Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Competitive method-based electrochemiluminescent assay with protein–nucleotide conversion for ratio detection to efficiently monitor the drug resistance of cancer cells†
Wen-Bin
Liang
 ab,
Ming-Zhen
Yang
ab,
Ming-Zhen
Yang
 b,
Ying
Zhuo
b,
Ying
Zhuo
 a,
Ying-Ning
Zheng
a,
Ying-Ning
Zheng
 a,
Cheng-Yi
Xiong
a,
Cheng-Yi
Xiong
 a,
Ya-Qin
Chai
a,
Ya-Qin
Chai
 *a and
Ruo
Yuan
*a and
Ruo
Yuan
 *a
*a
aKey Laboratory of Luminescent and Real-Time Analytical Chemistry (Southwest University), Ministry of Education, College of Chemistry and Chemical Engineering, Southwest University, Chongqing 400715, PR China. E-mail: yqchai@swu.edu.cn; yuanruo@swu.edu.cn
bDepartment of Clinical Biochemistry, Laboratory Sciences, Southwest Hospital, Third Military Medical University, 30 Gaotanyan Street, Shapingba District, Chongqing 400038, PR China
First published on 4th August 2016
Abstract
A simple and highly-efficient approach to monitor the expression of P-glycoprotein (P-gp) in cells was urgently needed to demonstrate the drug resistance of cancer cells. Herein, a competitive method-based electrochemiluminescent (ECL) assay with a single ECL indicator was proposed for the first time to efficiently estimate the concentration ratio of two proteins. By converting the different proteins to partially coincident nucleotide sequences via a sandwich type immunoassay on magnetic beads, the concentration ratio related ECL signals could be obtained via competitive nucleotide hybridization on an electrode surface. This method could thoroughly overcome the limitations of simultaneous ECL assays via multiple ECL indicators with inevitable cross reactions. At the same time, rolling circle amplification was employed to improve the detection performances, especially the detection limit and sensitivity. With P-gp and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a model, the proposed ECL assay was successfully employed to monitor the drug resistance of cancer cells. Compared with conventional technologies, improved sensitivity and accuracy were achieved with a correlation coefficient of 0.9928 and a detection limit of 0.52%. Success in the establishment of the competitive method-based ECL assay offered an efficient strategy to demonstrate the concentration ratio of two proteins and a potential approach for detecting other proteins and nucleotide sequences, revealing a new avenue for ultrasensitive biomolecule diagnostics, especially in cell function research.
Introduction
Currently, using anticancer drugs to regulate cell apoptosis continues to remain at the front line of treatment strategies against cancer.1 Unfortunately, a substantial proportion of treatments with anticancer drugs are ineffective due to the drug resistance of the cancer cells via various pathways. For instance, the most important and widely present drug resistance mechanism is based on adenosine 5′-triphosphate-dependent drug efflux pumps via P-glycoprotein (P-gp).2 It is reported that the state of drug resistance positively correlates to the expression of P-gp, and so it has been employed as an “artificial indicator” of drug resistance.3 At the early stage of drug resistance with a little increase in the expression of P-gp, the effectiveness of a drug could be recovered by reducing or stopping the medication. However, when the concentration of P-gp was high enough to be detected by the conventional technologies, such as western blotting and confocal laser scanning microscopy, the effectiveness of the drug couldn't be recovered at this stage.4–7 Considering the challenge and promising applications, it was urgent to develop an ultrasensitive approach to identify the expression of P-gp in cells and then demonstrate the drug resistance of those cancer cells, especially at that early stage.As a powerful analytical technique, electrochemiluminescent (ECL) assays have attracted considerable attention due to their low background, wide dynamic range, and cost-effectiveness, and especially for the high sensitivity that is obtained by simple approaches.8–11 Various ECL assays have been developed for the sensitive detection of proteins and nucleotide sequences, especially ECL assays with tris(2,2′-bipyridyl)ruthenium(II) (Ru(bpy)32+) and its derivatives as ECL indicators. Chen and co-workers have reported an ECL assay based on tris(2,2′-bipyridyl-4,4′-dicarboxylato)ruthenium(II) (Ru(dcbpy)32+) as the ECL indicator to label nucleotide sequences for ultrasensitive detection of the target nucleotide sequence.12 Zhu's group have developed an ECL assay to sensitively determine the activity of the activated cysteine protease via streptavidin-modified Ru(bpy)32+-doped silica as the signal indicator.13 Significant advantages have been obatined for the ultrasensitive detection of proteins, nucleotide sequences and cells, especially when combined with sensitizing approaches employing nucleotide amplification.14–18 However the approaches that employ just detection of a single target were not suitable to identify the expression of P-gp in cells accurately due to the difficulty in acquiring proteins from a single cell and calculating the cell numbers. In the available P-gp detection technologies, another stably expressed protein, such as β-actin or glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was essential to be used as a control.4–7,19–21 The expression of P-gp was investigated as the concentration ratio between P-gp and the stably expressed protein, such as GAPDH, which could be obtained by monitoring the concentrations of P-gp and GAPDH achieved via different ECL assays over multiple runs. Undoubtedly, it was time-consuming and laborious. It was a better choice to obtain these concentrations via an ECL assay using different ECL indicators on a single interface in a single run. However, inevitable limitations should be taken into consideration due to the potential cross reactions between these ECL indicators. Very recently, we have proposed a multiparameter analysis-based ECL assay to modestly overcome the limitations from the cross reactions between different ECL indicators.22 Although the multiparameter analysis with two ECL indicators was helpful to simultaneously determine the concentration of P-gp and the stably expressed protein, such as GAPDH, to obtain the concentration ratio between these proteins, it was laborious and time-consuming. This was due to the requirement of a massive amount of data to establish the multiparameter model and to eliminate the limitations from the cross reactions between the different ECL indicators. It was a highly valuable assay, but posed further challenges, namely to develop an efficient ECL strategy with a single ECL indicator to achieve the concentration ratio, which would thoroughly overcome the limitations from the cross reactions among multiple ECL indicators. In this regard, an efficient ECL approach with a single ECL indicator was designed based on a competitive method, which was accurate and highly valuable for the early determination of drug resistance of cancer cells via investigation of the concentration ratio. To the best of our knowledge, no related research has been reported to estimate the concentration ratio among different proteins based on an ECL assay until now.
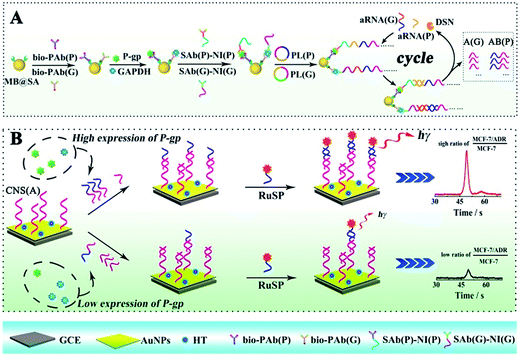
Herein, the competitive method-based ECL assay was proposed for the first time to demonstrate the concentration ratio between P-gp and GAPDH by converting these different proteins to partially coincident nucleotide sequences via a sandwich type immunoassay on magnetic beads to obtain the concentration ratio related ECL signals via competitive nucleotide hybridization on an electrode surface. Specifically, the biotinylated primary antibodies of P-gp and GAPDH were attached onto streptavidin coated magnetic beads (MB@SA), and the nucleotide initiator labeled secondary antibodies were captured in the presence of P-gp and GAPDH by specific antibody–antigen interactions. In order to amplify the responses, the nucleotide initiators were employed to further achieve rolling circle amplification (RCA), which could extend the initiator to a long repeated nucleotide sequence with cascade amplification.23–26 And then, these repeated nucleotide sequences from the RCA reactions were enzymatically cut into partially coincident nucleotide sequences, which could competitively react with the capture nucleotide sequences modified on the electrode surface. Just one of these partially coincident nucleotide sequences could capture the signal probes (ECL indicators with labeled nucleotide sequences). Thus, the ECL signals could be employed to demonstrate the competitive reactions and the concentration ratio of these nucleotide sequences and the related target proteins. The success in the establishment of the competitive method-based ECL assay offered an efficient strategy to demonstrate the concentration ratio simply. This method could be readily expanded to electrochemical, fluorescent and chemiluminescent assays for various kinds of proteins and nucleotide sequences, providing an efficient tool for early detection of drug resistance and revealing a new avenue for ultrasensitive biomolecule diagnostics, especially in cell function research.
Experimental methods
Preparation of the biotinylated primary antibodies
According to the manufacturer's instructions, the primary antibodies for P-gp and GAPDH were biotinylated based on the reaction between the N-hydroxysuccinimide (NHS) group on NHS–biotin and amine groups on the antibodies. Briefly, the appropriate NHS–biotin solution was added into a PBS solution with primary antibodies of P-gp (PAb(P), 1 mg mL−1), and the reaction was held at 37 °C for 2 h after complete mixing. Finally, the biotinylated PAb(P) (bio-PAb(P)) was obtained after removing the non-reacted NHS–biotin by careful desalinization. The same protocol was employed to prepare the biotinylated primary antibodies of GAPDH (bio-PAb(G)).Preparation of the nucleotide initiator labeled secondary antibodies
The secondary antibodies for P-gp and GAPDH were labeled with a nucleotide initiator based on bioconjugations via an EDC/NHS reaction. Briefly, 20 μL of secondary antibodies for P-gp (SAb(P), 1 mg mL−1) were firstly mixed with 100 μL of nucleotide initiator for P-gp (NI(P)) by continuous stirring for 10 min. After addition of 100 μL of reaction buffer including 0.20 M 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and 0.05 M sulfo-NHS, the mixture was stirred at room temperature for 2 h to enable full bioconjugation between NI(P) and SAb(P). Further dialysis with centrifugal filter devices (50 kD) was employed to remove the non-reacted NI(P) and related chemicals. The prepared NI(P) labeled SAb(P) was named as SAb(P)-NI(P). Based on the similar protocol as described above, the nucleotide initiator for GAPDH (NI(G)) was labeled onto the secondary antibodies for GAPDH (SAb(G)), and the product was named as SAb(G)-NI(G).Preparation of the ECL indicator labeled nucleotide sequences
Ru(dcbpy)32+ was employed as the ECL indicator in this work, which was labeled onto the nucleotide sequences as a signal probe based on the EDC/NHS reaction. Briefly, 100 μL of Ru(dcbpy)32+ solution (0.10 M) was mixed with 20 μL of nucleotide sequences (50 μM), and then 50 μL of EDC (0.20 M) and 50 μL of sulfo-NHS (0.05 M) was added into the mixture with continuous stirring at room temperature for 2 h for a complete reaction. Immediately, the reacted mixture was purified to remove the non-reacted Ru(dcbpy)32+ and related chemicals, and the obtained Ru(dcbpy)32+ labeled nucleotide sequences as signal probes (RuSP) were characterized via high performance liquid chromatography.Cell culture
Human breast cancer MCF-7 cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). MCF-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) that was supplemented with 10% foetal bovine serum, 1% non-essential amino acids and 10 μg mL−1 insulin. The doxorubicin-resistant MCF-7 cells (MCF-7/ADR) with a high expression of P-gp were developed from the parental MCF-7 cells that were subjected to persistent gradient exposure to doxorubicin through increasing doxorubicin concentration from 5 ng mL−1 until the cells acquired resistance to 1 mg mL−1 of doxorubicin. All of the cells were maintained in a humidified atmosphere with 5% CO2 at 37 °C.As a control, western blotting was performed as followed. Firstly, MCF-7 cells and MCF-7/ADR cells were seeded at different ratios for 24 h, and then the whole-cell lysates were extracted from the cells using a radio-immunoprecipitation assay buffer. Western blotting assays were performed as previously described.27 Whole-cell lysates were separated on an 8% SDS-PAGE gel and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were blocked for 1 h at 37 °C in 5% non-fat milk/TBST and were then incubated with primary antibodies overnight at 4 °C. The antibodies were used as follows: PAb(P) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and PAb(G) (1
1000) and PAb(G) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000). The membrane was then rinsed in TBST and incubated with various secondary antibodies for 2 h at 25 °C. Immunoreactive bands were visualized with a chemiluminescent horseradish peroxidase substrate (Millipore).
1000). The membrane was then rinsed in TBST and incubated with various secondary antibodies for 2 h at 25 °C. Immunoreactive bands were visualized with a chemiluminescent horseradish peroxidase substrate (Millipore).
Immunofluorescence with a confocal laser scanning microscope was also employed as a control in this study. Briefly, MCF-7 cells and MCF-7/ADR cells were seeded at different ratios for 24 h firstly. After washing with PBS, the cells were fixed in 4% paraformaldehyde for 30 min and blocked in immunol staining blocking buffer for 1 h at 37 °C. Following incubation with SAb(P) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100), the cells were washed with PBS containing 0.025% Triton X-100 3 times, and then incubated with rabbit anti-goat fluorescein isothiocyanate isomer I (FITC)-conjugated secondary antibodies (1
100), the cells were washed with PBS containing 0.025% Triton X-100 3 times, and then incubated with rabbit anti-goat fluorescein isothiocyanate isomer I (FITC)-conjugated secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) for 1 h at room temperature. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and the cells were observed by confocal laser scanning microscopy.
1000) for 1 h at room temperature. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and the cells were observed by confocal laser scanning microscopy.
Measurement procedure
The ECL assay was performed based on a sandwiched format immunoassay on magnetic beads and ECL determination on a biosensor as shown in Scheme 1. Before the ECL assay, the biosensor was fabricated as follows. Firstly, the glassy carbon electrode (GCE) was carefully polished with alumina slurries (0.3 and 0.05 μm) and ultrasonicated in deionized water to remove the physically adsorbed materials. After drying with nitrogen gas, the GCE was immersed in 2 mL of HAuCl4 solution (10 mg mL−1) for electrochemical deposition under a constant potential of −0.2 V for 30 s to obtain a porous gold nanoparticle (AuNP) modified electrode. And then, the AuNP modified GCE was dipped into the solution with the capture nucleotide sequences (CNS(A)) to modify the nucleotide sequences onto the AuNPs. Finally, 1-hexanethiol (HT) solution (0.1 mM) was employed to block the non-specific binding sites on the electrode to obtain the HT/CNS(A)/AuNP modified electrode used for the ECL assay. For the sandwiched format immunoassay on magnetic beads, the biotinylated primary antibodies of P-gp (bio-PAb(P)) and GAPDH (bio-PAb(G)) were attached onto the MB@SA based on the specific interaction between biotin and streptavidin. Then the P-gp, GAPDH and nucleotide sequence labeled detection antibodies of P-gp and GAPDH (SAb(P) and SAb(G), respectively) could be bound onto the magnetic beads via specific interactions between the antigens and antibodies by incubating at 37 °C for 40 min. The amount of nucleotide sequence labeled detection antibodies was positively related to the concentration of antigens. After washing twice with PBST to remove the non-reacted secondary antibodies and other related materials, the amplification via RCA was performed based on the reaction between the prepared padlocks (PL(P) and PL(G)) in TE buffer with 1 μL of Phi29 polymerase and 5 μL of dNTPs for 6 h at 37 °C. The obtained long nucleotide sequences with repeated sequences were then cleaved into very short nucleotide sequences (AB for P-gp (AB(P)) and A for GAPDH (A(G)), respectively) based on hybridization with assistant nucleotide sequences (aRNA(P) and aRNA(G)) via reaction in a Tris–HCl buffer at 95 °C for 5 min and 37 °C for 40 min, and specific cleavage with DSN via addition of 10 μL of DSN enzyme at 65 °C for 15 min and 50 μL of DSN stop solution at 65 °C for 5 min. The obtained nucleotide sequences (AB(P) and A(G)) were dropped onto the fabricated electrode surface and the reactions were maintained at 37 °C for 40 min for the competitive hybridization of AB(P) and A(G) to capture the nucleotide sequences (CNS(A)) modified on the electrode surface. The resultant biosensor was incubated with signal probes (RuSP) at 37 °C for 40 min. After every reaction, the electrode was carefully washed with Tris–HCl buffer. Finally, the ECL responses of these biosensors were investigated in PBS with 25 mM tripropylamine (TPrA).Results and discussion
Electrochemical characterization of the biosensor
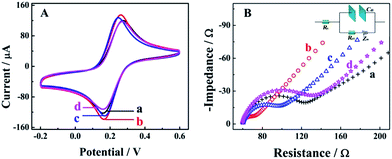
Cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) measurements were employed as useful electrochemical technologies to characterize the fabrication of the biosensor step by step (Fig. 1). As shown in the Fig. 1A for CV measurements in 0.1 M PBS with 5 mM [Fe(CN)6]3−/4− at the potential range from 0.2 to 0.6 V, there was a pair of well-defined redox peaks for [Fe(CN)6]3−/4− for the bare GCE (curve a), indicating good electron transfer on the electrode surface. When the AuNPs were electrodeposited onto the electrode surface, a higher peak current and smaller oxidation–reduction potential were observed due to the good electron transfer ability and conductivity of the AuNPs (curve b, AuNPs/GCE). An obviously decreased peak current could be observed after the modification of capture nucleotide sequences onto the AuNPs (curve c, CNS(A)/AuNPs/GCE), because the nonconductive nucleotide sequences would decrease the electron transfer. For the similar property of HT, the peak current was decreased furthermore after blocking with HT (curve d, HT/CNS(A)/AuNPs/GCE). Further characterization via EIS in 0.1 M PBS with 5 mM [Fe(CN)6]3−/4− at a frequency range from 5 × 10−2 to 1 × 106 Hz is shown in Fig. 1B in the form of a Nyquist plot. In the EIS measurements, the electron transfer resistance (Ret) was employed to demonstrate the electron transfer kinetics quantitatively, which was similar to the semicircle diameter in the EIS response in the form of a Nyquist plot and was calculated accurately based on the equivalent circuit with model impedance data (inset of Fig. 1B). As shown in curve a for the bare GCE, a normal EIS response was obtained with Ret of 53.2 Ω. After electrodeposition of the AuNPs onto the electrode surface (curve b, AuNPs/GCE), the Ret was decreased obviously to 19.6 Ω, indicating the good electron transfer on the electrode surface. When the capture nucleotide sequences were modified onto the AuNP surface (curve c, CNS(A)/AuNPs/GCE) and blocked with HT (curve d, HT/CNS(A)/AuNPs/GCE), the Ret was increased to 68.3 Ω and 96.7 Ω, respectively. These EIS results indicated the same tendency of electron transfer as that obtained by CV. All the results of these EIS and CV measurements confirmed the successful fabrication of the proposed biosensor.Mechanism of competitive method-based ECL assay
For the competitive detection of the ratio between P-gp and GAPDH, it is necessary to construct a competitive relationship between these entirely different proteins. Considering the favorable designability and various available amplification approaches of nucleotide sequences, the secondary antibodies of P-gp and GAPDH (SAb(P) and SAb(G)) were firstly labeled with two special nucleotide sequences respectively. Based on a sandwiched format immunoassay on magnetic beads, the amount of secondary antibodies was increased with the increasing concentrations of P-gp and GAPDH respectively, which could be demonstrated as cNI(P) ∝ cP and cNI(G) ∝ cG, where cNI(P) and cNI(G) are the concentrations of the nucleotide labels on the secondary antibodies of P-gp and GAPDH respectively, and cP and cG are the concentrations of P-gp and GAPDH respectively. Due to amplification via RCA reactions and specific cleavage via DSN, the nucleotide labels on the detection antibodies of P-gp and GAPDH could be amplified significantly to give two similar nucleotide sequences (named as A(G) and AB(P) respectively), where A(G) and AB(P) contained some of the same sequences as shown in Table S1† and the nucleotide sequence on AB(P) and A(G) could be employed for hybridization with the capture nucleotide sequence (named as CNS(A)) on the electrode surface. Thus, the concentrations of A(G) and AB(P) were positively related with the concentrations of the detection antibodies of GAPDH and P-gp, respectively, which could be demonstrated as cAB(P) ∝ cNI(P) ∝ cP and cA(G) ∝ cNI(G) ∝ cG, where cAB(P) and cA(G) are the concentrations of the amplified nucleotide sequences AB(P) and A(G), respectively. The obtained AB(P) and A(G) could be competitively reacted with CNS(A) on the electrode surface and section B on AB(P) could capture the signal probe (RuSP) to generate the ECL signal. Thus, the reactions could be demonstrated using the following kinetic equations:| CNS(A) + A(G) = CNS(A)A(G) | (1) |
| CNS(A) + AB(P) = CNS(A)AB(P) | (2) |
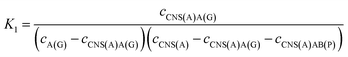 | (3) |
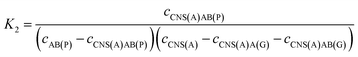 | (4) |
When the concentration of CNS(A) was much lower than that of A(G) and AB(P), the obtained AB(P) and A(G) would competitively react with CNS(A). Thus, the concentrations of CNS(A)A(G) (cCNS(A)A(G)) and CNS(A)AB(P) (cCNS(A)AB(P)) would be directly related to the ratio of cAB(P) and cA(G) and be illustrated in the following equations calculated from eqn (3) and (4):
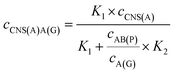 | (5) |
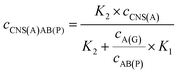 | (6) |
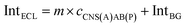 | (7) |
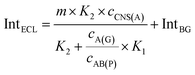 | (8) |
Electrochemical and ECL responses of the biosensor
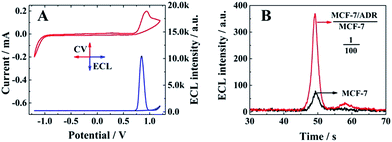
The electrochemical (red line) and ECL responses (blue line) of the biosensor are shown in Fig. 2A, when the ratio between MCF-7/ADR and MCF-7 was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. A significant ECL emission with the peak current at 0.85 V could be obtained with the oxidation of Ru(dcbpy)32+ and TPrA, indicating the presence of a signal probe (RuSP) on the biosensor surface due to the specific nucleotide hybridization. Furthermore, MCF-7 cells and MCF-7 cells with MCF-7/ADR cells were employed to confirm the relationship between the ECL emission and presence of MCF-7/ADR based on the competitive method-based ECL assay. As shown in Fig. 2B, there was a maximum ECL emission at about 370 a.u. for the MCF-7 cells with MCF-7/ADR cells (MCF-7/ADR
2. A significant ECL emission with the peak current at 0.85 V could be obtained with the oxidation of Ru(dcbpy)32+ and TPrA, indicating the presence of a signal probe (RuSP) on the biosensor surface due to the specific nucleotide hybridization. Furthermore, MCF-7 cells and MCF-7 cells with MCF-7/ADR cells were employed to confirm the relationship between the ECL emission and presence of MCF-7/ADR based on the competitive method-based ECL assay. As shown in Fig. 2B, there was a maximum ECL emission at about 370 a.u. for the MCF-7 cells with MCF-7/ADR cells (MCF-7/ADR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MCF-7 = 1
MCF-7 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100), which was significantly higher than that of MCF-7 cells without MCF-7/ADR cells, indicating that the proposed competitive method-based ECL assay could be employed to demonstrate the presence of cancer cells with drug resistance based on the detection of the concentration ratio between P-gp and GAPDH.
100), which was significantly higher than that of MCF-7 cells without MCF-7/ADR cells, indicating that the proposed competitive method-based ECL assay could be employed to demonstrate the presence of cancer cells with drug resistance based on the detection of the concentration ratio between P-gp and GAPDH.
Application of the proposed competitive method-based ECL assay
To estimate the performance of the as-proposed competitive method-based ECL assay for quantitative identification of the drug resistance of cancer cells, the ratio between P-gp and GAPDH in MCF-7 cells with a different ratio of MCF-7/ADR (MCF-7/ADR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MCF-7) was detected in the range from 1
MCF-7) was detected in the range from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128 to 1
128 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
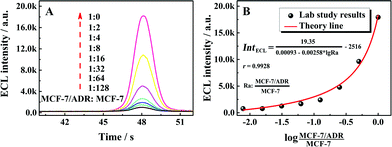
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0. As shown in Fig. 3A with the ECL–time responses of the proposed ECL assay, the ECL emissions were increased with the ratio of MCF-7/ADR
0. As shown in Fig. 3A with the ECL–time responses of the proposed ECL assay, the ECL emissions were increased with the ratio of MCF-7/ADR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MCF-7. The relationship between the ECL emissions and the ratio of MCF-7/ADR
MCF-7. The relationship between the ECL emissions and the ratio of MCF-7/ADR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MCF-7 could be fitted well with the demonstrated theory line according to the competitive strategy. The fitted theory line could be calculated as
MCF-7 could be fitted well with the demonstrated theory line according to the competitive strategy. The fitted theory line could be calculated as 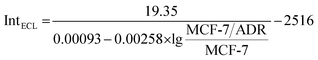 with a correlation coefficient of 0.9928 and a detection limit of 0.52%, where IntECL is the ECL intensity obtained by the competitive method-based ECL assay (Fig. 3B) and the detection limit was down to 52 MCF-7/ADR cells with drug resistance in 1 × 104 normal MCF-7 cells. At the same time, a limit of quantification of 0.86% was obtained, indicative of a good performance for quantitative determination with suitable precision and accuracy. Therefore, the as-proposed strategy could be employed to demonstrate the ratio of MCF-7/ADR
with a correlation coefficient of 0.9928 and a detection limit of 0.52%, where IntECL is the ECL intensity obtained by the competitive method-based ECL assay (Fig. 3B) and the detection limit was down to 52 MCF-7/ADR cells with drug resistance in 1 × 104 normal MCF-7 cells. At the same time, a limit of quantification of 0.86% was obtained, indicative of a good performance for quantitative determination with suitable precision and accuracy. Therefore, the as-proposed strategy could be employed to demonstrate the ratio of MCF-7/ADR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MCF-7 based on the ratio of P-gp to GAPDH obtained via the competitive method-based ECL assay, indicating the acceptable application of the as-proposed competitive method-based ECL assay to monitor the drug resistance of cancer cells quantitatively.
MCF-7 based on the ratio of P-gp to GAPDH obtained via the competitive method-based ECL assay, indicating the acceptable application of the as-proposed competitive method-based ECL assay to monitor the drug resistance of cancer cells quantitatively.
Additionally, as important properties for the ECL assay, the reproducibility, repeatability and stability of the assay were evaluated for duplicate measurements in this work. No more than 5% for the relative standard deviation (RSD) was obtained for all of the reproducibility, repeatability and long time stability studies, specifically 4.8%, 3.2% and 4.6% respectively, indicating the satisfactory application performance of the competitive method-based ECL assay.
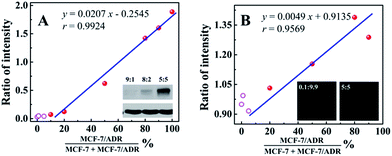
In order to evaluate the applicability of the competitive method-based ECL assay compared with the conventional technologies used to monitor the drug resistance of cancer cells, western blotting and immunofluorescence with confocal laser scanning microscopy were employed to detect MCF-7 cells with different ratios of MCF-7/ADR cells (Fig. 4). Detailed information and results of these studies are shown in the ESI.† As shown in Fig. 4A with the calibration curve for the relationship between the western blotting responses and the ratio of cancer cells with drug resistance, MCF-7/ADR, the ratio of intensity obtained from the gray level analysis was increased with the increasing ratio of cancer cells with drug resistance, with a correlation coefficient of 0.9924 and a detection limit of 13.99%. For the immunofluorescence results using confocal laser scanning microscopy (Fig. 4B), a better detection limit of 6.07% could be obtained. Whereas, a lower correlation coefficient (r = 0.9569) was obtained due to the narrow response range and low sensitivity. Compared with the conventional technologies used to identify the drug resistance of cancer cells, improved sensitivity and accuracy were achieved based on the as-proposed competitive method-based ECL assay, especially the correlation coefficient of 0.9928 and detection limit of 0.52%. This is indicative of the good performance and the potential application of the as-proposed ECL assay to monitor cancer cells with drug resistance and demonstrate their levels quantitatively.
Conclusions
In summary, a competitive method-based ECL assay was developed to demonstrate the concentration ratio of entirely different proteins and identify the drug resistance of cancer cells. The strategy was processed by a sandwich type immunoassay on magnetic beads to convert the proteins to partially coincident nucleotide sequences. Competitive nucleotide hybridization on the electrode surface would then result in an ECL signal related to the concentration ratio of the target proteins and the drug resistance of the cancer cells. With P-gp and GAPDH as a model, satisfactory performances were achieved to identify the drug resistance of the cancer cells based on the proposed ECL assay, which was fitted well with the theory line with a correlation coefficient of 0.9928 and a detection limit of 0.52%. The improved sensitivity and accuracy compared with the conventional technologies indicated the good performance and potential application of the as-proposed ECL assay to identify the drug resistance of cancer cells and demonstrate their levels quantitatively. Most importantly, the competitive method-based ECL assay offered an efficient strategy to demonstrate the concentration ratio simply, which could be readily expanded for various kinds of proteins and nucleotide sequences. This efficient strategy also provided an efficient tool for the early detection of drug resistance and revealed a new avenue for ultrasensitive biomolecule diagnostics, especially in cell function research.Abbreviation list
| AB(P) | Product AB for P-gp |
| A(G) | Product A for GAPDH |
| aRNA(G) | Assistant RNA for GAPDH |
| aRNA(P) | Assistant RNA for P-gp |
| AuNPs | Gold nanoparticles |
| bio-PAb(G) | Biotinylated primary antibodies for GAPDH |
| bio-PAb(P) | Biotinylated primary antibodies for P-gp |
| CNS(A) | Capture nucleotide sequences |
| CV | Cyclic voltammetry |
| CV% | Variation coefficients |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| DMEM | Dulbecco's modified Eagle's medium |
| dNTPs | Deoxynucleotides |
| DSN | Duplex-specific nuclease |
| EDC | 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride |
| ECL | Electrochemiluminescent |
| EIS | Electrochemical impedance spectroscopy |
| FITC | Fluorescein isothiocyanate isomer I |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| GCE | Glassy carbon electrode |
| HT | 1-Hexanethiol |
| HPLC | High performance liquid chromatography |
| MB@SA | Streptavidin coated magnetic beads |
| MCF-7/ADR | Doxorubicin-resistant MCF-7 cells |
| NHS–biotin | Biotin N-hydroxysuccinimide ester |
| PBS | Phosphate buffer saline |
| PBST | PBS buffer with Tween 20 |
| P-gp | P-Glycoprotein |
| PL(P) | Padlock for P-gp |
| PL(G) | Padlock for GAPDH |
| PVDF | Polyvinylidene difluoride |
| RCA | Rolling circle amplification |
| RSD | Relative standard deviations |
| RuSP | Ru(dcbpy)32+ labeled signal probe |
| Ru(bpy)32+ | Tris(2,2′-bipyridyl)ruthenium(II) |
| Ru(dcbpy)32+ | Tris(2,2′-bipyridyl-4,4′-dicarboxylato)ruthenium(II) |
| SAb(G)-NI(G) | Nucleotide initiator labeled secondary antibodies for GAPDH |
| SAb(P)-NI(P) | Nucleotide initiator labeled secondary antibodies for P-gp |
| sulfo-NHS | N-Hydroxysulfosuccinimide |
| TBST | Tris-buffered saline with Tween 20 |
| TE | Tris-buffered saline with EDTA |
Acknowledgements
This work was financially supported by the National Natural Science Foundation (NNSF) of China (81301518, 21575116, 51473136, 21275119 and 81302316), and the Fundamental Research Funds for the Central Universities (XDJK2015A002), China.References
- W. S. Bernard and P. W. Christopher, World cancer report 2014, World Health Organization, Lyon, 2014, pp. 18–20 Search PubMed.
- H. Thomas and H. M. Coley, Canc. Causes Contr., 2003, 10, 159–165 Search PubMed.
- T. Ozben, FEBS Lett., 2006, 580, 2903–2909 CrossRef CAS PubMed.
- C. F. Higgins, Nature, 2007, 446, 749–757 CrossRef CAS PubMed.
- R. T. Skeel and S. N. Khleif, Handbook of Cancer Chemotherapy, Lippincott Williams and Wilkins, London, 1999, pp. 3–20 Search PubMed.
- G. D. Kruh and M. G. Belinsky, Oncogene, 2003, 22, 7537–7552 CrossRef CAS PubMed.
- B. C. Baguley and D. J. Kerr, Anticancer Drug Development, Academic Press, London, 2002, pp. 1–9 Search PubMed.
- P. Wu, X. D. Hou, J. J. Xu and H. Y. Chen, Chem. Rev., 2014, 114, 11027–11059 CrossRef CAS PubMed.
- Z. Y. Liu, W. J. Qi and G. B. Xu, Chem. Soc. Rev., 2015, 44, 3117–3142 RSC.
- L. Z. Hu and G. B. Xu, Chem. Soc. Rev., 2010, 39, 3275–3304 RSC.
- W. J. Miao, Chem. Rev., 2008, 108, 2506–2553 CrossRef CAS PubMed.
- Q. M. Feng, Y. Z. Shen, M. X. Li, Z. L. Zhang, W. Zhao, J. J. Xu and H. Y. Chen, Anal. Chem., 2016, 88, 937–944 CrossRef PubMed.
- Y. P. Dong, G. Cheng, Y. Zhou and J. J. Zhu, Anal. Chem., 2016, 88, 1922–1929 CrossRef CAS PubMed.
- G. X. Li, X. X. Yu, D. P. Liu, X. Y. Liu, F. Li and H. Cui, Anal. Chem., 2015, 87, 10976–10981 CrossRef CAS PubMed.
- M. S. Wu, D. J. Yuan, J. J. Xu and H. Y. Chen, Chem. Sci., 2013, 4, 1182–1188 RSC.
- E. Puodziukynaite, J. L. Oberst, A. L. Dyer and J. R. Reynolds, J. Am. Chem. Soc., 2012, 134, 968–978 CrossRef CAS PubMed.
- Y. C. Yu, C. Lu and M. N. Zhang, Anal. Chem., 2015, 87, 8026–8032 CrossRef CAS PubMed.
- Y. P. Dong, G. Chen, Y. Zhou and J. J. Zhu, Anal. Chem., 2016, 88, 5469–5475 CrossRef CAS PubMed.
- E. Y. Park, E. S. Chang, E. J. Lee, H. W. Lee, H. G. Kang, K. H. Chun, Y. M. Woo, H. K. Kong, J. Y. Ko, H. Suzuki, E. Song and J. H. Park, Cancer Res., 2014, 74, 7573–7582 CrossRef CAS PubMed.
- M. M. Gottesman, T. Fojo and S. E. Bates, Nat. Rev. Cancer, 2002, 2, 48–58 CrossRef CAS PubMed.
- C. Martin, J. Walker, A. Rothnie and R. Callaghan, Br. J. Cancer, 2003, 89, 1581–1589 CrossRef CAS PubMed.
- W. B. Liang, C. C. Fan, Y. Zhuo, Y. N. Zheng, C. Y. Xiong, Y. Q. Chai and R. Yuan, Anal. Chem., 2016, 88, 4940–4948 CrossRef CAS PubMed.
- B. Schweitzer, S. Roberts, B. Grimwade, W. P. Shao, M. J. Wang, Q. Fu, Q. P. Shu, I. Laroche, Z. M. Zhou, V. T. Tchernev, J. Christiansen, M. Velleca and S. F. Kingsmore, Nat. Biotechnol., 2002, 20, 359–365 CrossRef CAS PubMed.
- C. Larsson, J. Koch, A. Nygren, G. Janssen, A. K. Raap, U. Landegren and M. Nilsson, Nat. Methods, 2004, 1, 227–232 CrossRef CAS PubMed.
- Y. Q. Cheng, X. Zhang, Z. P. Li, X. X. Jiao, Y. C. Wang and Y. L. Zhang, Angew. Chem., 2009, 121, 3318–3322 CrossRef.
- C. Russell, S. Roy, S. Ganguly, X. Y. Qian, M. H. Caruthers and M. Nilsson, Anal. Chem., 2015, 87, 6660–6666 CrossRef CAS PubMed.
- M. Z. Yang, F. Yuan, P. Li, Z. J. Chen, A. Chen, S. H. Li and C. M. Hu, Mol. Cell, 2012, 11, 54 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Characterization of the rolling circle amplifications, drug resistance assay with the conventional technologies and supplementary figures and tables. See DOI: 10.1039/c6sc02801b |
| This journal is © The Royal Society of Chemistry 2016 |