Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Differences in cisplatin distribution in sensitive and resistant ovarian cancer cells: a TEM/NanoSIMS study
Ronald F. S.
Lee
a,
Tina
Riedel
a,
Stéphane
Escrig
b,
Catherine
Maclachlan
c,
Graham W.
Knott
c,
Curt A.
Davey
de,
Kai
Johnsson
a,
Anders
Meibom
*bf and
Paul J.
Dyson
 *a
*a
aInstitute of Chemical Sciences and Engineering, Ecole Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland. E-mail: paul.dyson@epfl.ch; Tel: +41 21 693 98 54
bLaboratory for Biological Geochemistry, Ecole Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland. E-mail: anders.meibom@epfl.ch; Tel: +41 21 693 80 15
cInterdisciplinary Centre for Electron Microscopy, Ecole Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland
dSchool of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551
eNTU Institute of Structural Biology, Nanyang Technological University, 59 Nanyang Drive, Singapore 636921
fCenter for Advanced Surface Analysis, Institute of Earth Sciences, University of Lausanne, CH-1015, Lausanne, Switzerland
First published on 7th September 2017
Abstract
Cisplatin is a widely used anti-cancer drug, but its effect is often limited by acquired resistance to the compound during treatment. Here, we use a combination of transmission electron microscopy (TEM) and nanoscale-secondary ion mass spectrometry (NanoSIMS) to reveal differences between cisplatin uptake in human ovarian cancers cells, which are known to be susceptible to acquired resistance to cisplatin. Both cisplatin sensitive and resistant cell lines were studied, revealing markedly less cisplatin in the resistant cell line. In cisplatin sensitive cells, Pt was seen to distribute diffusely in the cells with hotspots in the nucleolus, mitochondria, and autophagosomes. Inductively coupled plasma mass spectrometry (ICP-MS) was used to validate the NanoSIMS results.
Significance to metallomicsWe studied the subcellular distribution of cisplatin in cancer cell lines sensitive and resistant to the drug using a combination of TEM/NanoSIMS. The study provides insights into the distribution of cisplatin in cancer cells, with implications for future design of metallodrugs. Our approach provides information on subcellular localization, cellular chemical environment and potentially metal ligand states from a single experimental run and could be applied to the study of other biological problems related to metallodrugs. |
Introduction
The anti-cancer properties of cisdiamineplatinum(II), better known as cisplatin, were discovered by Barnett Rosenberg in 1969.1 Revolutionizing the treatment of testicular cancers as cure rates soared from 10 to over 95%,2 cisplatin is used to treat many other types of cancers, notably ovarian, cervical, lung, bladder, head and neck cancers.3 Since its discovery, two other platinum(II) based drugs, carboplatin and oxaliplatin, have been approved for worldwide use, and others such as nedaplatin, heptaplatin and lobaplatin have been approved in Asia.4 All platinum(II) drugs are prodrugs which are thought to function via a series of steps that include cellular uptake, aquation in low chloride intracellular environments, followed by DNA binding, which induces DNA damage and lead to apoptosis.5One major challenge with the use of cisplatin is the development of resistance leading to chemotherapy failure and cancer recurrence. This resistance is thought to be mediated by a plethora of factors.6 Firstly, under-expression of membrane transporters7 or overexpression of drug efflux pumps8 contribute to reduced levels of Pt accumulation in the cell. Differential expression of proteins involved in protection against or susceptibility towards DNA damage such as heat shock9 and ribosomal proteins10 are also implicated. In addition, epigenetic changes such as in DNA methylation,11 structural changes in chromatin12,13 and histone modifications14 can confer resistance. Finally, involvement of different transcription factors15 and post-transcriptional gene regulating microRNA16 in cisplatin resistance has also been reported.
Developing strategies for overcoming resistance to cisplatin and related drugs is challenging due to the complexity of the resistance response. Thus, it is important to understand which factors contribute most to resistance in order to find ways to overcome the problem. Imaging of the distribution of cisplatin in cells could provide insights into the resistance mechanisms as some aspects of platinum resistance might be localized to certain cellular organelles, or reveal themselves as changes to intracellular concentration or distribution of the drug. However, imaging of metallodrugs is challenging as these compounds contain labile ligands and imaging modalities should allow the parent compound to be visualized with minimal modifications. In this respect, nano-scale secondary ion mass spectrometry (NanoSIMS) has emerged as a powerful tool to elucidate the distribution of metallodrugs. NanoSIMS allows high spatial resolution imaging (∼100 nm) of the metal centre and when used in tandem with isotopic labelling, provides valuable information on the ligands surrounding the metal centre.17,18
In recent years, NanoSIMS has been used to study cellular distribution of metallodrugs based on Au,19 Ru17 and Pt,20–22 with the cellular distribution of cisplatin being reported in colon cancer cells. Despite the high spatial resolutions of this technique, as far as we are aware no comparative study has been done to analyse with high spatial resolution differences in the distribution of Pt between cancer cells that are sensitive and resistant to the drug. Here, we explore the distribution of cisplatin in cisplatin sensitive (A2780) and cisplatin resistant (A2780CR) human ovarian cancer cells with NanoSIMS imaging. We show that there is a diffuse distribution of platinum observed in cisplatin sensitive cells with hotspots in the autophagosomes, mitochondria, nucleolus and in structures containing sulfur, and that there is a markedly lower accumulation of cisplatin in A2780CR cells.
Methods
Cell culture and preparation
Human ovarian carcinoma A2780 cisplatin sensitive and resistant [A2780 and A2780CR] cells (ATCC) were cultured in RPMI 1640 Glutamax medium (Invitrogen) supplemented with 10% fetal calf serum, penicillin 100 units per mL streptomycin 100 μg mL−1 (Invitrogen). Cells were incubated at 37 °C in a moist environment containing 5% CO2.Cell preparation for Nano-SIMS analysis
Cells were seeded at a density of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 24-well or 500
000 cells per well in 24-well or 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 6-well clear bottom plates fitted with 13 mm thermanox slips or sapphire disks. After 24 hours, cell media was aspirated and fresh media containing cisplatin (Tokyo Chemical Industries) 30 μM was added. Upon a 12 hour incubation, sapphire disks were removed from media and then high pressure frozen (Leica HPM100, Leica Microsystems), with excess 20% BSA solution in 0.01 M PBS (phosphate buffer solution) to avoid any air bubbles becoming trapped and the formation of ice crystals. The frozen cells were then embedded in resin at low temperature.23 Sapphire discs were placed on a frozen solution of 1% osmium, 0.5% uranyl acetate, 5% water in pure acetone. The samples were then warmed to room temperature in an ice bucket containing solid carbon dioxide blocks that was allowed to sublime over a period of 2 hours until room temperature was reached. At this point the solution was removed and replaced with dry acetone. After washing a further 2 times with acetone the samples were embedded in increasing concentrations of epoxy resin in acetone. At 100% concentration of resin the samples were then left overnight to fully infiltrate and then polymerised in a 60 °C oven for at least 12 hours. Samples were then glued to empty resin blocks, trimmed, and sections of alternating thickness of 500 nm and 50 nm cut sequentially from the face. The thicker sections were collected onto a glass coverslip stained with 1% toluidine blue and imaged with light microscopy, and NanoSIMS, and the 50 nm thick sections collected on to an electron microscopy slot grid ready for imaging with transmission electron microscopy at a final magnification of 1400 times (Tecnai Spirit, FEI Company, Netherlands).
000 cells per well in 6-well clear bottom plates fitted with 13 mm thermanox slips or sapphire disks. After 24 hours, cell media was aspirated and fresh media containing cisplatin (Tokyo Chemical Industries) 30 μM was added. Upon a 12 hour incubation, sapphire disks were removed from media and then high pressure frozen (Leica HPM100, Leica Microsystems), with excess 20% BSA solution in 0.01 M PBS (phosphate buffer solution) to avoid any air bubbles becoming trapped and the formation of ice crystals. The frozen cells were then embedded in resin at low temperature.23 Sapphire discs were placed on a frozen solution of 1% osmium, 0.5% uranyl acetate, 5% water in pure acetone. The samples were then warmed to room temperature in an ice bucket containing solid carbon dioxide blocks that was allowed to sublime over a period of 2 hours until room temperature was reached. At this point the solution was removed and replaced with dry acetone. After washing a further 2 times with acetone the samples were embedded in increasing concentrations of epoxy resin in acetone. At 100% concentration of resin the samples were then left overnight to fully infiltrate and then polymerised in a 60 °C oven for at least 12 hours. Samples were then glued to empty resin blocks, trimmed, and sections of alternating thickness of 500 nm and 50 nm cut sequentially from the face. The thicker sections were collected onto a glass coverslip stained with 1% toluidine blue and imaged with light microscopy, and NanoSIMS, and the 50 nm thick sections collected on to an electron microscopy slot grid ready for imaging with transmission electron microscopy at a final magnification of 1400 times (Tecnai Spirit, FEI Company, Netherlands).
Nano-SIMS analysis
NanoSIMS measurements were performed at the Laboratory of Biological Geochemistry, EPFL and the University of Lausanne. Prior to NanoSIMS imaging, the samples were gold-coated in order to avoid charging effects. Before acquiring an image, Cs+ ions were implanted into the surface of the sample in order to enhance the ionization of the element of interests. In our study, the electron multiplier detectors were set up to measure 12C2−, 12C14N−, 31P−, 34S−, and 194Pt− secondary ions, generated by bombarding the sample with a ∼4 pA Cs+ primary beam focused to a spot size of approximately 160 nm. In order to resolve possible isobaric interferences, the instrument was operated at a mass-resolving power (MRP) of about 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. For 194Pt−, due to the very low signal obtained on cells, peak-shape and mass resolving power was checked using a Pt-metal standard. Data acquisition was performed by scanning the Cs+ primary beam over areas of 34 × 34 μm with a 256 × 256 pixel image resolution. The per pixel dwell time of the primary ion beam was 10 ms. The final images are the accumulation of 120 layers obtained by sequential scanning and correspond to a cumulated acquisition time per pixel of 1.2 seconds. Between every layer, the focusing of the secondary ion beam was optimized and automatic peak centering was performed for 12C2− and 12C14N−. The Pt peak could not be centered due to the low count rates. However, post-analysis check revealed that there was no significant change in the peaks position during the acquisition time. The total acquisition time including the centering procedure was 22 h per image.
000. For 194Pt−, due to the very low signal obtained on cells, peak-shape and mass resolving power was checked using a Pt-metal standard. Data acquisition was performed by scanning the Cs+ primary beam over areas of 34 × 34 μm with a 256 × 256 pixel image resolution. The per pixel dwell time of the primary ion beam was 10 ms. The final images are the accumulation of 120 layers obtained by sequential scanning and correspond to a cumulated acquisition time per pixel of 1.2 seconds. Between every layer, the focusing of the secondary ion beam was optimized and automatic peak centering was performed for 12C2− and 12C14N−. The Pt peak could not be centered due to the low count rates. However, post-analysis check revealed that there was no significant change in the peaks position during the acquisition time. The total acquisition time including the centering procedure was 22 h per image.
Data extraction and image processing
Nano-SIMS image processing was performed using L'image (L. Nittler, Carnegie Institution of Washington). Over the ∼20 hours of image acquisition, the image drift of a 34 × 34 μm image was less than 7 pixels (i.e. less than 1 μm). The data reduction software can easily correct for such a drift by aligning the position of identified structures. Regions of interest (ROI's) were defined manually based on identifiable cell features on 31P− elemental maps. Images were accumulated from planes where accumulated counts per ROI were stable with 12C14N− used as the alignment mass. Raw platinum counts of different A2780 and A2780CR cells are shown in Table 1. All elements were normalized against 12C2, the images of which are essentially flat, normalizing small ionization variations across the sample surface. All graphs were plotted using Microsoft Excel 2013.| Cell | Cellular compartment | Total Pt counts |
|---|---|---|
| A | Cytoplasm | 3123 |
| Nucleus | 4023 | |
| Nucleolus | 156 | |
| B | Cytoplasm | 1945 |
| Nucleus | 1748 | |
| Nucleolus | 77 | |
| A′ | Cytoplasm | 1953 |
| Nucleus | 430 | |
| Nucleolus | 79 | |
| B′ | Cytoplasm | 1080 |
| Nucleus | 822 | |
| Nucleolus | 112 | |
| C′ | Cytoplasm | 1188 |
| Nucleus | 511 | |
| Nucleolus | N/A | |
Cell fractionation for ICP-MS analysis
A2780 and A2780CR were seeded at a concentration of 1 × 106 in 13 mL of media and allowed to grow to 70% confluency for 3 days. The media was exchanged by media containing cisplatin (30 μM) and the cells were incubated for 12 h. After internalization, cells were washed 3× with PBS and harvested using enzyme free cell dissociation buffer (Merck). The number of cells were estimated using a hemocytometer. Mitochondria were isolated using a mitochondria isolation kit for cultured cells (Thermo Scientific). Reagent A supplemented with protease inhibitor was added followed by incubation on ice for 2 min. Reagent B was added and incubated on ice for 5 min with vortexing every min. Then, reagent C was added and the cells were centrifuged at 700 × g for 10 min at 4 °C. The supernatant yielding the cytosolic and mitochondrial fractions were separated from the pellet containing nuclei and cellular debris. To obtain a more purified fraction of mitochondria with less lysosomal and peroxisomal contaminants, the supernatant was centrifuged at 3000 × g for 15 min. The pellet yielded the isolated mitochondria which were further purified by washing with reagent C and centrifuging at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C for 5 min. Nuclei were isolated using a nuclei isolation EZ prep kit (Sigma Aldrich). Cells were lysed by incubating with 1 mL of nuclei EZ lysis buffer on ice for 5 min and collected by centrifugation at 500 × g at 4 °C for 5 min. The crude nuclei pellet was washed in 1 mL of ice cold nuclei EZ lysis buffer and again collected by centrifugation. Nucleosomes and nucleoli were isolated using a Pierce Chromatin Prep Module (Thermo Scientific). Harvested cells were crosslinked with 1% formaldehyde in PBS (10 mL). After incubation at room temperature for 10 min the reaction was quenched by adding 1 mL of glycine solution (10×) and incubated for another 5 min. The cells were centrifuged at 3000 × g for 5 min at 4 °C and washed 2× with ice cold PBS supplemented with protease inhibitor. The crosslinked cells were lysed by incubating with Lysis Buffer 1 (100 μL) for 10 min and centrifuged at 9000 × g for 3 min. The supernatant yielding the cytosolic fraction was separated from the pellet containing nuclei and cell debris and kept for further ICP-MS analysis. The crude nuclei fraction was digested with micrococcal nuclease (25 U) in a 37 °C water bath for 15 min, mixing by inversion every 5 min. After incubating with 10 μL MNase stop solution for 5 min the nuclei were recovered by centrifugation at 9000 × g for 5 min. The nuclei were resuspended in Lysis Buffer 2 (50 μL), incubated on ice for 15 min, with vortexing for 15 s every 5 min, and centrifuged at 9000 × g for 5 min. The supernatant yielding the nucleosome fraction was separated from the pellet containing insoluble nuclear matrix proteins, nuclear membrane and the nucleoli fraction. The pellet was washed 2× with 0.35 M sucrose containing 0.5 mM MgCl2 and the purified nuclear membrane and nucleoli fraction was collected by centrifugation at 3500 × g for 5 min. The cells for total cell uptake and cellular fractions were digested with 69% HNO3 at room temperature for 24 hours and the Pt concentration was determined using ICP-MS. All samples were prepared in triplicate.
000 × g at 4 °C for 5 min. Nuclei were isolated using a nuclei isolation EZ prep kit (Sigma Aldrich). Cells were lysed by incubating with 1 mL of nuclei EZ lysis buffer on ice for 5 min and collected by centrifugation at 500 × g at 4 °C for 5 min. The crude nuclei pellet was washed in 1 mL of ice cold nuclei EZ lysis buffer and again collected by centrifugation. Nucleosomes and nucleoli were isolated using a Pierce Chromatin Prep Module (Thermo Scientific). Harvested cells were crosslinked with 1% formaldehyde in PBS (10 mL). After incubation at room temperature for 10 min the reaction was quenched by adding 1 mL of glycine solution (10×) and incubated for another 5 min. The cells were centrifuged at 3000 × g for 5 min at 4 °C and washed 2× with ice cold PBS supplemented with protease inhibitor. The crosslinked cells were lysed by incubating with Lysis Buffer 1 (100 μL) for 10 min and centrifuged at 9000 × g for 3 min. The supernatant yielding the cytosolic fraction was separated from the pellet containing nuclei and cell debris and kept for further ICP-MS analysis. The crude nuclei fraction was digested with micrococcal nuclease (25 U) in a 37 °C water bath for 15 min, mixing by inversion every 5 min. After incubating with 10 μL MNase stop solution for 5 min the nuclei were recovered by centrifugation at 9000 × g for 5 min. The nuclei were resuspended in Lysis Buffer 2 (50 μL), incubated on ice for 15 min, with vortexing for 15 s every 5 min, and centrifuged at 9000 × g for 5 min. The supernatant yielding the nucleosome fraction was separated from the pellet containing insoluble nuclear matrix proteins, nuclear membrane and the nucleoli fraction. The pellet was washed 2× with 0.35 M sucrose containing 0.5 mM MgCl2 and the purified nuclear membrane and nucleoli fraction was collected by centrifugation at 3500 × g for 5 min. The cells for total cell uptake and cellular fractions were digested with 69% HNO3 at room temperature for 24 hours and the Pt concentration was determined using ICP-MS. All samples were prepared in triplicate.
ICP-MS analysis
Platinum concentrations were measured on an ICP-MS instrument (Elan DRC II, Perkin Elmer) equipped with a Meinhard nebulizer and a cyclonic spray chamber. The ICP-MS instrument was tuned daily using a solution provided by the manufacturer containing 1 ppb each of Mg, In, Ce, Ba, Pb and U. 115In was used as an internal standard at a concentration of 1 ppb. External standards (ranging from 0.05 ppb to 20 ppb) were prepared gravimetrically in an identical matrix to the samples with single element standards (CPI International). Three repetitive measurements were performed for each sample.Results and discussion
Cell preparation samples prepared for NanoSIMS should be topographically flat, conducting, and able to resist high vacuum conditions.18 Thus, cell sample preparation for NanoSIMS usually involves dehydration of the sample followed by resin embedding before sectioning for NanoSIMS and other complementary analysis. Fixation can be achieved via either chemical or cryo-fixation methods and cryo-fixation via high pressure freezing and freeze substitution has been shown to improve sample integrity and intracellular ion composition for SIMS analysis,24,25 hence this method was used.A challenging aspect of metallodrug analysis in NanoSIMS is the inherent low sensitivity for certain transition metal elements, such as Pt, due to low secondary ion yields and the low amounts present in samples of interest when employing pharmacologically relevant drug doses. To overcome this limitation image acquisition times of ∼20 hours per NanoSIMS image are required and therefore semi-thin sections of ca. 500 nm are used to maintain sample integrity and thus keep the target immobile during analysis, during such long NanoSIMS analysis, ion beam sputtering removes <100 nm of the sample surface. However, thick semi-thin sections cannot be imaged with TEM, an otherwise useful complementary approach to NanoSIMS elemental mapping that provides ultrastructural details of cellular organelles, some of which are not unambiguously identifiable from NanoSIMS images alone. In this study, we circumvented this issue by consecutive sectioning of thin sections (∼50 nm) for TEM followed by semi-thin sections (∼500 nm) for NanoSIMS. Correlation between TEM and NanoSIMS images permit all identifiable cellular organelles larger than 1 μm in diameter to be clearly identified in NanoSIMS images.
Differences in cellular accumulation of cisplatin
The most active chemotherapy agents for ovarian cancers are the platinum drugs cisplatin and carboplatin.26 Ovarian tumors that are innately platinum resistant or recur after initial treatment with platinum drugs are associated with a poor prognosis.26 In this study we used the human ovarian cancer cell lines A2780 and A2780CR, the latter being a cisplatin resistant cell line, to understand the effects of such resistance on the cellular fate of the drug. A non-lethal cisplatin dose of 30 μM was applied to adherent cells in culture for 12 hours, followed by TEM and NanoSIMS imaging. The 194Pt maps of A2780 and A2780CR cells (Fig. 1A and B), clearly show the presence of platinum in drug treated cells as opposed to untreated cells (Fig. 1C). From C normalized counts of Pt in cisplatin sensitive and resistant cells there is an approximately 2.3 fold higher amount of Pt in sensitive cells (1.16 × 10−6vs. 5.00 × 10−7 counts respectively, taking a ROI of all cells in the image). From ICP-MS measurements (Table 2), we found a 3-fold higher accumulation of Pt in cisplatin sensitive vs. resistant cell lines correlating well with the NanoSIMS findings. This difference in accumulation is in accordance with various studies comparing cisplatin uptake in resistant and sensitive cell lines.27–30 Overall, this could relate to resistance being mediated by mechanisms of increased efflux and/or impaired uptake of cisplatin.6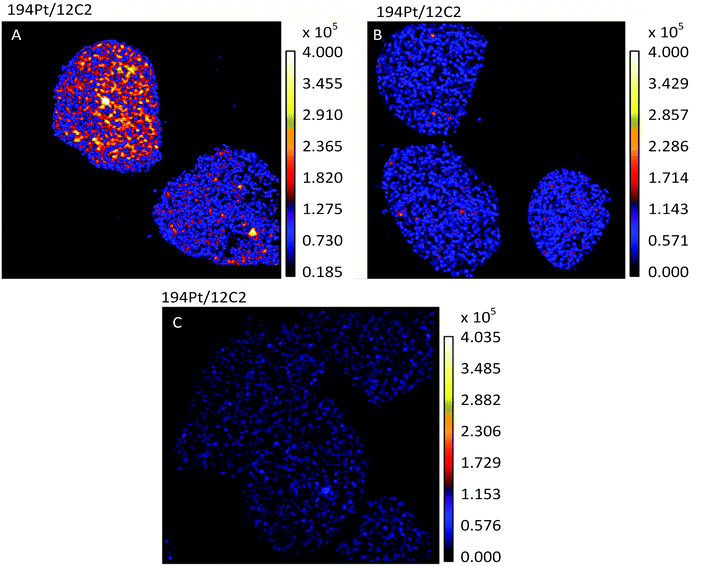 | ||
| Fig. 1 Secondary ion maps of 194Pt−/12C2− of cisplatin (A) sensitive (A2780), (B) resistant (A2780CR) cells treated with cisplatin (30 μM, 12 hours) and (C) untreated A2780 cells. | ||
| A2780 | A2780CR | |
|---|---|---|
| Total cellular uptake, pmol Pt/106 cells | 217 ± 17 | 68 ± 2 |
| Cytosol (% of total) | 29 ± 2 (13 ± 1) | 7.9 ± 0.3 (12 ± 1) |
| Mitochondria (% of total) | 9.2 ± 6.4 (4 ± 3) | 1.3 ± 0.3 (1.9 ± 0.4) |
| Nucleus (% of total) | 119 ± 16 (55 ± 7) | 19 ± 3 (28 ± 4) |
| Nucleolus + nuclear membrane + nuclear matrix (% of total) | 108 ± 15 (50 ± 7) | 14 ± 5 (20 ± 7) |
| Nucleosome (% of total) | 3.4 ± 0.5 (1.5 ± 0.2) | 0.7 ± 0.1 (1.0 ± 0.1) |
Subcellular distribution of cisplatin
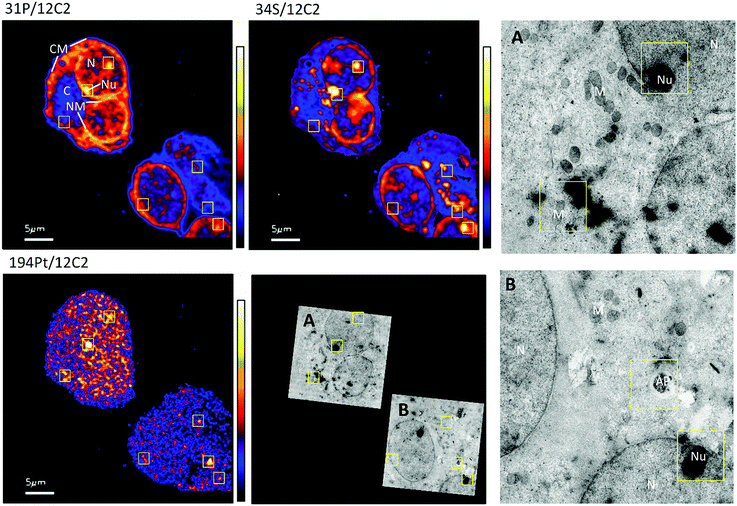
The distribution of platinum in the two cell lines was scrutinized to compare if resistance may be further correlated to intra-cellular distribution differences. NanoSIMS elemental maps of P provide clear visualization of the major cellular compartments, nucleus, nucleolus, cytoplasm and nuclear/cellular membranes (Fig. 2). The TEM images provided additional structural details of smaller organelles such as the mitochondria, lysosomes and autophagosomes (Fig. 2).In A2780 cells, platinum was seen to distribute diffusely throughout the cell with some areas having larger agglomerations of Pt signals. Across the different cellular compartments, around 50% of total Pt detections was located in the cytoplasm, with the remainder in the nucleus and 2% in the nucleolus (Fig. 3). In A2780CR cells there was in general a smaller proportion of Pt in the nucleus as compared to the cytoplasm. ICP-MS analysis revealed a similar distribution pattern in both cell lines with around 50% of Pt in the nucleus and 50% in other compartments in A2780 cells, and 28% of Pt in the nucleus with the remaining fraction in other compartments in the A2780CR cells. Note that during cellular fractionation, cell membrane proteins and other insoluble proteins are removed, thus the amount of Pt detected in the cytosol by ICP-MS may be reduced due to Pt attached to these insoluble materials. ICP-MS analysis also revealed an approximately 5-fold higher amount of Pt in the nucleosome fraction (containing chromatin) of A2780 compared to A2780CR cells (Table 2). The presence of cisplatin in the nucleus and more specifically the binding to the nucleosomes of a cell is consistent with a mechanism of action of cisplatin forming DNA adducts.31 However, in the nucleus of A2780 cells we observed concentrated Pt hotspots in the nucleolus (Fig. 2 and 3), which have also been observed in other studies of cisplatin distribution in cancer cells21,32 The nucleolus is responsible for ribosome synthesis and assembly and consists mainly of proteins and RNA.33 Although DNA is the main target of cisplatin, its high accumulation in the nucleolus also suggests a role of cisplatin in modulating protein production. Indeed, interactions of cisplatin with RNA have been demonstrated, with the drug preferentially blocking transcription of ribosomal RNA in HeLa cells.34 In addition, a crystal structure of cisplatin binding to 70S ribosome showed that it intercalates between the ribosome and mRNA impairing mRNA-translocation resulting in impaired protein synthesis.35
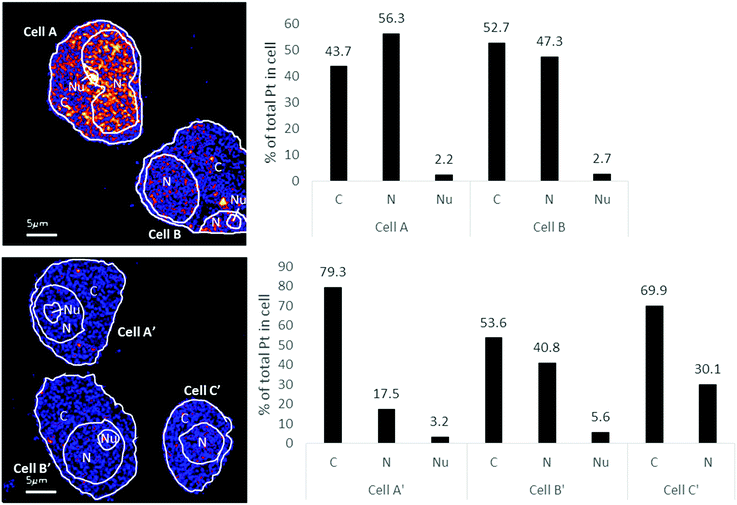 | ||
| Fig. 3 Distribution of Pt in different subcellular compartments in A2780 (top) & A2780CR (bottom) cells treated with cisplatin (30 μM, 12 hours). Regions of interest corresponding to C (cytoplasm), N (nucleus) and Nu (nucleolus) depicted on the 194Pt−/12C2− secondary ion map (left panel). Bar graph depicting percentage of Pt in subcellular compartment of cells (right panel). Data taken from cumulative counts of Pt from NanoSIMS imaging after pre-sputtering had established completely stable secondary ion counts. (Raw cumulative Pt counts in Table 1.) | ||
The diffuse Pt distribution pattern in A2780 cells shows that Pt is available to interact with many cellular organelles and proteins. From the TEM images we found platinum hotspots accumulating in both an autophagosome and in mitochondria of A2780 (Fig. 2). These findings were verified by ICP-MS analysis of the mitochondrial fraction of cisplatin treated A2780 and A2780CR cells confirming the presence of Pt (Table 2). The formation of autophagosomes is important for removing damaged organelles and molecules, which are then degraded by lysosomes.36 A previous study has shown that the formation of autophagosomes is involved in the detoxification of cisplatin,37 which correlates well with our findings. Cisplatin has also been shown to act in the mitochondria either by binding to mitochondrial DNA38,39 or inducing a mitochondrial-reactive oxygen species response,40 which contributes to its cytotoxicity. In the A2780CR cells small pockets of Pt were detected, mostly concentrated along sulfur rich hotspots in the cell (Fig. 4), which could be associated with sulfur containing molecules such as glutathione, metallothioneins and thioredoxins that detoxify metals in cells. Pt co-accumulation with S was similarly observed in A2780 (Fig. 2). Indeed, cisplatin resistant A2780CR cells are known to have higher levels of glutathione41 and metallothionein42 expression than A2780 cells.
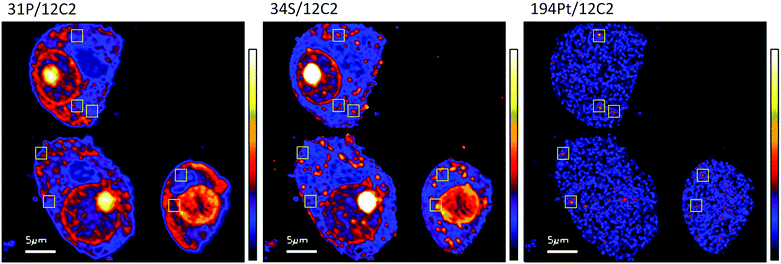 | ||
| Fig. 4 Secondary ion maps of 31P−/12C2−, 34S−/12C2−, 194Pt−/12C2− of A2780CR cells treated with cisplatin (30 μM, 12 hours). Yellow boxes are Pt enriched spots in cells. | ||
Conclusions
TEM and NanoSIMS were used to study the distribution of cisplatin in two types of human ovarian cancer cells that are sensitive and resistant to the drug. We observed a reduced accumulation of Pt in cells with acquired resistance to cisplatin compared to the cisplatin sensitive cells and the results were confirmed by ICP-MS. We also visualized, for the first time, Pt accumulation in mitochondria and autophagosomes, was previously shown following cell fractionation followed by ICP-MS,29 or inferred from phenotype studies.37 The importance of nucleolus-based targets for cisplatin was also highlighted in the sensitive ovarian cancer cells. In this respect, targeting Pt-based compounds to the nucleolus could be an interesting strategy to both determine the role played by targets in the nucleolus in cell death and to obtain more effective Pt-based drugs that are potentially less susceptible to acquired resistance or can be used in a later treatment stage.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the NCCR Chemical Biology, funded by the Swiss National Science Foundation and the Singapore Ministry of Education Academic Research Fund Tier 3 Programme (grant MOE2012-T3-1-001).Notes and references
- B. Rosenberg, L. VanCamp, J. E. Trosko and V. H. Mansour, Nature, 1969, 222, 385–386 CrossRef CAS PubMed.
- SEER Cancer Statistics Review 1975-2008-Previous Version – SEER Cancer Statistics, http://seer.cancer.gov/archive/csr/1975_2008/, (accessed June 13, 2016).
- Cisplatin | MIMS online, http://www.mims.co.uk/drugs/cancer/antineoplastics/cisplatin, (accessed April 4, 2016).
- T. C. Johnstone, K. Suntharalingam and S. J. Lippard, Chem. Rev., 2016, 116, 3436–3486 CrossRef CAS PubMed.
- T. C. Johnstone, K. Suntharalingam and S. J. Lippard, Philos. Trans. R. Soc., A, 2015, 373, 20140185 CrossRef PubMed.
- D.-W. Shen, L. M. Pouliot, M. D. Hall and M. M. Gottesman, Pharmacol. Rev., 2012, 64, 706–721 CrossRef CAS PubMed.
- A. K. Holzer, G. H. Manorek and S. B. Howell, Mol. Pharmacol., 2006, 70, 1390–1394 CrossRef CAS PubMed.
- D.-W. Shen, J. Ma, M. Okabe, G. Zhang, D. Xia and M. M. Gottesman, J. Cell. Physiol., 2010, 225, 822–828 CrossRef CAS PubMed.
- M. Landriscina, G. Laudiero, F. Maddalena, M. R. Amoroso, A. Piscazzi, F. Cozzolino, M. Monti, C. Garbi, A. Fersini, P. Pucci and F. Esposito, Cancer Res., 2010, 70, 6577–6586 CrossRef CAS PubMed.
- D.-W. Shen, X.-J. Liang, T. Suzuki and M. M. Gottesman, Mol. Pharmacol., 2006, 69, 1383–1388 CrossRef CAS PubMed.
- X. Chang, C. L. Monitto, S. Demokan, M. S. Kim, S. S. Chang, X. Zhong, J. A. Califano and D. Sidransky, Cancer Res., 2010, 70, 2870–2879 CrossRef CAS PubMed.
- Q.-E. Wang, C. Han, K. Milum and A. A. Wani, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2011, 708, 59–68 CrossRef CAS PubMed.
- S. V. Sharma, D. Y. Lee, B. Li, M. P. Quinlan, F. Takahashi, S. Maheswaran, U. McDermott, N. Azizian, L. Zou, M. A. Fischbach, K.-K. Wong, K. Brandstetter, B. Wittner, S. Ramaswamy, M. Classon and J. Settleman, Cell, 2010, 141, 69–80 CrossRef CAS PubMed.
- N. Miyamoto, H. Izumi, T. Noguchi, Y. Nakajima, Y. Ohmiya, M. Shiota, A. Kidani, A. Tawara and K. Kohno, J. Biol. Chem., 2008, 283, 18218–18226 CrossRef CAS PubMed.
- G. Hirano, H. Izumi, A. Kidani, Y. Yasuniwa, B. Han, H. Kusaba, K. Akashi, M. Kuwano and K. Kohno, Mol. Cancer Res., 2010, 8, 864–872 CrossRef CAS PubMed.
- R. Hamano, H. Miyata, M. Yamasaki, Y. Kurokawa, J. Hara, J. H. Moon, K. Nakajima, S. Takiguchi, Y. Fujiwara, M. Mori and Y. Doki, Clin. Cancer Res., 2011, 17, 3029–3038 CrossRef CAS PubMed.
- R. F. S. Lee, S. Escrig, M. Croisier, S. Clerc-Rosset, G. W. Knott, A. Meibom, C. A. Davey, K. Johnsson and P. J. Dyson, Chem. Commun., 2015, 51, 16486–16489 RSC.
- P. Hoppe, S. Cohen and A. Meibom, Geostand. Geoanal. Res., 2013, 37, 111–154 CrossRef CAS.
- L. E. Wedlock, M. R. Kilburn, J. B. Cliff, L. Filgueira, M. Saunders and S. J. Berners-Price, Met. Integr. Biometal Sci., 2011, 3, 917–925 RSC.
- L. E. Wedlock, M. R. Kilburn, R. Liu, J. A. Shaw, S. J. Berners-Price and N. P. Farrell, Chem. Commun., 2013, 49, 6944 RSC.
- A. A. Legin, A. Schintlmeister, M. A. Jakupec, M. Galanski, I. Lichtscheidl, M. Wagner and B. K. Keppler, Chem. Sci., 2014, 5, 3135–3143 RSC.
- A. A. Legin, S. Theiner, A. Schintlmeister, S. Reipert, P. Heffeter, M. A. Jakupec, J. Mayr, H. P. Varbanov, C. R. Kowol, M. Galanski, W. Berger, M. Wagner and B. K. Keppler, Chem. Sci., 2016, 7, 3052–3061 RSC.
- K. L. McDonald and R. I. Webb, J. Microsc., 2011, 243, 227–233 CrossRef CAS PubMed.
- K. L. Moore, E. Lombi, F.-J. Zhao and C. R. M. Grovenor, Anal. Bioanal. Chem., 2012, 402, 3263–3273 CrossRef CAS PubMed.
- Q. Jin, T. Paunesku, B. Lai, S.-C. Gleber, S. Chen, L. Finney, D. Vine, S. Vogt, G. Woloschak and C. Jacobsen, J. Microsc., 2017, 265, 81–93 CrossRef CAS PubMed.
- C. W. Helm and J. C. States, J. Ovarian Res., 2009, 2, 2 CrossRef PubMed.
- X.-J. Liang, D.-W. Shen, K. G. Chen, S. M. Wincovitch, S. H. Garfield and M. M. Gottesman, J. Cell. Physiol., 2005, 202, 635–641 CrossRef CAS PubMed.
- K. G. Chen, J. C. Valencia, B. Lai, G. Zhang, J. K. Paterson, F. Rouzaud, W. Berens, S. M. Wincovitch, S. H. Garfield, R. D. Leapman, V. J. Hearing and M. M. Gottesman, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9903–9907 CrossRef CAS PubMed.
- M. Groessl, O. Zava and P. J. Dyson, Metallomics, 2011, 3, 591–599 RSC.
- M. D. Hall, C. T. Dillon, M. Zhang, P. Beale, Z. Cai, B. Lai, A. P. J. Stampfl and T. W. Hambley, JBIC, J. Biol. Inorg. Chem., 2003, 8, 726–732 CrossRef CAS PubMed.
- Z. H. Siddik, Oncogene, 2003, 22, 7265–7279 CrossRef CAS PubMed.
- M. U. Khan and P. J. Sadler, Chem.-Biol. Interact., 1978, 21, 227–232 CrossRef CAS PubMed.
- M. C. Alliegro, S. Hartson and M. A. Alliegro, J. Biol. Chem., 2012, 287, 6702–6713 CrossRef CAS PubMed.
- P. Jordan and M. Carmo-Fonseca, Nucleic Acids Res., 1998, 26, 2831–2836 CrossRef CAS PubMed.
- S. V. Melnikov, D. Söll, T. A. Steitz and Y. S. Polikanov, Nucleic Acids Res., 2016, 44, 4978–4987 CrossRef CAS PubMed.
- E.-L. Eskelinen and P. Saftig, Biochim. Biophys. Acta, 2009, 1793, 664–673 CrossRef CAS PubMed.
- L. Bao, M. C. Jaramillo, Z. Zhang, Y. Zheng, M. Yao, D. D. Zhang and X. Yi, Mol. Med. Rep., 2015, 11, 91–98 CrossRef CAS PubMed.
- J. L. Podratz, A. M. Knight, L. E. Ta, N. P. Staff, J. M. Gass, K. Genelin, A. Schlattau, L. Lathroum and A. J. Windebank, Neurobiol. Dis., 2011, 41, 661–668 CrossRef CAS PubMed.
- Z. Yang, L. M. Schumaker, M. J. Egorin, E. G. Zuhowski, Z. Guo and K. J. Cullen, Clin. Cancer Res., 2006, 12, 5817–5825 CrossRef CAS PubMed.
- R. Marullo, E. Werner, N. Degtyareva, B. Moore, G. Altavilla, S. S. Ramalingam and P. W. Doetsch, PLoS ONE, 2013, 8, e81162 Search PubMed.
- S. Okuno, H. Sato, K. Kuriyama-Matsumura, M. Tamba, H. Wang, S. Sohda, H. Hamada, H. Yoshikawa, T. Kondo and S. Bannai, Br. J. Cancer, 2003, 88, 951–956 CrossRef CAS.
- P. Surowiak, V. Materna, A. Maciejczyk, M. Pudełko, E. Markwitz, M. Spaczyński, M. Dietel, M. Zabel and H. Lage, Virchows Arch., 2007, 450, 279–285 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |