exhibited peaks at 3300–3400 cm−1 and 1800–1500 cm−1 corresponding to the N–H stretching vibrations and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations, respectively. In addition, the peaks at 1400–1500 cm−1 corresponded to –CH3 bending vibrations (ESI,† Fig. S4–S8 and Table S1).
O stretching vibrations, respectively. In addition, the peaks at 1400–1500 cm−1 corresponded to –CH3 bending vibrations (ESI,† Fig. S4–S8 and Table S1).
Effect of concentration
The influence of concentration on the morphology of the self assembly patterns the nano/microstructures obtained was further examined by varying the concentration of the solution. Out of the solvents used as discussed in the last section, DMSO was chosen for this study, as rod clusters were formed in DMSO at 10 mM concentration. The formation of rod-like structures, though embedded together, seems to have initiated at a relatively low concentration of 1 mM with the length of these rods observed in the nanometer range [Fig. 5(a)]. As the concentration was increased to 5 mM, it was observed that distinct structures with morphology similar to plate- and rod-like structures begin to form although the overall structure is still not uniform over the analysed surface [Fig. 5(b)]. With a further increase in concentration to 10 mM, the rod clusters appear to form with the length ranging in few microns. As discussed earlier, the driving force for the formation of such structures may be the hydrogen bonding interaction between GABA derivative 3 and DMSO [Fig. 5(c)]. Further, it was observed that these rod clusters appear to be aggregated at a higher concentration (15 mM) leading to larger sized non-uniform structures [Fig. 5(d)]. Hence, it can be stated from these observations that the variation in concentration plays an important role in manipulating the self assembled patterns of GABA derivative 3 leading to the change in structural morphology at different concentrations.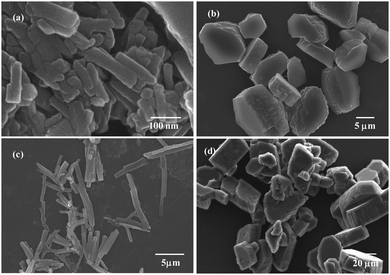 | ||
| Fig. 5 FE-SEM images of DMSO solutions of GABA derivative 3 at concentrations: 1 mM (a) 5 mM (b) 10 mM (c) and 15 mM (d). | ||
Effect of temperature
As documented earlier, temperature variation leads to the manipulation of the self assembly patterns. Based on this, experiments were carried out in vacuo by varying the temperature conditions in order to understand the effect of this variation on the self assembly of compound 3. Initially, as reported earlier, rod clusters were obtained at 25 °C for a 10 mM solution of GABA derivative 3 in DMSO [Fig. 6(a)]. As the temperature of the solution was increased to 50 °C, rod-like structures seem to form solid plate-like structures [Fig. 6(b)]. This change in morphology can be visualised from the increased size of these plate structures at 50 °C as compared to the distinct rod-like morphology at 25 °C. With the increase in temperature to 100 °C and further to 150 °C, the self assembled structures appear to be aggregated to a larger extent forming large sized aggregated rods as well as plates [Fig. 6(c) and (d)]. Thus, it can be safely interpreted that the observed change in morphology resulting from the increasing temperature is mainly due to diminishing interactions between the studied compound and solvent. This observation also re-emphasized the role of solvent and the optimum temperature conditions required to manipulate the morphology of the self assembly of the chemical moiety in question. In other words, it can be stated that GABA derivative 3 forms rod clusters at room temperature in DMSO and these rods tend to aggregate as the temperature of the initial solution is further increased up to 150 °C.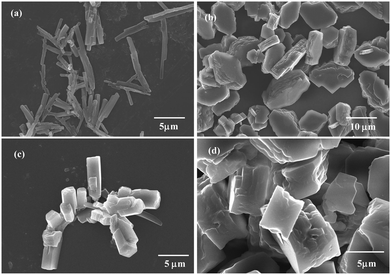 | ||
| Fig. 6 FE-SEM images of GABA derivative 3 obtained after thermal treatment for 1 h in a vacuum oven at a constant temperature of 25 °C (a), 50 °C (b), 100 °C (c), and 150 °C (d). | ||
Conclusions
Aside from lithography and e-beam lithography techniques, self assembly provides a promising route to fabricate nanostructures like nanowires, nanotubes, nanorods and other hierarchical nanostructures. Herein, we have successfully demonstrated that GABA derivative 3 undergoes self assembly into a variety of structures ranging from well-defined rods to sheets, whose morphology is dependent on the solvent, concentration and temperature. The structural elucidation using computational methods revealed the presence of intermolecular hydrogen bonding, van der Waals interactions and electrostatic interactions with solvent molecules in the self assembly process. In DMSO, clusters of rods were formed, while these clusters were observed to have aggregated in methanol. In acetonitrile, well-defined rods were obtained, whereas non polar solvents such as toluene resulted in the formation of sheets. It was also observed that the variation in concentration and temperature plays a significant role in the organisation of molecules in a well-defined manner. These rod-shaped biocompatible templates can be fine tuned and tailored for casting and fabrication of functional materials. For instance, rod-shaped viruses (M13 bacteriophage and tobacco mosaic virus) have been used as templates to fabricate a variety of nanostructures.29 Such a template design can be achieved from the small molecule based hierarchical self assembly. The morphological variety of template designs derived from the applied conditions can be used to fabricate various nano/micro scaled magnetic and semiconductor nanomaterials. Overall, the results are immensely important in gaining significant new insights into the fabrication of variable architectures from simple self assembling organic molecules and could pave the way for a better understanding of the self assembly process at the molecular level.Materials and methods
Materials
All the starting materials were purchased from Sigma-Aldrich and Avra Chemicals and were used as such. All the solvents used for the self assembly were of analytical grade having >99% purity. Analytical thin-layer chromatography (TLC) was performed using aluminum plates precoated with silica gel (0.25 mm, 60 A pore-size) impregnated with a fluorescent indicator (254 nm). Visualization on TLC was achieved by the use of UV light (254 nm), and staining with iodine vapors. Proton nuclear magnetic resonance spectra (1H NMR) were recorded on a Bruker AVANCE III 400 (400 MHz). Proton chemical shifts are expressed in parts per million (ppm, δ scale). The following abbreviations were used to describe peak patterns where appropriate: br = broad, s = singlet, d = doublet, t = triplet, q = quadruplet, m = multiplet, and coupling constants, J, were reported in units of Hertz (Hz). The formation of the compound was confirmed by electron spray ionization mass spectrometry (ESI-MS) and m/z peaks were recorded. A Thermo Scientific Nicolet iS50 FT-IR spectrophotometer was employed to collect infrared spectra (4000–500 cm−1) of the samples in the attenuated total reflection mode.Synthesis and characterization of Boc-4-aminobutyric acid dicyclohexylurea [tert-butyl-4-(1,3-dicyclohexylureido-4-oxobutyl)carbamate]
To a solution of 4-aminobutyric acid (1) (4.85 mmol) in 1,4-dioxane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (3
water (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), sodium hydroxide (4 N) solution was added to adjust the pH to between 10 and 11. The reaction mixture was then stirred for 15 minutes after the addition of the first fraction of di-tert-butyl dicarbonate. Again, the solution was made alkaline by the addition of 4 N NaOH. The reaction mixture was further stirred for 24 hours after the addition of the second fraction of di-tert-butyl dicarbonate. Then, the reaction mixture was concentrated completely, dissolved in methanol and stirred overnight. Further, it was concentrated completely followed by the minimum addition of water (pH = 11–12). Then, to the reaction mixture, a saturated solution of KHSO4 was added to adjust the pH to 3. Further, the reaction mixture was concentrated completely followed by addition of t-butanol and filtration. The filtrate was collected and concentrated in vacuo to obtain a viscous product with 90% yield (2). To a solution of Boc-4-aminobutyric acid (2) (3.54 mmol) in dichloromethane (DCM, 15 ml), dicyclohexylcarbodimide (DCC, 7.23 mmol) was added and the mixture was stirred for 48 h at room temperature. The reaction mixture was further extracted with ethylacetate (50 ml) and dicyclohexylurea (DCU) was filtered off. The organic layer was washed with 1 M sodium carbonate (3 × 50 ml) and brine (2 × 50 ml), dried over anhydrous sodium sulfate and evaporated in vacuo followed by purification using column chromatography to obtain a solid product (3) in 62% yield.
2), sodium hydroxide (4 N) solution was added to adjust the pH to between 10 and 11. The reaction mixture was then stirred for 15 minutes after the addition of the first fraction of di-tert-butyl dicarbonate. Again, the solution was made alkaline by the addition of 4 N NaOH. The reaction mixture was further stirred for 24 hours after the addition of the second fraction of di-tert-butyl dicarbonate. Then, the reaction mixture was concentrated completely, dissolved in methanol and stirred overnight. Further, it was concentrated completely followed by the minimum addition of water (pH = 11–12). Then, to the reaction mixture, a saturated solution of KHSO4 was added to adjust the pH to 3. Further, the reaction mixture was concentrated completely followed by addition of t-butanol and filtration. The filtrate was collected and concentrated in vacuo to obtain a viscous product with 90% yield (2). To a solution of Boc-4-aminobutyric acid (2) (3.54 mmol) in dichloromethane (DCM, 15 ml), dicyclohexylcarbodimide (DCC, 7.23 mmol) was added and the mixture was stirred for 48 h at room temperature. The reaction mixture was further extracted with ethylacetate (50 ml) and dicyclohexylurea (DCU) was filtered off. The organic layer was washed with 1 M sodium carbonate (3 × 50 ml) and brine (2 × 50 ml), dried over anhydrous sodium sulfate and evaporated in vacuo followed by purification using column chromatography to obtain a solid product (3) in 62% yield.
1H NMR (400 MHz, CDCl3) δ: 1.40–1.42 (s, 9H, –C(CH3)3), 1.08–1.30 (m, 11H, cyclohexyl), 1.60–1.80 (m, 11H, cyclohexyl), 2.37 (t, 2H, –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2–CH2–), 2.29–2.31 (t, 2H, –C
2–CH2–), 2.29–2.31 (t, 2H, –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CO–), 3.07–3.10 (m, 2H, –NH–C
2CO–), 3.07–3.10 (m, 2H, –NH–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2–), 3.60–3.69 (m, 1H, cyclohexyl, –C
2–), 3.60–3.69 (m, 1H, cyclohexyl, –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –NH–).
–NH–).
General procedure for self assembly
A 10 mM solution of compound 3 was prepared in 1 ml of solvent used (DMSO, acetonitrile, methanol and toluene). For concentration studies, 1 mM, 5 mM, 10 mM and 15 mM of compound 3 were dissolved in 1 ml of DMSO separately. Similarly, to understand the effect of temperature, a 10 mM solution of compound 3 was dissolved in 1 ml of DMSO and then placed in a vacuum oven for 1 hour at 50 °C, 100 °C and 150 °C separately.Field emission scanning electron microscopic (FE-SEM) analysis
Field emission scanning electron microscopy (FE-SEM) was used to investigate the morphology of the self assembled structures. In general, freshly prepared solutions of GABA derivative 3 (10 mM) were first sonicated and then taken on glass cover slips and evaporated. A platinum coating was applied on the top of the samples to make them conductive for analysis followed by FE-SEM observation using a Hitachi, SU8010 electron microscope, operating at 10–15 kV.Confocal experimentation
Fresh solutions of GABA derivative 3 were prepared and confocal images were recorded using a confocal laser scanning microscope (Zeiss, LSM 510).Computational studies
All quantum chemical calculations pertaining to compound 3 were performed by applying the HF method, with the 3-21G basis set using Gaussian 03 software (Gaussian 03; Gaussian Inc.: Wallingford, CT, USA, 2004). The geometry was optimized by minimizing energies with respect to all geometrical parameters without imposing any molecular symmetry constraints. Frequency calculations showed the absence of any imaginary frequency modes which confirmed that the optimised structure is an energy minimum.X-ray diffraction studies
X-ray diffraction (XRD) patterns were obtained by using Cu-Kα radiation where nickel metal was used as a beta filter, equipped with an X'celerator solid-state detector. Powdered samples were placed on a sample holder and data were collected for 2θ values between 0 and 65°.Acknowledgements
This work was supported by the Science Engineering & Research Board (SERB) of India (grant no. SB/SO/BB/0040/2013). SKB thanks the INSPIRE, Department of Science and Technology (DST), India for her research fellowship. PS and MB thank University Grants Commission (UGC), India for their research fellowships.Notes and references
- R. V. Georgakalis, F. Pellarini, M. Prato, D. M. Guldi, M. M. Frenco and F. Zerbetto, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5075–5080 CrossRef PubMed.
- A. Lomander, W. Hwang and S. Zhang, Nano Lett., 2005, 5, 1255–1260 CrossRef CAS PubMed.
- D. Missirlis, A. Chworos, C. J. Fu, H. A. Khant, D. V. Krogstad and M. Tirrell, Langmuir, 2011, 27, 6163–6170 CrossRef CAS PubMed.
- F. Taraballi, M. Campione, A. Sassella, A. Veseovi, A. Paleari, W. Hwang and F. Gelain, Soft Matter, 2009, 5, 660–668 RSC.
- G. Demirel and F. Buyukserin, Langmuir, 2011, 27, 12533–12538 CrossRef CAS PubMed.
- S. Zhang, Nat. Biotechnol., 2003, 21, 1171–1178 CrossRef CAS PubMed.
- R. Roytman, L. A. Abramovich, K. S. A. Kumar, T. C. Kuan, C. C. Lin, E. Gazit and A. Brik, Org. Biomol. Chem., 2011, 9, 5755–5761 CAS.
- C. M. Moyon, V. Venkatesh, K. V. Krishna, F. Bonachera, S. Verma and A. Bianco, Chem. – Eur. J., 2015, 21, 11681–11686 CrossRef PubMed.
- P. Koley, M. Sakurai and M. Aono, J. Mater. Sci., 2015, 50, 3139–3148 CrossRef CAS.
- P. Koley and A. Pramanik, J. Mater. Sci., 2014, 49, 2000–2012 CrossRef CAS.
- R. J. Brea, C. Reiriz and J. R. Granja, Chem. Soc. Rev., 2010, 39, 1448–1456 RSC.
- A. R. Hirst, B. Huang, V. Castelletto, I. W. Hamley and D. K. Smith, Chem. – Eur. J., 2007, 13, 2180–2188 CrossRef CAS PubMed.
- S. Cavalli, F. Albericio and A. Keos, Chem. Soc. Rev., 2010, 39, 241–263 RSC.
- P. Tamamis, L. A. Abramovich, M. Reches, K. Marshall, P. Sikorski, L. Serpell, E. Gazit and G. Archontis, Biophys. J., 2009, 96, 5020–5029 CrossRef CAS PubMed.
- M. Reches and E. Gazit, Nano Lett., 2004, 4, 581–585 CrossRef CAS.
- P. Koley and A. Pramanik, Adv. Funct. Mater., 2011, 21, 4126–4128 CrossRef CAS.
- X. Lu, X. Wang, Q. Liao and H. Fu, J. Phys. Chem. C, 2015, 119, 22108–22113 CAS.
- S. T. Stupp and L. C. Palmer, Chem. Mater., 2014, 26, 507–518 CrossRef CAS.
- A. F. Eftaiha, A. D. Hendsbee, J. P. Sun and I. G. Hill, Mater. Lett., 2015, 157, 252–255 CrossRef CAS.
- S. Rosenne, E. Grinvald, E. Shirman, L. Neeman, S. Dutta, O. B. Eli, R. B. Zvi, E. Oksenberg, P. Milko, V. Kalchenko, H. Weissman, D. Oron and B. Rybtchinski, Nano Lett., 2015, 15, 7232–7237 CrossRef CAS PubMed.
- A. Bogomolova, S. Keller, J. Klingler, M. Sedlak, D. Rak, A. Sturcova, M. Hruby, P. Stepanek and S. K. Filippov, Langmuir, 2014, 30, 11307–11308 CrossRef CAS PubMed.
- S. Parween, B. Chandanshive, H. C. Kotamarthi and D. Khushalani, Soft Matter, 2013, 9, 10141–10145 RSC.
- S. Kar, B. H. Huang, K. W. Wu, C. R. Lee and Y. Tai, Soft Matter, 2014, 10, 8075–8082 RSC.
- S. Parween, A. Mishra, S. Ramakumar and V. S. Chauhan, J. Mater. Chem., 2014, 2, 3096–3106 RSC.
- R. W. Olsen, Neurochem. Res., 2014, 39, 1924–1941 CrossRef CAS PubMed.
- D. L. Crittenden, M. Chebib and M. J. T. Jordan, J. Phys. Chem. A, 2005, 109, 4195–4201 CrossRef CAS PubMed.
- H. Cui, M. J. Webber and S. I. Stupp, Biopolymers, 2010, 94, 1–18 CrossRef CAS PubMed.
- P. Kumaraswamy, R. Lakshmanan, S. Sethuraman and U. M. Krishnan, Soft Matter, 2011, 7, 2744–2754 RSC.
- C. E. Flynn, S. W. Lee, B. R. Peelle and A. M. Belcher, Acta Mater., 2003, 51, 5867–5880 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: 1H NMR and mass spectra, X-ray diffraction pattern, and FTIR spectra. See DOI: 10.1039/c6qm00007j |
| This journal is © the Partner Organisations 2017 |