Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Metal-responsive structural transformation between artificial DNA duplexes and three-way junctions†
Yusuke
Takezawa
,
Shuhei
Yoneda
,
Jean-Louis H. A.
Duprey
,
Takahiro
Nakama
and
Mitsuhiko
Shionoya
*
Department of Chemistry, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan. E-mail: shionoya@chem.s.u-tokyo.ac.jp
First published on 16th February 2016
Abstract
DNA three-way junctions (3WJs) are essential structural motifs for DNA nanoarchitectures and DNA-based materials. We report herein a metal-responsive structural transformation between DNA duplexes and 3WJs using artificial oligonucleotides modified with a 2,2′-bipyridine (bpy) ligand. A mixture of bpy-modified DNA strands and natural complementary strands were self-assembled exclusively into duplexes without any transition metal ions, while they formed 3WJs in the presence of NiII ions. This transformation was induced by the formation of an interstrand NiII(bpy)3 complex, which served as a template for the 3WJ assembly. Altering the amount and identity of the metal ion regulated the 3WJ induction efficiency. Removal of the metal using EDTA quantitatively regenerated the duplexes. The metal-dependent structural conversion shown here has many potential applications in the development of stimuli-responsive DNA materials.
Introduction
DNA branched structures have been proven to be an essential structural motif for developing DNA nanoarchitectures, whose structures are programmed based on the sequence-specificity of DNA hybridization.1 Since Seeman published a milestone paper demonstrating the construction of immobile DNA junctions,2 DNA branched structures, especially three-way junctions (3WJs), have been widely utilized as building blocks of two- and three-dimensional nanoscale structures1 as well as soft materials.3 In addition, 3WJs have been employed as scaffolds for molecular assembly4 and as reaction spaces.5 In light of the functional versatility of DNA 3WJ structures, it would be highly advantageous to be able to stabilize and induce these motifs.6 This would endow DNA-based materials with stimuli-responsiveness.Metal–ligand coordination is one of the most exploited molecular interactions to develop stimuli-responsive supermolecules and materials.7 This is also the case for DNA architectures, in which a variety of interstrand metal complexes have been covalently incorporated.8 Notable examples include the introduction of artificial metal-mediated base pairs into DNA duplexes.9 This approach has yielded a wide range of metal-responsive functional DNA, whose structure,10 catalytic activity,11 as well as electrical conductivity12 can be regulated. In contrast, conjugation of metal complexes with other DNA structural motifs, including triplexes,13 quadruplexes,14 and junctions,15,16 has not been widely explored for the purpose so far. We have previously developed an artificial metallo-DNA 3WJ,15 which was composed of three oligonucleotides containing an unnatural bipyridine-modified nucleoside. Upon addition of NiII ions, the 3WJ was thermally stabilized by the formation of a tris(bipyridine) metal complex that crosslinked the three strands. A similar metallo-DNA 3WJ was thereafter exploited for constructing higher-order structures by others,16 suggesting its potential usefulness as a component of DNA-based materials.
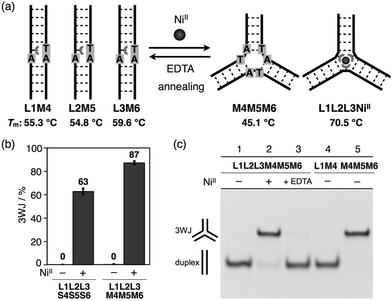
In this study, we have investigated the structural transformation between DNA duplexes and 3WJ structures in response to metal coordination (Scheme 1). Such a structural reorganization involving metallo-DNA conjugates is of great use because the structural motif would be readily embedded into higher-order DNA architectures. The transformation was demonstrated with six DNA strands, whose sequences were designed so as to form both duplexes and 3WJs (Table 1). Three strands (L1, L2, and L3) have a bpy ligand at the middle, and the others (S4, S5, and S6) are complementary to the bpy-modified strands. We expected that the addition of transition metal ions would induce a structural transformation to 3WJs through the formation of a tris(bipyridine) metal complex at the junction core.
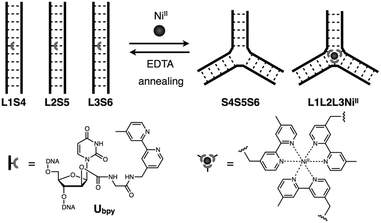 | ||
| Scheme 1 Schematic representation of the metal-responsive structural transformation between artificial DNA duplexes and three-way junctions. | ||
| DNA | Sequences (5′ to 3′)b |
|---|---|
| a See the ESI for other strands. b X = Ubpy for L1, L2, and L3, X = T for S1, S2, and S3. | |
| L1/S1 | GAA GGA ACG XAC ACT CGC AG |
| L2/S2 | GTT CCA CGC XAC GTT CCT TC |
| L3/S3 | CTG CGA GTG XAG CGT GGA AC |
| S4 | CTG CGA GTG TAC GTT CCT TC |
| S5 | GAA GGA ACG TAG CGT GGA AC |
| S6 | GTT CCA CGC TAC ACT CGC AG |
Results and discussion
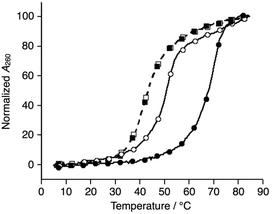
We synthesized a novel bpy-modified nucleoside, which possesses a bpy ligand at the 2′-α position (Ubpy). The novel Ubpy nucleoside was efficiently incorporated into DNA strands to provide bpy-modified oligonucleotides L1, L2, and L3 (see the ESI†). As described later, a DNA 3WJ containing three 2′-α-modified Ubpy nucleotides (i.e.L1L2L3) showed a higher thermal stabilization upon metal coordination compared to a 3WJ with 2′-β-modified nucleotides used in the previous study.15We began our investigation by determining the thermal stability of the hybridization products (Tables 2 and S3†). Bpy-modified 3WJ L1L2L3 showed a sigmoidal melting curve with a Tm = 51.7 °C in the absence of metal ions (Fig. 1). When one equiv. of NiII ion was added, the Tm of L1L2L3 was increased substantially to 70.5 °C (ΔTm = +18.8 °C). Such a significant metal-dependent stabilization was not observed for other 3WJs containing fewer than three bpy ligands, i.e.S1S2S3 (ΔTm = +0.5 °C), S1S2L3 (+0.7 °C), or S1L2L3 (+5.8 °C) (Fig. S3 and Table S2†). Thus, the stabilization was attributed to the formation of an interstrand NiII(bpy)3 complex at the core of the 3WJ, similarly to the previously reported metallo-3WJ.15 The formation of the metallo-3WJ L1L2L3·NiII was further confirmed by mass spectrometry (Fig. S4†). In contrast, Tm values of the duplexes (L1S4, L2S5, and L3S6), containing one bpy ligand, were not affected by the addition of NiII ions (Table S3†), precisely because of the inability to form an interstrand bipyridine–metal complex.
| Metal-freeb | 1 eq. of NiII | ||
|---|---|---|---|
| T m/°C | T m/°C | ΔTm/°C | |
| a Average of at least 3 runs after annealing. Standard errors are also listed. b In the presence of 10 equiv. of EDTA. | |||
| Three-way junctions | |||
| L1L2L3 | 51.7 ± 0.3 | 70.5 ± 0.4 | +18.8 |
| S1S2S3 | 42.1 ± 0.3 | 42.6 ± 0.5 | +0.5 |
| S4S5S6 | 43.3 ± 0.5 | 43.0 ± 0.4 | −0.3 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Mixture of six strands | |||
| L1, L2, L3, S4, S5, and S6 | 62.5 ± 0.3 | 43.7 ± 0.7 | −18.8 |
| 69.3 ± 0.6 | +6.8 | ||
Comparison of the melting temperatures demonstrated that the duplexes (L1S4, L2S5, and L3S6, Tm = 62.0, 60.3, and 65.1 °C, respectively) were more stable than the 3WJs (L1L2L3 and S4S5S6, Tm = 51.7 and 43.3 °C, respectively) under metal-free conditions. This suggests that duplex formation is favored in the absence of metal ions. Conversely, addition of one equiv. of NiII ions made the bpy-modified 3WJ the most stable structure (L1L2L3·NiII). This raised the possibility that formation of 3WJs can be induced by metal coordination.
Subsequently, melting profiles of a mixture of all six strands (L1, L2, L3, S4, S5, and S6) were analyzed (Fig. 2). In the absence of metal ions, the melting curve showed a one-step transition with a Tm of 62.5 °C, which is almost an average of the Tm values of the duplexes, L1S4, L2S5, and L3S6. This indicated that the duplexes were predominantly formed from the six DNA strands. When one equiv. of NiII ions was added to the DNA mixture, its melting curve seemed to change to a two-step transition. The Tm value of each transition (43.7 °C and 69.3 °C) is in good agreement with those of the unmodified 3WJ S4S5S6 (43.0 °C) and the metallo-3WJ L1L2L3·NiII (70.5 °C), respectively. This melting profile implied preferential formation of the 3WJs (L1L2L3·NiII and S4S5S6) in the presence of NiII ions.17 Notably, the melting curve of the mixture of six natural strands (S1, S2, S3, S4, S5, and S6) was not changed upon NiII addition, indicating that NiII(bpy)3 complexation altered the hybridization behavior of the DNA mixture.
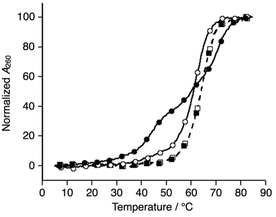 | ||
| Fig. 2 Melting curves of a mixture of the six DNA strands in the absence (white) and presence of NiII ions (black); (circles) L1, L2, L3, S4, S5, and S6; (squares) S1, S2, S3, S4, S5, and S6. The conditions were the same as for Fig. 1. | ||
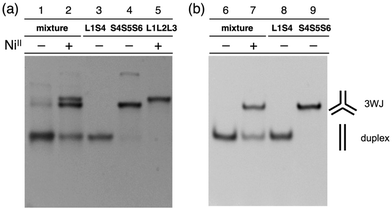
The hybridization products were then evaluated by native polyacrylamide gel electrophoresis (PAGE). Fig. 3a shows an image of the gel stained with SYBR Gold dye. The DNA mixture without metal ions showed nearly a single band corresponding to the DNA duplexes (lane 1). Upon addition of one equiv. of NiII ions, two new bands with lower mobility appeared (lane 2). These bands were ascribable to the 3WJs, S4S5S6 and L1L2L3·NiII, indicating that NiII addition induced the formation of the two 3WJs. To quantify the amount of the 3WJs formed, native PAGE analysis was conducted with a FAM-labeled S4 strand (FAM-S4), and then the band intensities of the FAM-labeled products were compared (Fig. 3b). The results showed that the DNA duplexes were exclusively formed in the absence of the metal ions (lane 6) while 3WJs were formed in ca. 60% yield17 in the presence of NiII ions (lane 7). Such 3WJ formation was not observed when natural DNA strands were used in place of some of the bpy-modified strands (e.g. a mixture of S1, L2, L3, S4, S5, and S6; Fig. S7 and S8†). This reinforces the conclusion that the 3WJ formation was induced by the interstrand NiII(bpy)3 complexation.
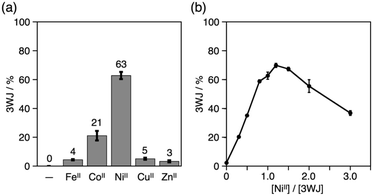
The yield of the 3WJs varied in response to the identity and the amount of metal ions added (Fig. S9 and S10†). The yield dropped to ca. 20% when CoII ions were substituted for NiII, while other ions showed almost no 3WJ induction effects (Fig. 4a). This presumably correlates with the large formation constant of the NiII(bpy)3 complex (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β3 = 20.2).15,18 The efficiency of the NiII-mediated 3WJ formation increased in proportion to the NiII concentration in the range of 0 to 1.2 equiv. and began to decline with excess additions (Fig. 4b). This stoichiometric behavior is almost consistent with a 3
β3 = 20.2).15,18 The efficiency of the NiII-mediated 3WJ formation increased in proportion to the NiII concentration in the range of 0 to 1.2 equiv. and began to decline with excess additions (Fig. 4b). This stoichiometric behavior is almost consistent with a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexation of the bpy ligand and NiII ions. These results represent a coordination-driven feature of the structural transformation using the bpy-modified DNA strands.
1 complexation of the bpy ligand and NiII ions. These results represent a coordination-driven feature of the structural transformation using the bpy-modified DNA strands.
With the goal of promoting a metal-responsive duplex-to-3WJ transformation, we redesigned the natural counter strands M4, M5, and M6, such that the two nucleobases in the middle were mutated from TA to AT (Fig. 5 and Table S1†). The resulting duplexes showed lower thermal stability (Tm = 55.3, 54.8, and 59.6 °C for L1M4, L2M5, and L3M6, respectively) than the original duplexes (L1S4etc.) (Table S4†) owing to the existence of mismatch pairs (Ubpy–T and A–A). In contrast, the 3WJ M4M5M6 contains no mismatches and thus showed a similar thermal stability (Tm = 45.1 °C) to the original 3WJ S4S5S6 (43.3 °C). Native PAGE analysis (Fig. 5c) revealed that alteration of the relative thermal stabilities led to more efficient 3WJ induction formation (ca. 90%).19 In addition, subsequent treatment with a chelating agent (EDTA) followed by annealing, to remove the NiII ions, led to the complete regeneration of the metal-free duplexes (lane 3). These results indicate that the NiII-responsive structural transformation under these conditions is both nearly quantitative and reversible.
Conclusions
In conclusion, we have demonstrated a metal-dependent structural transformation between DNA duplexes and 3WJs using bpy-modified artificial oligonucleotides. While the mixture of bpy-modified strands and complementary strands exclusively forms duplexes under metal-free conditions, NiII addition induces the formation of 3WJs. This transformation occurs as a result of the formation of an interstrand NiII(bpy)3 complex. In addition, the removal of NiII ions by EDTA regenerates the duplexes, confirming the metal-ion responsiveness of the structural rearrangement. Stimuli-responsive switching of 3WJ structures has gained much attention as a versatile tool to functionalize DNA materials.20 Therefore, we believe that the metal-responsive 3WJ transformation presented here will provide an exciting advance in DNA nanotechnology and create new opportunities in DNA-based materials science.Experimental section
Oligonucleotide synthesis
Oligodeoxynucleotides were synthesized on an Applied Biosystems 394 DNA synthesizer by standard phosphoramidite chemistry. The synthesis of the phosphoramidite derivative of Ubpy is presented in the ESI.† The DNA synthesis was carried out on a 1 μmol scale in DMTr-on mode with standard reagents purchased from Glen Research. The coupling time of the nucleosides was extended to 15 min. The products were deprotected in 25% NH3 solution at 55 °C for 8 h. The oligomers were firstly purified and detritylated using a PolyPak II cartridge (Glen Research) and further purified by reverse-phase HPLC (Waters XBridge C18 column, 0.1 M TEAA (pH 7.0)/MeCN gradient, 60 °C) (Fig. S1†). All DNA strands were identified by MALDI-TOF mass spectrometry (see ESI†). The amount of the oligomers was determined based on the UV absorbance at 260 nm. The molar extinction coefficients (ε260) of the bpy-modified DNA strands (L1, L2, and L3) were estimated15 by the sum of the ε260 value of the bpy group and that of corresponding unmodified oligonucleotides calculated by the nearest-neighbor method. Some of the unmodified oligonucleotides purified by HPLC were purchased from Japan Bio Services and used without further purification.Melting analysis
All samples were prepared by mixing the DNA strands (1.0 μM) in 10 mM MOPS buffer (pH 7.0) containing 100 mM NaCl. After addition of NiSO4·7H2O (Soekawa) or EDTA, the solutions were heated to 85 °C and cooled slowly to 5 °C at the rate of 1.0 °C min−1. Absorbance at 260 nm was monitored by a UV-1700 spectrophotometer (Shimadzu) equipped with a TMSPC-8 temperature controller while the temperature was raised from 5 °C to 85 °C at the rate of 0.2 °C min−1. A drop of mineral oil was laid on the sample to prevent evaporation. Normalized absorbance shown in the figures was calculated as follows:| Normalized A260 = {A260(t °C) − A260(5 °C)}/{A260(85 °C) − A260(5 °C)} × 100. |
The melting temperature (Tm) was determined as an inflection point of a melting curve using the LabSolutions Tm analysis software (Shimadzu) with a 17-point adaptive smoothing program. Average Tm values of at least 3 independent runs are shown in Tables S2–S4 in the ESI.†
PAGE analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). After running at 120 V for 3 h in the cool incubator (4 °C), the gels were observed using an Alpha imager mini (LMS) with a blue-LED transilluminator (Optocode). For unlabelled samples, the gels were stained with SYBR Gold (Invitrogen). Quantification of each product was accomplished by comparing the band intensities of a 3WJ S4S5S6 (or M4M5M6) with that of a duplex L1S4 (or L1M4), in which S4 (or M4) was labeled with FAM. Averages of at least three independent experiments are shown in figures.
1). After running at 120 V for 3 h in the cool incubator (4 °C), the gels were observed using an Alpha imager mini (LMS) with a blue-LED transilluminator (Optocode). For unlabelled samples, the gels were stained with SYBR Gold (Invitrogen). Quantification of each product was accomplished by comparing the band intensities of a 3WJ S4S5S6 (or M4M5M6) with that of a duplex L1S4 (or L1M4), in which S4 (or M4) was labeled with FAM. Averages of at least three independent experiments are shown in figures.
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research (S) (No. 21225003) for M. S. and a Grant-in-Aid for Young Scientists (B) (No. 23750181) for Y. T. from JSPS of Japan. Y. T. also thanks the Foundation Advanced Technology Institute for an ATI Research Grant.Notes and references
- F. Zhang, J. Nangreave, Y. Liu and H. Yan, J. Am. Chem. Soc., 2014, 136, 11198–11211 CrossRef CAS PubMed , and references therein.
- (a) N. C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed; (b) N. R. Kallenbach, R.-I. Ma and N. C. Seeman, Nature, 1983, 305, 829–831 CrossRef CAS.
- Y. G. Li, Y. D. Tseng, S. Y. Kwon, L. D'Espaux, J. S. Bunch, P. L. Mceuen and D. Luo, Nat. Mater., 2004, 3, 38–42 CrossRef CAS PubMed.
- (a) A. L. Benvin, Y. Creeger, G. W. Fisher, B. Ballou, A. S. Waggoner and B. A. Armitage, J. Am. Chem. Soc., 2007, 129, 2025–2034 CrossRef CAS PubMed; (b) M. Probst, D. Wenger, S. M. Biner and R. Häner, Org. Biomol. Chem., 2012, 10, 755–759 RSC.
- M. H. Hansen, P. Blakskjær, L. K. Petersen, T. H. Hansen, J. W. Højfeldt, K. V. Gothelf and N. J. V. Hansen, J. Am. Chem. Soc., 2009, 131, 1322–1327 CrossRef CAS PubMed.
- (a) A. Oleksi, A. G. Blanco, R. Boer, I. Uson, J. Aymami, A. Rodger, M. J. Hannon and M. Coll, Angew. Chem., Int. Ed., 2006, 45, 1227–1231 CrossRef CAS PubMed; (b) C. Ducani, A. Leczkowska, N. J. Hodges and M. J. Hannon, Angew. Chem., Int. Ed., 2010, 49, 8942–8945 CrossRef CAS PubMed; (c) B. M. Laing and R. L. Juliano, ChemBioChem, 2015, 16, 1284–1287 CrossRef CAS PubMed.
- (a) A. J. McConnell, C. S. Wood, P. P. Neelakandan and J. R. Nitschke, Chem. Rev., 2015, 115, 7729–7793 CrossRef CAS PubMed; (b) S. Tashiro and M. Shionoya, Chem. Lett., 2013, 42, 456–462 CrossRef CAS.
- (a) E. Stulz and G. H. Clever, DNA in supramolecular chemistry and nanotechnology, Wiley, Chichester, 2015 Search PubMed; (b) H. Yang, K. L. Metera and H. F. Sleiman, Coord. Chem. Rev., 2010, 254, 2403–2415 CrossRef CAS.
- (a) Y. Takezawa and M. Shionoya, Acc. Chem. Res., 2012, 45, 2066–2076 CrossRef CAS PubMed; (b) P. Scharf and J. Müller, ChemPlusChem, 2013, 78, 20–34 CrossRef CAS; (c) Y. Takezawa, K. Nishiyama, T. Mashima, M. Katahira and M. Shionoya, Chem.–Eur. J., 2015, 21, 14713–14716 CrossRef CAS PubMed; (d) T. Kobayashi, Y. Takezawa, A. Sakamoto and M. Shionoya, Chem. Commun., 2016, 52, 3762–3765 RSC.
- S. Johannsen, N. Megger, D. Böhme, R. K. O. Sigel and J. Müller, Nat. Chem., 2010, 2, 229–234 CrossRef CAS PubMed.
- J. Liu and Y. Lu, Angew. Chem., Int. Ed., 2007, 46, 7587–7590 CrossRef CAS PubMed.
- S. Liu, G. H. Clever, Y. Takezawa, M. Kaneko, K. Tanaka, X. Guo and M. Shionoya, Angew. Chem., Int. Ed., 2011, 50, 8886–8890 CrossRef CAS PubMed.
- (a) K. Tanaka, Y. Yamada and M. Shionoya, J. Am. Chem. Soc., 2002, 124, 8802–8803 CrossRef CAS PubMed; (b) Y. Takezawa, W. Maeda, K. Tanaka and M. Shionoya, Angew. Chem., Int. Ed., 2009, 48, 1081–1084 CrossRef CAS PubMed; (c) T. Ihara, T. Ishii, N. Araki, A. W. Wilson and A. Jyo, J. Am. Chem. Soc., 2009, 131, 3826–3827 CrossRef CAS PubMed.
- D. M. Engelhard, R. Pievo and G. H. Clever, Angew. Chem., Int. Ed., 2013, 52, 12843–12847 CrossRef CAS PubMed.
- J.-L. H. A. Duprey, Y. Takezawa and M. Shionoya, Angew. Chem., Int. Ed., 2013, 52, 1212–1216 CrossRef CAS PubMed.
- C. Stubinitzky, A. Bijeljanin, L. Antusch, D. Ebeling, H. Hölscher and H.-A. Wagenknecht, Chem.–Eur. J., 2014, 20, 12009–12014 CrossRef CAS PubMed.
- The two-step melting curve shown in Fig. 2 was fitted to a curve simulated on an assumption that the 3WJs were formed in 70% yield and the duplexes in 30% yield (Fig. S6†). This result is consistent with the yield of the 3WJs (ca. 60%) determined by native PAGE analysis (Fig. 3b).
- R. M. Smith and A. E. Martell, Critical Stability Constants, Plenum Press, New York, 1975, vol. 2 Search PubMed.
- Exclusive duplex formation was also observed without transition metal ions. For other mutations, see Fig. S11†.
- (a) I. T. Seemann, V. Singh, M. Azarkh, M. Drescher and J. S. Hartig, J. Am. Chem. Soc., 2011, 133, 4706–4709 CrossRef CAS PubMed; (b) J. M. Thomas, H.-Z. Yu and D. Sen, J. Am. Chem. Soc., 2012, 134, 13738–13748 CrossRef CAS PubMed; (c) S. A. Barros and D. M. Chenoweth, Angew. Chem., Int. Ed., 2014, 53, 13746–13750 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Full experimental procedures, thermal denaturation experiments, ESI-MS and NMR spectra, and other experimental results. See DOI: 10.1039/c6sc00383d |
| This journal is © The Royal Society of Chemistry 2016 |