Nanomaterials for biocatalyst immobilization – state of the art and future trends
Eliane P. Cipolattiab,
Alexsandra Valérioa,
Rosana O. Henriquesa,
Denise E. Moritza,
Jorge L. Ninowa,
Denise M. G. Freire b,
Evelin A. Manoelbc,
Roberto Fernandez-Lafuente*d and
Débora de Oliveira*a
b,
Evelin A. Manoelbc,
Roberto Fernandez-Lafuente*d and
Débora de Oliveira*a
aChemical and Food Engineering Department, Federal University of Santa Catarina (UFSC), P.O. Box 476, Florianópolis, SC 88040-900, Brazil. E-mail: debora.oliveira@ufsc.br
bBiochemistry Department, Chemistry Institute, Federal University of Rio de Janeiro, Av. Athos da Silveira Ramos, 149, Ilha do Fundão, Cidade Universitária, 21949-909 Rio de Janeiro, RJ, Brazil
cIntegrated Research Laboratory in Biotechnology, Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil
dDepartamento de Biocatálisis, Instituto de Catálisis-CSIC, C/Marie Curie 2, Campus UAM-CSIC, Cantoblanco, 28049 Madrid, Spain. E-mail: rfl@icp.csic.es
First published on 26th October 2016
Abstract
Nanotechnology is an area that has been growing over the years, being possible nowadays to find numerous materials constructed at nanoscale. In addition, many applications have been attributed to these “new” materials. In this review is presented a brief overview of nanoparticles used for the immobilization of enzymes. Considering the extensive universe of immobilization in nanoparticles, some were chosen to be exposed here, such as chitosan, graphene, silica, polymers, magnetic, nanoflowers, among others. Advantages, disadvantages and limitations of nanoimmobilization also be discussed. Some applications of nanoimmobilized enzymes are presented, like as biodiesel, flavor synthesis ester and biosensors. The purpose of this paper is to provide an overview of what is being studied in relation to nanoparticles for enzymes immobilization, and some discussions about them, aimed at assisting researchers in future studies and reviews.
1. Introduction
Enzyme immobilization arises as an answer to the necessity of reusing the expensive enzymes in industrial processes.1,2 However, nowadays, to permit the re-use of the enzyme is not the only objective of the immobilization, as it has been revealed itself as a powerful tool to improve many enzyme properties. Among them, stability (via multipoint covalent attachment, multi-subunit immobilization of multimeric proteins or via generation of favorable enzyme environments) is expected to increase after a proper immobilization.3–6 Other enzyme properties may be also improved, e.g., activity, selectivity or specificity (by altering the conformation of the enzyme after immobilization), resistance to inhibitors, etc.3–6 Thus, the immobilization system (support, activation method and immobilization conditions) should be designed to maximize our objective that not always will be the improvement of all the features that may be improved (e.g., enzymes having perfect specificity or enzymes from thermophilic microorganisms having very high thermo-stability). Immobilization may, in certain cases, be associated to enzyme purification and in this way compensate the costs related to the immobilization step.7The linking between the supports and the enzyme can be done by adsorption, covalent bond, ionic, encapsulation, among other more sophisticated techniques of immobilization.6c,7,30 The link by adsorption is the simplest technique and allows immobilizing enzymes on solid supports through low energy connections, such as van der Waals or hydrophobic, hydrogen bonds and ionic, among others.124,165c But if the researcher's interest is also related to the support, this is a very interesting technique for allowing the enzymes desorption with application of detergents gradients.35a It may also be formed a covalent bond between the enzyme and a water-insoluble support, or by crosslinking with the matrix.124 The enzyme-support covalent bond formation is strong and irreversible, with a greater operating stability, but when observed denaturation of the enzyme, the support is disposed of together.30
Indeed, it is important to investigate the “efficiency of contribution” of immobilized enzyme on the process of immobilized enzymes to determine the total productivity on a kilogram of product per kilogram of biocatalyst, and this can be indirectly measured by the determination of the number of reuse over cycles.8 The same purpose is related to the so-called “nanoimmobilization”. Immobilized enzymes on nanoparticles can show a broader range of pH and temperature usage, higher thermal stability besides providing changes in selectivity and specificity compared to the native enzymes.9,10
In this context, nanoparticles have been extensively studied for enzymes immobilization.11–15 Recently, the high level of publications in the literature shows the growing interest in the use of nanoscale particles for enzyme immobilization is mainly due to the inherent characteristics of these particles. Ansari and Husain16 reported some characteristics related to the nanoparticles: (i) enzyme nanoparticles can be easily synthesized in high solid content without surfactants and toxic reagents, (ii) homogeneous and well defined core–shell nanoparticles with a thick enzyme shell can be obtained, and (iii) designed according to the researcher necessity.
Advances in nanoscience stimulated the interest in the study of the particles properties in nanometric sizes.17,18 Different materials are used in nanometric sizes in several areas of science, like fine chemicals and medicine. As some examples: polymers,19,20 silica,21,22 gold,23,24 diamond,21 graphene,25,26 magnetics.27,28 When using nanomaterials as supports for enzyme immobilization some basic parameters in the immobilization processes also should be considered, these are the basis for the selection and subsequent use of derivatives: immobilization yield, specific activity, recovered activity, effective catalyst utilization, minimal enzyme deactivation and the cost-effective of the operations.12,29
As will be shown in this review, there are numerous immobilization techniques, and numerous ways to stabilize the proposed structures (would be impossible to list them all). This paper attempts to provide an overview of the use of the nanosupports in the enzyme immobilization. The authors believe that keeping in mind the previous concepts related to immobilization, allied with the knowledge of some existing materials, will help researchers to choose the best method to use in their work, and possibly some ideas and suggestions for new immobilization techniques and new nanomaterials can arise after reading.
2. Nanomaterials
Based on the advances of nanotechnology, many works have been developed in order to immobilize enzymes onto surfaces of nanoscale materials such as nanoparticles, nanotubes, mesoporous materials and nanofibrous membranes,30–32,165c. Therefore, much has been discussed about the use of nanomaterials and its advantages in the interaction with the biocatalyst. Table 1 shows some advantages and disadvantages of nanomaterials compared to standard porous supports. We will briefly comment some of the points.| Advantages | Disadvantages |
|---|---|
| Mass transfer resistance | Cost of the fabrication process |
| Effective enzyme loading | Large scale application |
| High surface area | Separation of the reaction medium |
| Diffusional problems minimization | — |
Nanomaterials use to be no porous, that makes that all enzyme molecules are located in the surface of the particle. This way, internal diffusion limitations are not produced. The enzymes immobilized on non-porous nanomaterials may be multipointly attached to the support to increase its stability and still be able to act in very large or even insoluble materials as long as the enzyme is properly oriented on the support surface;5 this is not possible using conventional porous supports.6b However, the immobilization in the external surface of the support raises some problems that need to be considered. Now the enzyme is not protected from interactions with hydrophobic interfaces like gas bubbles, also the enzyme molecules in one particle may interact with the enzyme molecules in other particle (permitting proteolysis).33 This may be solved if the immobilized support is coated with a polymer that prevent this deleterious interactions, avoiding enzyme inactivation in stirred systems.6b,33,34
Diffusion limitations use to be a problem that decrease enzyme expressed activity or even enzyme stability.35 However, not always the diffusion limitations are an undesired problem.33,36 For example, the pH gradient occurring in hydrolysis of penicillin G by immobilized penicillin G acylase improve enzyme stability.36 This effect is not possible when using nanoparticles. Other case where diffusion problems are positive is in coimmobilized enzymes. The second enzyme, when coimmobilized, acts on high concentration of the product of the first enzyme due to these gradients, and in some cases the apparent activity of the combi-enzyme catalyst may be greatly improved.35,36 For example, this permitted a much more effect recycling of NADH in a three enzymatic system, overpassing even the sue of equivalent amounts of free enzymes.36b Again, this effect is not possible using non porous nanomaterials. Thus, nanomaterials have many advantages, but in certain cases, the lack of some of the effects of immobilization on standard porous supports may generate the lack of some desired effects.
During the current work some examples of nanoparticles use as support for enzyme immobilization are exposed, and in most of these cases, the result is positive, but one of the concerns is the application in large scale. Another problem is the separation process of these nanoparticles from the reaction medium at the end of reaction process, that may be very complex and sometimes very expensive decreasing the application.
Nanoparticles usually are in a range of 1–100 nm composed of several hundreds of atoms,37 or, as in the case of produced by miniemulsion may reach 500 nm.38 Particles with diameters smaller than 1 nm are generally referred to clusters of particles. Nanoparticles with diameter up to 10 nm are particularly interesting owing it can be considered as almost fully surface, due all atoms are on surface or near to the surface.37,39
Nanostructures have been reported as supports for enzyme immobilization by different links including enzyme adsorption, covalent attachment, enzyme encapsulation, and sophisticated methods combinations.40 The literature provides a large number of works in terms of enzyme immobilization, commercial or not. We can relate different supports used for this purpose, as well as, coatings and surface functionalization to make the support most effective in the reaction of interest. We begin with a brief table, just to give us an initial idea about the immobilization world. Table 2 provides an overview of nanomaterials what is taking place in recent years regarding the enzyme immobilization on nanostructures.
| Enzyme/origin | Methodology | Support | Immobilization yield (%) | Reuse (times) | Reactions possible for application | Other improvement | Reference |
|---|---|---|---|---|---|---|---|
| a Fe3O4 carrier modified by 3-aminopropyl triethoxysilane and covalently linked by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and N-hydroxysuccinimide. | |||||||
| Aspergillus niger β-glucosidase | Entrapment | Polymeric materials (polyurethane, latex and silicone) | 93.0 | 6 (75% of residual activity) | Cellobiose hydrolysis | Increase in optimum temperature and activation energy for cellobiose hydrolysis | 29 |
| Catalase from bovine liver | Physical adsorption | Graphene oxide–Fe3O4 | — | 10 (74% of initial activity) | — | Increase in stability | 41 |
| Candida rugosa lipase (CRL) | Adsorption | ZnO nanowires | — | 12 (89.9% of initial activity) | Esterification of phytosterols with oleic acid | Better thermal stability and pH adaptability in comparison to free CRL | 42 |
| Aspergillus terreus lipase | Encapsulation | Alginate-g-PEG/cyclodextrin particles | — | 20 (40%) | Enantioselective hydrolysis of ketoprofen vinyl ester | Higher ability to resolve (R,S)-ketoprofen vinyl ester into (S)-ketoprofen vinyl ester and (R)-ketoprofen | 43 |
| Thermomyces lanuginosus lipase | Covalent | Fe3O4-AP-ENa | 86.5 | 5 (>80% of original activity) | Transesterification of refined palm oil | Promising biocatalyst for biodiesel synthesis | 44 |
| Candida rugosa lipase (CRL) | Adsorption | Multi-walled carbon nanotubes | — | 6 (50% reduction in catalytic efficiency) | Synthesis of geranyl propionate | Improved the activity and stability of CRL | 45 |
| Deoxyriboaldolase (expressed by E. coli BL21(pET30)) | Covalent | Glutaraldehyde-(3-aminopropyl) triethoxysilane nano-magnet material | 75.0 | — | — | The immobilized enzyme exhibited a wider range of reaction temperatures than the free enzyme | 46 |
| Phenylalanine-dehydrogenase | Entrapment | Polytaurine matrix | — | — | Electrochemical biosensor for detection of phenylalanine | The good stability, wide linear concentration range, low detection limit, and a distinct advantage of polishing in the event of surface fouling | 20 |
| Superoxide dismutase, bovine | Covalent | Iron oxide nanoparticles (nano Fe3O4) coated on a gold electrode surface | — | — | Biosensor for superoxide (O2˙−) | Rapid estimation of superoxide anions | 47 |
| β-Galactosidase from Kluyveromyces lactis | Adsorption | Multi-walled carbon nanotubes | — | 6 (90% of relative activity) | Lactose hydrolysis | Greater biocatalytic activity at higher galactose concentration | 48 |
| α-Amylase from Aspergillus oryzae | Covalent/adsorption | Magnetite prepared with gum acacia and using glutaraldehyde | 60.0 | 6 (∼70% of initial activity) | Hydrolysis of starch | Thermal stability | 49 |
| β-Glucosidase from Aspergillus niger | Covalent | Iron oxide nanoparticles | 93.0 | 16 (50% of enzyme activity) | Biofuel production | Improved thermostability | 50 |
| Candida Antarctica exp. Pichia pastoris | Adsorption | Core–shell polymeric nanoparticules | xxx | Xx | Biofuel production and pharmaceutical compounds | Higher ability to resolve (D,L)-1,3,5 mioinositol triphosphate into (L)-and (D) isomers | Cunha et al. 2015 Pinto et al. 2016 |
Before we begin to study nanomaterials separately, it is worth remembering that the goals to immobilize an enzyme, is in nanometric supports or on a larger scale, are to improve the stability of the enzyme, maintain or improve the activity and reuse.1–7,35 It should be noted that it is very important to control the enzyme–support interactions in order to understand the possible applications and modifications that can be made, and the control the orientation of the enzyme may be very relevant (large substrates, area involved in the immobilization).5 There are some cases where the use of nanoparticles as a support is almost mandatory, for example, large or insoluble substrates, in cases that avoid using immobilized enzymes on porous supports.6b
This review presents some data on the use of the following materials in nanometric scale: polymers, chitosan, magnetic nanoparticles, sílica, zircônia, gold, graphene, zinc oxide, hybrid organic–inorganic nanoflowers, and some nanoimmobilized enzymes applications.
2.1 Polymers
In the last years, the use of enzymes in industrial processes became possible due to the increase in the scale up production and the development of genetic engineering techniques. However, some factors still limit enzymes application on large scale, such as low stability, selectivity, and activity of many biocatalysts.51 In this way, the immobilization process can be an alternative to the low stability, resistant to solvents, temperature and pH, make possible the increase of enzyme concentration in the reaction medium and the biocatalyst reuse.1–7,35The supports for enzyme immobilization should present some desired characteristics such as high ability to interact with enzymes (without significantly changing its activity), chemical and mechanical stability/resistance, hydrophobic or hydrophilic surfaces, defined porous morphology, possibility to medium-long term storage and low costs.19,51,52 The support can be a synthetic organic polymer, a biopolymer or an inorganic polymer.53 Polymeric supports such as poly(styrene), poly(methyl-methacrylate), poly(acrylates), poly(acrylamide), poly(urea-urethane) are widely used for enzyme immobilization.53 Moreover, the use of inorganic supports, such as silica gel, aluminum oxide, apatite, and glass, is also common due to the high mechanical and thermal stability, non-toxic and resistant to attack by microorganisms and solvents.53
For the polymeric supports, different methods for enzyme immobilization can be used and are reported in the literature.19,52–55 Besteti et al.52 reported a combined semi-batch and suspension-emulsion polymerizations for polymer supports synthesis used for lipase B from Candida antarctica (CALB) immobilization, using styrene and methyl methacrylate as monomers. It was reported that the obtained polymer particles had core–shell particle structure, with specific areas and average pore sizes with comparable values with that presented by commercial support Accurel MP 1000, ranging respectively from 0.9 to 36.7 m2 g−1 and 141.2 to 354 Å. It was also shown that produced particles could be used for CALB immobilization, leading to higher immobilization efficiency and enzyme activity than obtained by Accurel MP 1000.
Poly(methyl-methacrylate) (PMMA) is appointed as a promising support for enzyme immobilization. In study reported by Valério et al.,19 CalB was immobilized on PMMA nanoparticles obtained by miniemulsion polymerization. The authors evaluated the influence of the initiator type, the enzyme nature, and the crodamol concentrations on CalB enzyme immobilization. The authors confirmed by transmission electron microscopy (TEM) images the morphology of PMMA–CalB enzyme nanoparticles with PMMA core (darker region) and CalB enzyme on the surface (brighter region), as shown in Fig. 1 (right superior insert). The kinetic properties of immobilized CalB enzyme in PMMA nanoparticles were evaluated in terms of monomer conversion, particle size, zeta potential, and relative activity. The immobilized enzyme on PMMA showed a relative activity of 40% after 20 recycle rounds, while free CalB enzyme showed a relative enzyme activity of 5% after 20 recycle rounds, as shown in Fig. 1.
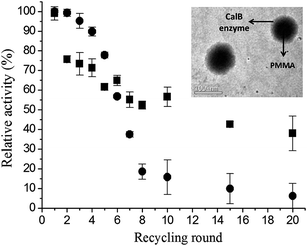 | ||
| Fig. 1 Recycling study of CalB enzyme: ■ PMMA–CalB enzyme and ● free CalB enzyme. TEM images of PMMA–CalB enzyme nanoparticles synthesized using 5 wt% crodamol, 10 wt% CalB enzyme and KPS as initiator (right superior insert).19 This figure has been reproduced from ref: A. Valério, G. Nicoletti, E. P. Cipolatti, J. L. Ninow, P. H. H. Araújo, C. Sayer and D. de Oliveira, Appl. Biochem. Biotechnol., 2015, 175, 2961–2971 with permission from Pan Stanford Publishing. | ||
Cipolatti et al.55 reported the synthesis of PEGylated poly(urea-urethane) nanoparticles as a new alternative to the already methods used as support to Candida antarctica (CalB lipase immobilization by miniemulsion polymerization). The authors reported that it was possible to obtain a high esterification activity (21 U mg−1). The nanoparticles size was 158 ± 5 nm by using the proposed methodology. In addition, it was reported that thermal stability of the immobilized enzyme improved (Fig. 2). After 4 h of incubation time, the relative activity of immobilized enzymes was 67, 25 and 14.8% at 40, 50, and 60 °C, respectively. On the other hand, the free enzyme relative activity was lower than 10% at all temperatures. The authors state that the temperature increase may produce a slight change in the enzyme conformation,3 producing a higher activity, as observed after 6 h of incubation time at 50 and 60 °C (44 and 36%, respectively), whereas at 40 °C the observed value of relative activity was 31.9%.
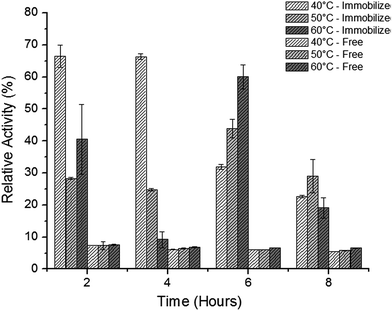 | ||
| Fig. 2 Thermal stability at 40, 50 and 60 °C of free and immobilized CalB lipase in PEGylated poly(urea-urethane) nanoparticles.55 This figure has been reproduced from ref: E. P. Cipolatti, A. Valério, G. Nicoletti, E. Theilacker, P. H. H. Araújo, C. Sayer, J. L. Ninow and D. de Oliveira, J. Mol. Catal. B Enzym., 2014, 109, 116–121 with permission from Pan Stanford Publishing. | ||
2.2 Chitosan
Chitosan, poly[β-(1-4)-linked-2-amino-2-deoxy-D-glucose], is a biopolymer derived from deacetylation of chitin.15 Chitosan can be considered as an attractive support for enzyme immobilization due the presence of reactive surface groups such as amino and hydroxyl.15 Other favorable characteristics of chitosan is your biocompatibility and biodegradability, associate with low cost, since it is the second most abundant biopolymer in the earth, after cellulose.15,56–58 Particles of chitosan as support for enzyme immobilization can be produced in macro, micro or nanosize scale by precipitation, emulsion cross-linking and ionic gelation methods, respectively.15,59Chitosan nanoparticles can also be prepared in water-in-oil microemulsion as reported by Wu et al.60 that showed the synthesis of nanoparticles with 7 nm of diameter and a loading capacity of 156 mg of Candida rugosa lipase per g on the chitosan nanoparticles. Due to the advantages described above, some authors have reported the use of chitosan to modify magnetic particles.61,62 Zang et al.61 linked covalently cellulase on chitosan coated on Fe3O4 nanoparticles (Fig. 3). The derivative obtained had a loading capacity of 123 mg g−1 and an activity of 5.23 IU mg−1 cellulase.
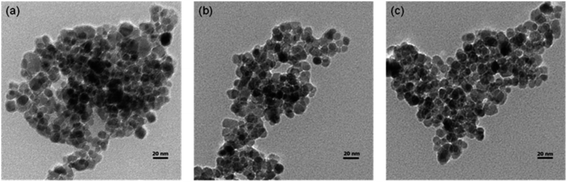 | ||
| Fig. 3 TEM images of Fe3O4 (a), Fe3O4–chitosan (b) and immobilized cellulase (c).61 This figure has been reproduced from ref: L. Zang, J. Qiu, X. Wu, W. Zhang, E. Sakai and Y. Wei, Ind. Eng. Chem. Res., 2014, 53, 3448–3454 with permission from American Chemical Society. | ||
Chitosan based nanomaterials have superior physical and chemical properties such as high surface area, porosity, tensile strength, conductivity, photo-luminescent as well as increased mechanical properties compared to pure chitosan.15 Chitosan shows special properties such as viscosity, solubility in different solvents, mucoadhesivity, polyoxysalt formation, polyelectrolyte behavior, ability to form films, metal chelations, optical, and structural characteristics. Furthermore, it is widely used as support for enzyme immobilization due to its different geometric configurations, such as powders, flakes, hydrogels, membranes, nanofibers and nanoparticles.15,31,63,64
2.3 Polymer nanofibers
Polymer nanofibers have high potential for enzyme immobilization, in situ formation of nanofiber reinforcement composites, biosensors, and biocatalysis/separation. Compared to typical membranes, nanofibers have smaller size (denoting large specific area) of the fiber, higher porosity (higher enzyme loading per unit mass with reduced diffusion resistance), higher conductivity and simple fabrication13,65Zhu and Sun,31 reported covalent immobilization of lipase from Candida rugosa on nanofiber membranes of poly(vinyl alcohol-co-ethylene) activated with glutaraldehyde (PVA-co-PE). The derivative obtained in this study achieved high enzyme activity (676.19 U g−1 of the membrane). From scanning electron microscope images (Fig. 4), the authors confirmed the morphology of the proposed support, with a diameter range of 50–350 nm, and the morphology after use in catalysis reaction (Fig. 4c), confirming the stability of the structure. Additionally the authors showed that pH tolerance, thermal and storage stability of the immobilized lipase on PVA-co-PE nanofibers were improved.
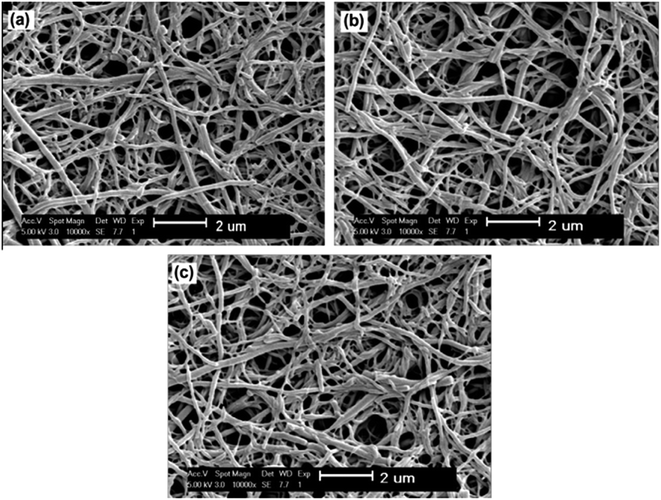 | ||
| Fig. 4 SEM images of (a) original PVA-co-PE nanofibrous membrane, (b) lipase immobilized on PVA-co-PE nanofibrous membrane and (c) lipase immobilized on PVA-co-PE nanofibrous membrane after catalytic reactions.31 This figure has been reproduced from ref: J. Zhu and G. Sun, React. Funct. Polym., 2012, 72, 839–845 with permission from Elsevier. | ||
In a study performed by Ghosh and coauthors,13 L-asparaginase was immobilized on polyaniline nanofibers. The enzyme activity and stability was enhanced after immobilization process. The maximum enzyme activity was 65 U μg−1 of protein. According to the authors, using 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8
1, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 20
1 and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of support to enzyme ratio, the crystalline size was 8.3, 11.07 and 116.6 nm, respectively.
1 of support to enzyme ratio, the crystalline size was 8.3, 11.07 and 116.6 nm, respectively.
2.4 Magnetic nanoparticles
Much has been said about magnetic nanoparticles, mainly due to its mechanical strength and ease recovery from reaction medium (by apply a magnetic field).66 Since 1970s, magnetic particles have increasingly been used in the area of bioscience and medicine.67 A very interesting review written by Netto et al.68 addresses some aspects of the superparamagnetic nanoparticles application as efficient supports for enzyme immobilization. Nude magnetic nanoparticles no effectively interact with protein particle (enzyme), and a surface modification is required. In the literature some modifications used on the magnetic nanoparticles surface modification are reported as for example by crosslinking with glutaraldehyde, coating with polymers,69 coupled with compounds like as agarose,56 use of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDAC),70 or as previously mentioned using chitosan.61Substantial progress in size and morphology control of magnetic nanoparticles has been reported by developing methods such as co-precipitation, thermal decomposition and/or reduction, micelle synthesis, and hydrothermal synthesis.18 A recent method, developed by Pospiskova and Safarik71 allowed the magnetic modification at low temperatures using nano and micro magnetic iron oxides particles prepared by microwave-assisted system, led to the magnetization crosslinked trypsin and lipase powder. Chen et al.56 studied the immobilization of β-glucosidase on magnetic Fe3O4 nanoparticles (MNPs) coupled with agarose (AMNPs) synthesized by co-precipitation via alkaline condition and span-80 surfactants in organic solvent. The derivative could be reused by 15 cycles, retaining more than 90% of original enzyme activity. Additionally, increased enzyme thermostability.
The use of functionalized magnetic nanoparticles was also presented by Soozanipour et al.72 The authors immobilized xylanase by covalent bonding using silica-coated modified magnetite nanoparticles by cyanuric chloride activation. Fig. 5 shows a scheme of the method used in the work, Fe3O4@SiO2 represents silica-encapsulated magnetic nanoparticles, that was modified with (3-aminopropyl) trimethoxysilane (APTES), after, cyanuric chloride (CC) was added to facilitate covalent binding with xylanase.
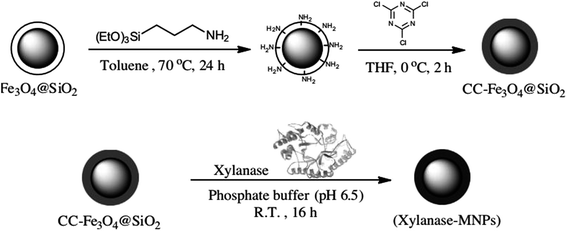 | ||
| Fig. 5 Schematic representation of the method for covalently immobilization of xylanase on functionalized magnetic nanoparticles.72 This figure has been reproduced from ref: A. Soozanipour, A. Taheri-Kafrani and A. Landarani Isfahani, Chem. Eng. J., 2015, 270, 235–243 with permission from Elsevier. | ||
Raita et al.44 immobilized Thermomyces lanuginosus lipase in different forms (Fig. 6). Magnetic nanoparticles to immobilize the lipase were synthesized from four different forms with variation in covalent linkages and protein crosslinking. Fe3O4 or Fe3O4–APTES ((3-aminopropyl)triethoxysilane) nanoparticles were covalently coupled with lipase via EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide) and/or NHS (N-hydroxysuccinimide) as activating agent6d with or without protein cross-linking by GA (glutaraldehyde), which is a bi-functional protein crosslinker.6c Fe3O4-E lipase, hydroxyl groups of Fe3O4 nanoparticles were activated by EDC and subsequently reacted with lipase carboxyl groups, and enzyme molecules on the biocatalyst surface were further crosslinked by GA to obtain Fe3O4-E/G. Fe3O4-AP-EN lipase was prepared by linking lipase carboxyl groups to amino groups of Fe3O4–APTES activated by EDC and NHS and then further crosslinked to obtain Fe3O4-AP-EN/G lipase. Considering the results obtained by the authors, they efficiently developed a method for Fe3O4-AP-EN-lipase preparation with superior properties to be used as biocatalyst for biodiesel synthesis.
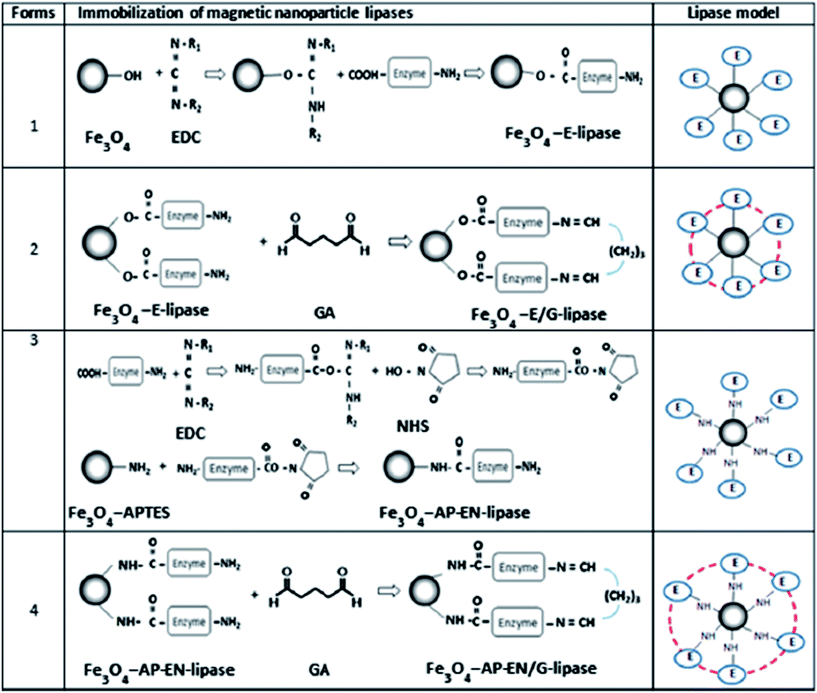 | ||
| Fig. 6 Schematic diagram of magnetic nanoparticle lipase immobilization methods.44 This figure has been reproduced from ref: M. Raita, J. Arnthong, V. Champreda and N. Laosiripojana, Fuel Process. Technol., 2015, 134, 189–197 with permission from Elsevier. | ||
Nanocrystalline and nanoporous metal oxide surfaces are also reported as a novel matrices for enzyme immobilization.9 Metallic nanoparticles has a higher magnetization than their oxidic counterparts do, however, its toxicity and high reactivity may preclude its application in areas such as biomedicine and biotechnology. To solve these problems, these nanoparticles can be coated with polymers or silica.72 However, magnetic nanoparticles coated with polymers are most unstable at high temperature, since the intrinsic instability of the polymers is further adversely affected by the catalytic properties of the nanoparticles.73
2.5 Silica
Silica (SO2) is also a material that has potential as support for enzyme immobilization.53 Silica can be classified according to their physiochemical and morphological characteristics, such as natural or synthetic, micro-, meso- or macroporous, amorphous or crystalline, or with polar properties, with efficient adsorption sites for enzyme immobilization.74,75,165a,b Mesoporous silica, nowadays often referred to nanosilicas, is studied and has several advantages as supports for enzyme immobilization.53 This particles have uniform pore diameters (2–40 nm), very high surface areas (300–1500 m2 g−1) and pore volumes (ca. 1 mL g−1), and are inert and stable at elevated temperatures.53 It should be considered that morphology and particle size of mesoporous silica materials could also have a pronounced effect on the protein and enzyme immobilization.76Silica surface can be easily functionalized, and various compounds can be used for this purpose, as: polyethyleneglycol (PEG), polyvinyl alcohol (PVA), casein, gelatin, albumin (egg or bovine), ionic liquids, among others.53,77 Mohammadi et al.78 immobilized Rhizomucor miehei lipase (RML) covalently on silica nanoparticles (MCM-41 and SBA-15), functionalized by glycidyloxypropyl trimethoxysilane. Two different techniques for RML immobilization were used: (1) reaction of the protein with surfaces containing epoxy groups, promoting random immobilization of RML, and (2) immobilization of RML on partially modified epoxy functionalized nanoparticles in order to promote oriented protein immobilization.5 The enzyme derivatives were used in selective hydrolysis of fish oil. The authors showed that 15% of epoxy group modification in oriented immobilization procedure decreased the number of covalent linkage between enzyme and support resulting in a derivative with lower stability. In contrast, the authors affirmed that the remarkable improvement in selectivity of fish oil hydrolysis compensates undesirable decrease of their stabilities.
Deka et al.76 used cubic mesoporous silica FDU-12 functionalized with tunable content of carboxylic acid (–COOH) groups for lysozyme immobilization (from hen egg white). The synthesized particles showed size in a range of 200–400 nm. The authors obtained high lysozyme adsorption (895 mg g−1), with good enzymatic activity at different pH values. Furthermore, the toxicological safety, stability and the possibility of immobilized enzyme reuse together with the advantages of nanoparticles, make this type of support attractive to industry.74
An interesting study was published by Kuwahara et al.164a where the authors investigated the use of Candida antarctica lipase A (CalA) embedded within silica nanoparticles with oil-filled core–shell structure (Cal-A@OSN) in transesterification reactions. The authors affirmed that this proposed structure showed high catalytic performance both in water and in organic media with increased stability and recyclability. Additionally, the methodology is simple, and it appears as a promising alternative, especially in relation to its reuse capacity. A deeper study about the Cal-A@OSN was published by the same group,164b and the influence of the silicate support and the performances of the immobilized enzymes were evaluated confirmed the importance of this heterogeneous biocatalyst.
2.6 Magnetic cross-linked aggregates (mCLEAs)
Magnetic nanoparticles are used also to facilitate the handling of some biocatalyst that have mechanical properties no very adequate for the industrial handling, like the case of crosslinking enzyme aggregates (CLEAS).6b Some authors have proposed the inclusion of magnetic nanoparticles inside the CLEAS to facilitate their handling, in some cases this strategy also permitted to improve the enzyme properties if the nanoparticle interacts with the enzyme molecules.79–81 This technology is appointed as allowing the production of a robust catalyst in a simple way, carrier-free immobilization and even with the possibility of using semipurified enzyme.82,83Although CLEAs are already broadly applied in many enzymatic processes, it presents some handling difficulties for the biocatalyst recovery.6b CLEAs tends to form stable suspensions that are difficult separated from the reaction medium by centrifugation or filtration, and increase the size of the aggregates.80 Furthermore it imply in some problems as internal mass transfer.6b In this context the combination of magnetic nanoparticles and CLEAs can work around the problem by ease the separation, allowing the use of aggregates with reduced sizes.79–81 Additionally, the high surface area of nanoparticles increases the enzyme loading, improving its applicability and stability in continuous processes such as biodiesel production.84
Cruz-Izquierdo et al.85 developed a method for synthesizing magnetic cross-linked aggregates (mCLEAs) from magnetic nanoparticles aminated by glutaraldehyde.6c This methodology was carried using lipases from Candida antarctica and Aspergillus niger; and α-amylase from Bacillus sp. The method permitted preparing mCLEAs from any kind of enzymes by a simple protocol. Bhattacharya and Pletschke86 described the use of mCLEAs and calcium-mCLEAs as an effective solution for bioconversion of lignocellulosic materials. The use of CLEAs is restricted in the bioconversion of lignocellulosic substrates due the reactional media is composed for insoluble raw substrate, preventing the enzyme recovery by conventional methods. Thus mCLEAs from xylanases was applied in the continuous hydrolysis of lignocellulose. The mCLEAS were easily recovered from the reactional media and a high stability was observed. The reaction was conducted during 136 h at 50 °C and, after this time, the magnetic aggregates was successfully recovery showing 80% activity, against 50% activity for traditional CLEAs.86
2.7 Zirconia
Zirconia is a polymorphic bioinert material that is seen as an attractive support for enzyme immobilization due high thermal, pH and solvent stability.77,87–90 This material has hydroxyl groups on the surface and can occur in different forms depending on the temperature changes: monoclinic, tetragonal and cubic.77,87–90Guncheva et al.77 synthesized nanostructures from zirconia (nanoZrO2-CeO2 and nanoZrO2-B) for immobilization of Candida rugosa lipase. The immobilized enzyme preserved 20% of initial activity after six consecutive tributyrin hydrolysis reaction recycles. Chen et al.91 immobilized lipase from Pseudomona cepacia on zirconia nanoparticles modified with carboxylic acid to use in resolution of (R,S)-1-phenylethanol through acylation in isooctane. Immobilized lipase on stearic acid-modified ZrO2 gave the best performance, increasing by about 10.5 and 16.6 times the initial activity obtained with lipase loaded onto unmodified ZrO2 and crude lipase powder, respectively. Masuda et al.92 studied the immobilization of formaldehyde dehydrogenase (FDH) onto mesoporous silica (pore size = 12.3 nm). The authors affirmed that the enzyme immobilized on the mesoporous zirconia material synthesized using [poly(ethylene glycol)–poly(propylene glycol)–poly(ethylene glycol) (Pluronic P123, EO20PO70EO20)], zirconium(IV) n-propoxide (ca. 75% in 1-propanol), acetylacetone, 1,3,5-trimethylbenzene, and ethanol, exhibited higher activity than the enzyme immobilized on mesoporous silica material due to the increase in substrate affinity resulting from interparticle pore space.
2.8 Gold
Gold also deserves to be mentioned as support for biocatalysts immobilization. There is a growing interest in gold nanoparticles in catalysis, although these are not considered practical supports mainly due to economic issues.93,94 Some examples of immobilized enzymes on gold nanoparticles are: α-amilase from Bacillus subtilis,94 Thermomyces lanuginosus xylanase,23 peroxidase from P. chrysosporium,24 cellulase from Trichoderma reesei,95 superoxide dismutase (bovine).47 The choice of using gold nanoparticles as supports should be based on the final application, and the use of this high cost support in the production of compounds with low aggregate value does not make sense and it should be considered that gold is a metallic catalyst.According to Yan et al.,23 gold nanoparticles (NPG) has some unique characteristics compared with other nanoparticles: (i) can be easily used and recovered while has a high surface area; (ii) has an open and bicontinuous porous network structure, which favors strong adsorption and can afford high enzyme loading; (iii) structural unit is tunable in a wide range from a few nanometers to many microns, which fits for a wide range of enzyme molecules sizes and function; (iv) excellent biocompatibility; (v) processed under organic- and surfactant-free conditions, NPG has extremely clean surfaces, which exclude the possible of interference effects on enzymes from unwanted molecules or ions.
Venditti et al.96 studied the immobilization of Candida rugosa lipase (CRL) on hydrophilic gold nanoparticles functionalized with 2-diethylaminoethanethiol hydrochloride (DEA) (Au-DEA@CRL) and with sodium 3-mercapto-1-propanesulfonate (3MPS) (Au-3MPS@CRL). In their work, the authors showed a simplified and very interesting scheme showing gold nanoparticles functionalization, making it easy to understand (Fig. 7). The authors point out that these derivatives could be promising candidates for applications in industrial processes, with enzyme activity improvement especially for Au-DEA@CRL, showing better results in terms of enzyme loading percentage (65–72%) and residual lipolytic activity (95%), while the Au-3MPS@CRL showed 53–61% and 45%, respectively. Additionally, the derivative with DEA proved to be more stable, compared to free CRL, in a temperature range of 20–55 °C and in a pH range of 5–8.
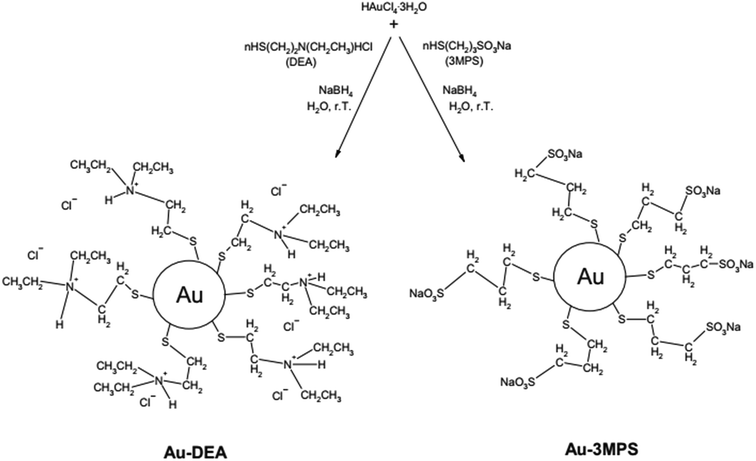 | ||
| Fig. 7 Schematic synthesis of AuNPs stabilized with DEA and 3MPS.96 This figure has been reproduced from ref: I. Venditti, C. Palocci, L. Chronopoulou, I. Fratoddi, L. Fontana, M. Diociaiuti and M. Vittoria, 2015, 131, 93–101 with permission from Elsevier. | ||
2.9 Graphene
Graphene films is the first material one atom thick isolated in nature.97 This is one structure extracted from graphite, with a monolayer of carbon atoms packed into a dense honeycomb crystal structure, as unrolled single-wall carbon nanotubes or as a giant flat fullerene molecule.98,99 The first reports of graphene isolation date from 2004.99 Since then, there is a growing study of the use of graphene in the immobilization of enzymes mainly aimed for use in biosensors.100,101 Considering that the direct electron communication between electrode and enzyme active center is critical in the development of “reagentless” biosensors, biomedical devices and biofuel cells in order to achieve high performance, efficiency and simplicity, the nanographene appears as ideal support, it presents extraordinary electron transport property and high specific surface area.101Graphene-based nanomaterials can interact with biomolecules mostly through electrostatic, van der Waals forces, π–π stacking, or hydrophobic interactions.25 Several strategies are proposed in the literature for enzyme immobilization using functionalized graphene nanoparticles. The synthesis of graphene by graphene oxide resulting in a graphene with large content of oxygen functional groups consisting of epoxide, peroxide, carbonyl (aldehyde, ketone and quinone), and carboxyl groups.101
Jiang et al.102 studied the immobilization of trypsin on dendrimer grafted graphene oxide nanosheets, by covalent binding, using glutaraldehyde6c as coupling agent. The authors affirmed that the enzymatic reactor developed might provide a promising tool for high throughput proteome identification. Protein could be efficiently digested, after only 15 min, with sequence coverage comparable to that obtained by conventional over-night in-solution digestion.
Other enzymes such as cellulase have already been immobilized on graphene supports. Nano magnetoresponsive support of graphene was developed through a supramolecular assembly of oppositely charged quenched polyelectrolytes and maghemite–magnetite nanoparticles on 2D graphene supports. The enzyme Accellerase-1000 was covalently immobilized, showing a marked improvement bio-receptivity of graphene supports. Additionally, it was possible to reuse the enzyme for 5 cycles, maintaining 55% of initial activity.103 Another works studied graphene with glucose oxidase for applications in biosensors.104,105 The authors point out that immobilized enzymes in nanographene structures differ from traditional derivatives in terms of catalytic efficiency, operational stability, and application potential.25
2.10 Zinc oxide
Zinc oxide (ZnO) with different nanostructures by same or different fabrication techniques has been widely used for enzyme immobilization in recent years.106,107 Wet chemical route is quite a popular method to fabricate various ZnO nanostructures, such as nanoparticles, nanorods and nanosheets. It has been proposed to use these ZnO nanostructures as platform for cholesterol oxidase (ChOx) immobilization via physical adsorption. Nano-ZnO is nontoxic, biological compatibility, with high catalytic efficiency, strong adsorption ability, fast electron transfer rate and relative easy preparation, and can be consider a favorable material for immobilization of biomolecules.106,1072.11 Hybrid organic–inorganic nanoflowers
Recently, a new type of nanomaterial has been described in the literature as a potential material for the immobilization of enzymes.108 Flower-like nanomaterials (nanoflowers) are nanostructures from hybrid organic–inorganic materials synthesized from an inorganic part as a metal ion such copper, manganese, or calcium, and an organic part like proteins and DNA. As long as inorganic flower-shaped structures have been used a long time for application in catalysis and analytical science, the organic–inorganic nanoflowers are not long ago reported.108 Hybrid nanoflowers (HNFs) have demonstrating some advantages due the conventional immobilization methods, like their simplicity of synthesis, a greater surface area than spherical nanoparticles, and a higher stability and catalytic activity when compared to free enzymes and immobilized enzymes. The five most important kinds of HNFs are protein-copper, calcium-protein, protein-manganese, copper-DNA, and capsular nanoflowers.108One of the firsts protocols for hybrid structures from copper/enzymes were proposed for Ge et al.108e, that studied the nanoflowers formation when accidentally added CuSO4 to phosphate buffered saline containing bovine serum at pH 7.4 at room temperature. After three days a precipitate was formed and analyzed, showed microstructures like flowers and a protocol using copper(II) ions and another enzymes as α-lactalbumin, laccase, carbonic anhydride and lipases was immobilized replaced the BSA (Fig. 8). The authors reported improvements of 650% increase for lacasse nanoflower activity compared with free laccase in solution; 260% increased activity for carbonic anhydrase nanoflower compared with free enzyme in the hydration of CO2; and 95% for lipases nanoflowers compared to the activity of free lipase.
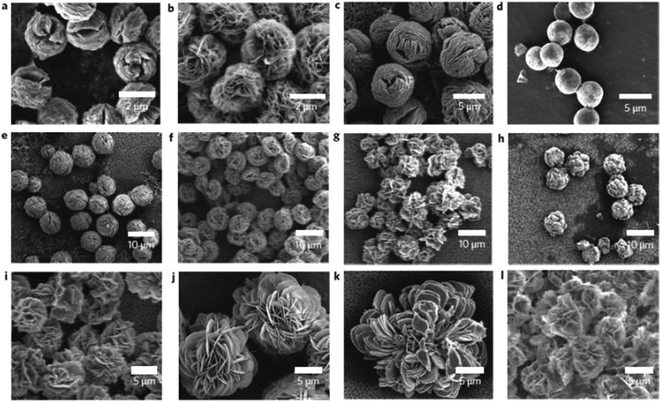 | ||
| Fig. 8 SEM images of hybrid nanoflowers (a–l), Column 1, α-lactalbumin; column 2, laccase; column 3, carbonic anhydrase; column 4, lipase; at protein concentrations of 0.5 mg ml−1 (a–d), 0.1 mg ml−1 (e–h) and 0.02 mg ml−1 (i–l)108a. This figure has reproduced from ref: J. Ge, J. Lei and R. N. Zare, Nat. Nanotechnol., 2012, 7, 428–43 with permission from Nature Publishing Group. | ||
Another application as dye adsorption carrier and catalase immobilization were studied for Wang et al.109 They described the synthesis of chitosan/calcium pyrophosphate microflowers made by a one-pot synthesis using a combination of ionotropic gelation with biomimetic mineralization. The chitosan–tripolyphosphate (CS-TPP) nanocomplexes were firstly synthesized through ionotropic gelation, while the excess of TPP was partly hydrolyzed into P2O74− ions. After, a solution of CaCl2 was applied to induce in situ mineralization of Ca2P2O7 and to direct the growth of the microflowers. The final structures showed a composition of 23% CS-TPP nanocomplexes and 77% of Ca2P2O7 crystals. These microstructures were applied for the removal of Congo red from water and they had a high adsorption capacity of 520 mg g−1 for Congo red dye. In the catalase immobilization the enzymatic derivate retained 85% catalytic activity compared with the free enzyme and a reusability of 10 cycles retained 60% their initial activity.
Immobilization process is not the assurance of immobilized enzyme success but the knowledge of the dynamic interaction between enzyme and solid support interfaces is an important key to help the development and control of reaction rates using immobilized enzyme. Nanotechnology has opened a new frontier in the development of polymeric supports for enzyme immobilization. A wide range of reports are described in the academic literature and it is possible to see that enzyme immobilization in a polymeric matrix can provide good enzyme stability, as well good support to cycles of reuse (see the Reference list). In this sense, the choice of a support may not be based only on its cost but should also be consider the opportunity that it will give to the selection of optimal operating conditions range or to decide upon the feasibility of different process options.
3. Nanoimmobilized enzymes applications
Immobilized enzymes on nanostructures have numerous applications.110,111 With the goal of showing the perspectives of obtaining these biocatalysts, some products of scientific and industrial interest are described below.3.1 Modification of cellulose and other polysaccharides in precipitated systems
Lignocellulose, main component materials such as wood and agricultural residues, forestry, urban, is formed primarily by three types of polymers: cellulose, hemicellulose and lignin. Its hydrolysis to produce bioethanol (from glucose fermentation) is growing.112 This way, the study of the immobilization of microbial enzymes responsible for the degradation of plant cell wall components becomes very interesting,113,114 as immobilization permits the improvement of enzyme features.1–7 However, cellulose is a solid material and therefore, conventional porous supports may be not used in this instance. In these cases, the high external surface area of the nanocatalysts, which allows for better interaction with the substrate are almost the only alternative to immobilize enzymes in preexisting solids.6bAbraham and coauthors112 studied the immobilization of cellulase from Trichoderma reesei on a magnetic nanosupport by covalent binding achieved and used to investigate the hydrolysis of a synthetic carboxymethyl cellulose (CMC) and a natural pretreated substrate hemp hurd biomass (HHB). Immobilization of cellulase can facilitate enzyme recycling in a sequential batch-wise process. The immobilized enzyme was stable for up to seven consecutive cycles at 60 °C of CMC hydrolysis for 30 min. The immobilized cellulase provided successful hydrolysis of 83% with CMC and 93% with hemp hurd biomass.
Commercial cellulases is usually a complex mixture of a variety of hydrolytic enzymes (C1 enzyme, Cx enzyme, and β-glucoside enzyme), and it can be immobilized efficiently in several nanosupports aiming numerous applications, like: aminated Fe3O4 nanoparticles for decomposition of corncob,115 silica through the assistance of L-cysteine functionalized gold nano-particle for the hydrolysis of waste bamboo chopsticks powder,116 polyvinyl alcohol/Fe2O3 magnetic nanoparticle for degrade cellulose117 among others.
Other interesting use of nanobiocatalyst is the clarification of juices. Considering the importance of degradation of starch and pectin in the juice processing, an amino functionalized magnetic nanoparticle was used to co-immobilize all enzymes involved in the reaction (alpha-amylase, cellulase, pectinase and cellulase).115 In this study, the authors stabilized the structure of the immobilized enzymes with glutaraldehyde,6c and indicated this magnetic nanobiocatalyst as promising industrial one due to their high thermal stability and possibility of recycling (eight cycles).115
3.2 Biodiesel production
Biodiesel, a mixture of fatty acid alkyl esters (FAAE), is a biodegradable fuel derived from renewable sources such as vegetable oils and animal fats.118,119 The production of this biofuel by immobilized enzymes is an interesting technological alternative because it meets the demand for cleaner processes and is more selective compared to traditional chemical catalysts using NaOH, KOH or sodium methoxide. Another advantage of using lipases as catalyst for biodiesel production is that different alcohols can be applied as feedstock, such as methanol, ethanol, propanol, isopropanol, butanol and isobutanol, and also free fatty acids may be present.118,119The use of immobilized lipases in biodiesel production is a known methodology, since it shows great potential for industrial application. In the literature, numerous lipase immobilization strategies for use in biodiesel production are described. Reactors like stirred tank, packed-bed, airlift and other heterogeneous reactors are used in transesterification reactions using immobilized enzymes for biodiesel production.120
Cellulases and lipases are the primary candidates for large-scale implementation of enzymatic biofuel production.121 MacArio et al.122 reported the application of encapsulated liposome and lipase from Rhizomucor miehei on hybrid-nanospheres with 90% of immobilization efficiency for biodiesel production. The authors used commercial triolein (60%) as substrate, prepared heterogeneous biocatalysts (10 wt% with respect to triolein) and methanol (molar ratio oil![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol 1
methanol 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6). The reaction system was stirred at 350 rpm and kept at 37 °C. The immobilized enzyme kept the activity after 5 reactions cycle, with biodiesel yield between 89% and 98%.
6). The reaction system was stirred at 350 rpm and kept at 37 °C. The immobilized enzyme kept the activity after 5 reactions cycle, with biodiesel yield between 89% and 98%.
Raita et al.44 also reported the use of lipase for biodiesel production. The authors used immobilized Thermomyces lanuginosus lipase on magnetic nanoparticle using different covalent linkage (as seen in Fig. 6). For the standard reaction, 250 mg of refined palm oil (RPO) and methanol was reacted in a molar ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 MeOH/FFAs in the presence of 1
1 MeOH/FFAs in the presence of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) t-BuOH to RPO. The magnetic nanoparticle lipase was added at 20% (w/w based on RPO). The reaction was kept at 50 °C for 6–24 h at 40 rpm. Central composite design was used to optimize the reaction, which identified the following optimal parameters: 23.2% w/w enzyme loading and 4.7
1 (v/v) t-BuOH to RPO. The magnetic nanoparticle lipase was added at 20% (w/w based on RPO). The reaction was kept at 50 °C for 6–24 h at 40 rpm. Central composite design was used to optimize the reaction, which identified the following optimal parameters: 23.2% w/w enzyme loading and 4.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol to FFAs molar ratio with 3.4% water content in the presence of 1
1 methanol to FFAs molar ratio with 3.4% water content in the presence of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) tert-butanol to palm oil, leading to 97.2% FAME yield after incubation at 50 °C for 24 h. The biocatalyst was recycled for at least 5 consecutive recycles with 80% of activity remaining.
1 (v/v) tert-butanol to palm oil, leading to 97.2% FAME yield after incubation at 50 °C for 24 h. The biocatalyst was recycled for at least 5 consecutive recycles with 80% of activity remaining.
Although the use of immobilized enzymes nanostructures is promising, the improvement in the activity and stability of enzymes for hydrolysis and esterification reactions, the use of nanomaterial-bound enzyme-catalyzed biofuel production is necessary. The literature also point that the use of co-immobilization of multienzymes in nanomaterials could facilitate the application of various enzymes in hydrolyzing complex substrates for biofuel production.121,123
3.3 Synthesis of flavor esters
Esters are important organic compounds obtained by chemical synthesis (esterification, transesterification or interesterification) or derived from some natural products. It is known that enzymatic processes are conducted at mild conditions of pressure, temperature and pH compared to processes using inorganic catalysts.124,125 The advantages far commented in the use of immobilized enzymes on nanomaterials are also mentioned for esters production, and lipases are the most used enzymes in this area. An important advantage is that the esters synthesized by fermentation or using enzyme as a catalyst can be considered as natural, becoming commercially attractive.125,126 The synthesis of an important flavor and fragrance ester compound used in food, pharmaceutical and cosmetic industries was reported by Gupta et al.127 They studied Thermomyces lanuginosus lipase (TLL) immobilized on electrospun polyacrylonitrile nanofiber membrane (PANNFM) for geranyl acetate synthesis using geraniol, acyl donor, and vinyl acetate as substrates in an organic media. TLL enzyme was immobilized by physical adsorption and covalent bonding on the support. The optimum conditions for immobilization in both cases were 90–150 min, 45 °C, and protein concentration of 2 mg mL−1, achieving conversion values of 90% in the physical adsorption case and 66% for covalent bonding technique showing higher operational stability.Mahmood and coauthors27 synthetized ethyl isovalerate derivative from valeric acid, mainly found in fruits (one of the principal component of blueberry). They immobilized Candida rugosa lipase on gum arabic coated magnetic Fe3O4 nanoparticles (GAMNP). For this purpose, the enzyme surface was initially coated with different surfactants to stabilize enzyme in its open form, and then immobilized on the support. The authors affirm that this immobilization protocol improves enzyme activity and stability for enhanced ethyl isovalerate synthesis.
Guncheva et al.128 studied the synthesis of isoamyl acetate (banana flavor) by of Candida rugosa lipase immobilization on nanostructured tin dioxide (nano-SnO2-CRL). The immobilization parameters were compared with the same enzyme on polypropylene (PP-CRL). According to the results nano-SnO2-CRL has shown a specific activity eight times higher than that found for PP-CRL. The obtained results showed that the use of nanostructured tin dioxide result in a derivative more tolerant toward the reaction medium and can be applied in synthetic reactions in the presence of organic solvents.
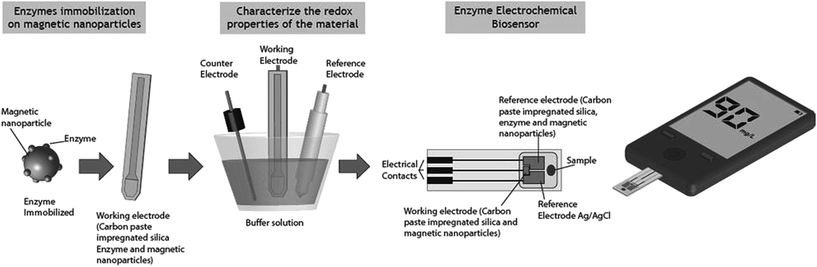
3.4 Biosensors
Biosensors also called bioelectrodes in the 80s, enzymatic electrodes or biocatalytic membrane electrodes, has attracted the interest of the scientific community for the selection of the most important analytical technologies and clearly the progress in the miniaturization of the materials.129 Only in 2007, after US investment in approximately $11 billion in research and development of a variety of applications (biodefense, medical and pharmaceutical research, food and beverage and environmental monitoring) is that the biosensors were properly valued. The biosensor was first proposed in 1962 by Clark and Lyons, an enzymatic amperometric biosensor used for glucose detecting via enzyme glucose oxidase (GOx).130,131 Because of the versatility of biosensors, there was a significant increase in their use in a varied field of science in recent decades.Biosensors are analytical tools that use a bioactive element (enzymes, antibodies, DNA, microorganism, fabric, organelles) and an electrical transducer for the detection or quantification of substances in various fields of knowledge, for example: disease diagnosis and environmental monitoring.130–133 The purpose of the biosensor is to produce an electrical signal that is proportional in magnitude or frequency to the concentration of analyte. The biolayer including bioreceptor element is immobilized on the biosensor substrate, usually nanoparticle, clay or polymers.134–136 The immobilization plays an important role in determining the overall performance of a biosensor. An interesting review published by Holzinger and coauthors137 report some particles used in biosensors, such as gold nanoparticles, semi-conductor quantum dots, polymer nanoparticles, carbon nanotubes, nanodiamonds, and graphene.
Significant progress was achieved in synthetic approaches to prepare nanomaterials with desired properties, such as controllable size, shape, surface charge and physicochemical characteristics. These features make it possible to integrate nanomaterials to biosensors for any required function, a fact that has led to an increased use of nanomaterials in biosensors, especially the electrochemical.138–143
When the analyte contacts the bioreceptor immobilized on the surface of the biosensor produces a physical-chemical modification (e.g. changing the concentration of protons, gas emissions, emission or absorption of light, heat release, increased receptor mass and/or alteration of the analyte oxidation state), which are read and processed by the converter into a measurable signal (e.g. variation in current, potential, heat resistance, refractive index, capacitance, etc.) that can be identified by an electric transducer. The resulting electrical signal is then acquired and processed, and then the data acquisition system informs the user whether analyte is detected or not and its sample concentration (Fig. 9).144–146
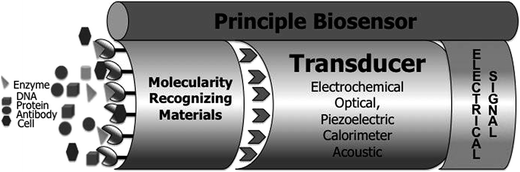
The biological element recognition or bioreceptor is the most important component of the biosensor device. The bioreceptor is the key for specificity and is classified according to several different groups as shown in the Fig. 10. Its function is to transmit selectivity for the biosensor. Generally, the major classes of biosensors are distinguished from another by the process of nature and in terms of its biological or biochemical component, e.g. biocatalytic (enzyme), immune (antibody) and nucleic acid (DNA).145,147–153
Among the various biosensors, enzyme electrochemical used in the diagnostic area is the most commercialized, powered by glucose sensors for medical and food purposes. Although other major markets are being envisioned, such as nucleic acid biosensor for DNA detection, detection of lactate, cholesterol, ethanol, mycotoxins and heavy metals.148,154–157
Many studies have been devoted to improve the electron transfer rates to increase the electrochemical efficacy of enzymes.139,148,152,153 However, many have not been focused on stabilizing the enzyme activity, which is critical point for successful use of enzymatic electrodes142,158,.
Zhang et al. (2010)159 constructed a biosensor by electrochemical adsorption of glucose oxidase in microporous polyacrylonitrile. However, the sensitivity and stability of the obtained biosensor were not satisfactory, probably due to the hydrophobicity of the host organic polymer.160 Recently, an ultra-sensitive cholesterol biosensor was developed using ZnO nanostructure, in which cholesterol oxidase (ChOx) was immobilized to the surface of modified electrode via physical adsorption followed by covering of Nafion solution. Such biosensor exhibited a very high and reproducible sensitivity of 61.7 μA cm−2 mM with a Michaelis–Menten constant (KM) of 2.57 mM and fast response time of 5 s.161,162
There are many challenges faced towards practical applications of biosensors. For example, the construction of a biosensor with a low cost is still essential when considering commercial devices. The main field of application of biosensors is still the medical diagnostic devices for commercial. Biosensors in other areas, such as food and ecology industry, needed to be explored more deeply. There are also challenges to find ways to improve the performance criteria including high sensitivity, wider linear range, lower limit of detection, rapid response and repetitive. The research now still retains continuing to investigate the most effective ways to build electrochemical biosensors based on enzymes with more perfect performance.
The development of biosensors based on enzyme immobilization appeared to solve various problems, such as loss of enzyme (especially for enzymes with a high cost), maintenance of enzyme stability and shelf life of the biosensor, and additionally to reduce the enzymatic response time and provide disposable devices capable of being easily used in stationary systems or continuous flow systems.163 Face new challenges and believe in advancing the development of nanotechnology also in the area of nanoscale optical fibers can be the solution to many problems faced today, especially the biosensor response time.
4. Trends
Enzyme immobilization remains an area of great interest both, in scientific community or in industrial sector. The advantages combined to choose the ideal technique to enzyme immobilization, and their interaction with the substrate must be taken into consideration in applying the method of immobilization, as well as the reaction that it will be applied and the costs involved in the final product. In addition, with the growing attention paid to cascade enzymatic reaction and in vitro synthetic biology, it is possible that co-immobilization of multi-enzymes could be achieved on these nanoparticles.9 With the advancement of the media development of techniques, many materials and combinations of materials have emerged as promising options in immobilization of enzymes.164c,d,eThe universe of possibilities to enzyme immobilization is so vast that it is impossible to cite all existing types of particles and procedures used in immobilization. We highlight some of them in this review. Although there are many works, the interest is growing, which can be seen in search engines due the special interesting features related to the nanoparticles. Improve biocatalyst features in order to increase productivity in the final reaction and process cost reduction is main goals of immobilization. Combine an efficient catalyst in a process that stimulates the production can improve the obtaining interest product. The authors believe that the choice of nanoparticle as support to be used depends directly of the reaction in which it will be used. In addition, acquaintance of the enzyme properties in question, enzyme stability and process characteristics, as pH and temperature, are very important for further application. Considering the aspects cited in this review, researchers should consider the use of immobilized enzyme on nanoparticles in their reactions.
Acknowledgements
The authors thank the financial support and scholarships from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). MINECO (Spanish Goberment) (projects numbers CTQ2013-41507-R and CTQ2016-78587-R) is also gratefully recognized.References
- R. DiCosimo, J. McAuliffe, A. J. Poulose and G. Bohlmann, Chem. Soc. Rev., 2013, 42, 6437–6474 RSC.
- R. A. Sheldon and S. van Pelt, Chem. Soc. Rev., 2013, 42, 6223–6235 RSC.
- F. Secundo, Chem. Soc. Rev., 2013, 42, 6250–6261 RSC.
- H. Vaghari, H. Jafarizadeh-Malmiri, M. Mohammadlou, A. Berenjian, N. Anarjan, N. Jafari and S. Nasiri, Biotechnol. Lett., 2015, 38, 223–233 CrossRef PubMed.
- (a) K. Hernandez and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2011, 48, 107–122 CrossRef CAS PubMed; (b) O. Barbosa, R. Torres, C. Ortiz, A. Berenguer-Murcia, R. C. Rodrigues and R. Fernandez-Lafuente, Biomacromolecules, 2013, 2013(14), 2433–2462 CrossRef PubMed.
- (a) R. Fernandez-Lafuente, Enzyme Microb. Technol., 2009, 45, 405–418 CrossRef CAS; (b) C. Garcia-Galan, A. Berenguer-Murcia, R. Fernandez-Lafuente and R. C. Rodrigues, Adv. Synth. Catal., 2011, 353, 2885–2904 CrossRef CAS; (c) O. Barbosa, C. Ortiz, A. Berenguer-Murcia, R. Torres, R. C. Rodrigues and R. Fernandez-Lafuente, RSC Adv., 2014, 4, 1583–1600 RSC; (d) R. C. Rodrigues, O. Barbosa, C. Ortiz, A. Berenguer-Murcia, R. Torres and R. Fernandez-Lafuente, RSC Adv., 2014, 4, 38350–38374 RSC.
- O. Barbosa, C. Ortiz, Á. Berenguer-Murcia, R. Torres, R. C. Rodrigues and R. Fernandez-Lafuente, Biotechnol. Adv., 2015, 33, 435–456 CrossRef CAS PubMed.
- T. M. D. S. Bezerra, J. C. Bassan, V. T. D. O. Santos, A. Ferraz and R. Monti, Process Biochem., 2014, 50, 417–423 CrossRef.
- S. A. Ansari and Q. Husain, Biotechnol. Adv., 2012, 30, 512–523 CrossRef CAS PubMed.
- M. L. Verma, M. Puri and C. J. Barrow, Crit. Rev. Biotechnol., 2016, 36, 108–119 CrossRef CAS PubMed.
- C. Hu, N. Wang, W. Zhang, S. Zhang, Y. Meng and X. Yu, J. Biotechnol., 2015, 194, 12–18 CrossRef CAS PubMed.
- E. P. Cipolatti, M. J. A. Silva, M. Klein, V. Feddern, M. M. C. Feltes, J. V. Oliveira, J. L. Ninow and D. de Oliveira, J. Mol. Catal. B: Enzym., 2014, 99, 56–67 CrossRef CAS.
- S. Ghosh, S. R. Chaganti and R. S. Prakasham, J. Mol. Catal. B: Enzym., 2012, 74, 132–137 CrossRef CAS.
- B. Sahoo, S. K. Sahu and P. Pramanik, J. Mol. Catal. B: Enzym., 2011, 69, 95–102 CrossRef CAS.
- Z. X. Tang, J. Q. Qian and L. E. Shi, Mater. Lett., 2007, 61, 37–40 CrossRef CAS.
- S. A. Ansari and Q. Husain, Biotechnol. Adv., 2012, 30, 512–523 CrossRef CAS PubMed.
- Y. Cao, L. Wen, F. Svec, T. Tan and Y. Lv, Chem. Eng. J., 2016, 286, 272–281 CrossRef CAS.
- L. Lu, M. Zhao and Y. Wang, World J. Microbiol. Biotechnol., 2007, 23, 159–166 CrossRef CAS.
- A. Valério, G. Nicoletti, E. P. Cipolatti, J. L. Ninow, P. H. H. Araújo, C. Sayer and D. de Oliveira, Appl. Biochem. Biotechnol., 2015, 175, 2961–2971 CrossRef PubMed.
- E. Omidinia, N. Shadjou and M. Hasanzadeh, Mater. Sci. Eng., C, 2014, 42, 368–373 CrossRef CAS PubMed.
- G. A. Kovalenko, A. B. Beklemishev, L. V. Perminova, A. L. Mamaev, N. A. Rudina, S. I. Moseenkov and V. L. Kuznetsov, J. Mol. Catal. B: Enzym., 2013, 98, 78–86 CrossRef CAS.
- F. Nabati, M. Habibi-Rezaei, M. Amanlou and A. Moosavi-Movahedi, J. Mol. Catal. B: Enzym., 2011, 70, 17–22 CrossRef CAS.
- X. Yan, X. Wang, P. Zhao, Y. Zhang, P. Xu and Y. Ding, Microporous Mesoporous Mater., 2012, 161, 1–6 CrossRef CAS.
- H. Qiu, Y. Li, G. Ji, G. Zhou, X. Huang, Y. Qu and P. Gao, Bioresour. Technol., 2009, 100, 3837–3842 CrossRef CAS PubMed.
- I. V. Pavlidis, M. Patila, U. T. Bornscheuer, D. Gournis and H. Stamatis, Trends Biotechnol., 2014, 32, 312–320 CrossRef CAS PubMed.
- A. A. Gokhale, J. Lu and I. Lee, J. Mol. Catal. B: Enzym., 2013, 90, 76–86 CrossRef CAS.
- I. Mahmood, I. Ahmad, G. Chen and L. Huizhou, Biochem. Eng. J., 2013, 73, 72–79 CrossRef CAS.
- Y. Liu, S. Jia, Q. Wu, J. Ran, W. Zhang and S. Wu, Catal. Commun., 2011, 12, 717–720 CrossRef CAS.
- M. Rizwan, A. Buthe, M. Hamid and P. Wang, Food Chem., 2016, 190, 1078–1085 CrossRef PubMed.
- E. P. Cipolatti, M. J. a Silva, M. Klein, V. Feddern, M. M. C. Feltes, J. V. Oliveira, J. L. Ninow and D. De Oliveira, J. Mol. Catal. B: Enzym., 2014, 99, 56–67 CrossRef CAS.
- J. Zhu and G. Sun, React. Funct. Polym., 2012, 72, 839–845 CrossRef CAS.
- A. Takimoto, T. Shiomi, K. Ino, T. Tsunoda, A. Kawai, F. Mizukami and K. Sakaguchi, Microporous Mesoporous Mater., 2008, 116, 601–606 CrossRef CAS.
- L. Betancor, M. Fuentes, G. Dellamora-Ortiz, F. López-Gallego, A. Hidalgo, N. Alonso-Morales, C. Mateo, J. M. Guisán and R. Fernández-Lafuente, J. Mol. Catal. B: Enzym., 2005, 32, 97–101 CrossRef CAS.
- L. Betancor, F. López-Gallego, A. Hidalgo, N. Alonso-Morales, M. Fuentes, R. Fernández-Lafuente and J. M. Guisán, J. Biotechnol., 2004, 110, 201–207 CrossRef CAS PubMed.
- (a) C. Mateo, J. M. Palomo, G. Fernandez-Lorente, J. M. Guisan and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2007, 40, 1451–1463 CrossRef CAS; (b) R. C. Rodrigues, C. Ortiz, Á. Berenguer-Murcia, R. Torres and R. Fernández-Lafuente, Chem. Soc. Rev., 2013, 42, 6290–6307 RSC.
- (a) J. M. Guisan, A. Alvaro, C. M. Rosell and R. Fernandez-Lafuente, Biotechnol. Appl. Biochem., 1994, 20, 357–369 CrossRef CAS PubMed; (b) J. Rocha-Martín, B. L. Rivas, R. Muñoz, J. M. Guisán and F. López-Gallego, ChemCatChem, 2012, 4, 1279–1288 CrossRef.
- G. B. Sergeev and T. I. Shabatina, Surf. Sci., 2002, 500, 628–655 CrossRef CAS.
- K. Landfester, N. Bechthold, F. Tiarks and M. Antonietti, Macromolecules, 1999, 32, 5222–5228 CrossRef CAS.
- D. Shi, H. Yang, S. Ji, S. Jiang, X. Liu and D. Zhang, Procedia Eng., 2015, 102, 1555–1562 CrossRef CAS.
- J. Kim, J. W. Grate and P. Wang, Chem. Eng. Sci., 2006, 61, 1017–1026 CrossRef CAS.
- D. Yang, X. Wang, J. Shi, X. Wang, S. Zhang, P. Han and Z. Jiang, Biochem. Eng. J., 2015, 105, 273–280 CrossRef.
- C.-Y. Shang, W.-X. Li and R.-F. Zhang, Mater. Res. Bull., 2015, 68, 336–342 CrossRef CAS.
- C. Hu, N. Wang, W. Zhang, S. Zhang, Y. Meng and X. Yu, J. Biotechnol., 2015, 194, 12–18 CrossRef CAS PubMed.
- M. Raita, J. Arnthong, V. Champreda and N. Laosiripojana, Fuel Process. Technol., 2015, 134, 189–197 CrossRef CAS.
- N. R. Mohamad, N. A. Buang, N. A. Mahat, Y. Y. Lok, F. Huyop, H. Y. Aboul-Enein and R. Abdul Wahab, Enzyme Microb. Technol., 2015, 72, 49–55 CrossRef CAS PubMed.
- H. Fei, G. Xu, J.-P. Wu and L.-R. Yang, J. Mol. Catal. B: Enzym., 2014, 101, 87–91 CrossRef CAS.
- K. Thandavan, S. Gandhi, S. Sethuraman, J. B. B. Rayappan and U. M. Krishnan, Sens. Actuators, B, 2013, 176, 884–892 CrossRef CAS.
- S. A. Ansari, R. Satar, S. Chibber and M. J. Khan, J. Mol. Catal. B: Enzym., 2013, 97, 258–263 CrossRef CAS.
- V. Swarnalatha, R. A. Esther and R. Dhamodharan, J. Mol. Catal. B: Enzym., 2013, 96, 6–13 CrossRef CAS.
- M. L. Verma, R. Chaudhary, T. Tsuzuki, C. J. Barrow and M. Puri, Bioresour. Technol., 2013, 135, 2–6 CrossRef CAS PubMed.
- P. Villeneuve, J. M. Muderhwa, J. Graille and M. J. Haas, J. Mol. Catal. B: Enzym., 2000, 9, 113–148 CrossRef CAS.
- M. D. Besteti, A. G. Cunha, D. M. G. Freire and J. C. Pinto, Macromol. Mater. Eng., 2014, 299, 135–143 CrossRef CAS.
- (a) Z. Zhou and M. Hartmann, Chem. Soc. Rev., 2013, 42, 3894–3912 RSC; (b) M. Hartmann and X. Kostrov, Chem. Soc. Rev., 2013, 42, 6277–6289 RSC; (c) E. Magner, Chem. Soc. Rev., 2013, 42, 6213–6222 RSC; (d) C. Garcia-Galan, O. Barbosa, K. Hernandez, J. C. S. Dos Santos, R. C. Rodrigues and R. Fernandez-Lafuente, Molecules, 2014, 19, 7629–7645 CrossRef PubMed.
- G. Nicoletti, E. P. Cipolatti, A. Valério, N. G. Carbonera, N. S. Soares, E. Theilacker, J. L. Ninow and D. de Oliveira, Bioprocess Biosyst. Eng., 2015, 1739–1748 CrossRef CAS PubMed.
- E. P. Cipolatti, A. Valério, G. Nicoletti, E. Theilacker, P. H. H. Araújo, C. Sayer, J. L. Ninow and D. de Oliveira, J. Mol. Catal. B: Enzym., 2014, 109, 116–121 CrossRef CAS.
- H. Chen, J. Zhang, Y. Dang and G. Shu, J. Chem. Pharm. Res., 2014, 6, 612–616 Search PubMed.
- M. P. Klein, M. R. Nunes, R. C. Rodrigues, E. V. Benvenutti, T. M. H. Costa, P. F. Hertz and J. L. Ninow, Biomacromolecules, 2012, 13, 2456–2464 CrossRef CAS PubMed.
- R. S. Juang, F. C. Wu and R. L. Tseng, Bioresour. Technol., 2001, 80, 187–193 CrossRef CAS PubMed.
- E. Biró, A. S. Németh, C. Sisak, T. Feczkó and J. Gyenis, J. Biochem. Biophys. Methods, 2008, 70, 1240–1246 CrossRef PubMed.
- Y. Wu, Y. Wang, G. Luo and Y. Dai, Bioresour. Technol., 2010, 101, 841–844 CrossRef CAS PubMed.
- L. Zang, J. Qiu, X. Wu, W. Zhang, E. Sakai and Y. Wei, Ind. Eng. Chem. Res., 2014, 53, 3448–3454 CrossRef CAS.
- R. G. López, M. G. Pineda, G. Hurtado, R. D. de León, S. Fernández, H. Saade and D. Bueno, Int. J. Mol. Sci., 2013, 14, 19636–19650 CrossRef PubMed.
- S. K. Shukla, A. K. Mishra, O. A. Arotiba and B. B. Mamba, Int. J. Biol. Macromol., 2013, 59, 46–58 CrossRef CAS PubMed.
- J. Zhu and G. Sun, React. Funct. Polym., 2012, 72, 839–845 CrossRef CAS.
- M. Asgher, M. Shahid, S. Kamal and H. M. N. Iqbal, J. Mol. Catal. B: Enzym., 2014, 101, 56–66 CrossRef CAS.
- C. G. C. M. Netto, H. E. Toma and L. H. Andrade, J. Mol. Catal. B: Enzym., 2013, 85–86, 71–92 CrossRef CAS.
- M. Shinkai and A. Ito, Adv. Biochem. Eng./Biotechnol., 2004, 91, 191–220 CrossRef CAS PubMed.
- C. G. C. M. Netto, H. E. Toma and L. H. Andrade, J. Mol. Catal. B: Enzym., 2013, 85–86, 71–92 CrossRef CAS.
- X. Y. Jiang, S. Bai and Y. Sun, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 852, 62–68 CrossRef CAS PubMed.
- W. Xie and N. Ma, Biomass Bioenergy, 2010, 34, 890–896 CrossRef CAS.
- I. Pospiskova and K. Safarik, J. Magn. Magn. Mater., 2015, 380, 197–200 CrossRef.
- A. Soozanipour, A. Taheri-Kafrani and A. Landarani Isfahani, Chem. Eng. J., 2015, 270, 235–243 CrossRef CAS.
- J. Lu, R. R. Weerasiri and I. Lee, Biotechnol. Lett., 2013, 35, 181–188 CrossRef CAS PubMed.
- C. M. F. Carvalho, N. B. Lima and A. S. Soares, Quim. Nova, 2015, 38, 399–409 Search PubMed.
- P. Liu, J. Pang, H. Yin and S. Ai, Anal. Chim. Acta, 2015, 879, 34–40 CrossRef CAS PubMed.
- J. R. Deka, D. Saikia, Y. Lai, C. Tsai, W. Chang and H. Kao, Microporous Mesoporous Mater., 2015, 213, 150–160 CrossRef CAS.
- M. Guncheva, K. Paunova, M. Dimitrov and D. Yancheva, J. Mol. Catal. B: Enzym., 2014, 108, 43–50 CrossRef CAS.
- M. Mohammadi, Z. Habibi, S. Dezvarei, M. Yousefi, S. Samadi and M. Ashjari, Process Biochem., 2014, 49, 1314–1323 CrossRef CAS.
- S. Talekar, V. Ghodake, T. Ghotage, P. Rathod, P. Deshmukh, S. Nadar, M. Mulla and M. Ladole, Bioresour. Technol., 2012, 123, 542–547 CrossRef CAS PubMed.
- W. Kopp, T. P. da Costa, S. C. Pereira, M. Jafelicci Jr, R. C. Giordano, R. F. C. Marques, F. M. Araújo-Moreira and R. L. C. Giordano, Process Biochem., 2014, 49, 38–46 CrossRef CAS.
- K. Jafari Khorshidi, H. Lenjannezhadian, M. Jamalan and M. Zeinali, J. Chem. Technol. Biotechnol., 2016, 91, 539–546 CrossRef CAS.
- S. Shaarani, J. Jahim, R. A. Rahman, A. Idris, A. Munir, A. Murad and R. Illias, J. Mol. Catal. B: Enzym., 2016, 133, 65–76 CrossRef CAS.
- Y. Liu, C. Guo and C. Z. Liu, Chem. Eng. J., 2015, 280, 36–40 CrossRef CAS.
- W.-W. Zhang, X.-L. Yang, J.-Q. Jia, N. Wang, C.-L. Hu and X.-Q. Yu, J. Mol. Catal. B: Enzym., 2015, 115, 83–89 CrossRef CAS.
- Á. Cruz-Izquierdo, E. a Picó, C. López, J. L. Serra and M. J. Llama, PLoS One, 2014, 9, e115202 Search PubMed.
- A. Bhattacharya and B. I. Pletschke, Enzyme Microb. Technol., 2014, 55, 159–169 CrossRef CAS PubMed.
- S.-K. Hsu, H.-C. Hsu, W.-F. Ho, C.-H. Yao, P.-L. Chang and S.-C. Wu, Thin Solid Films, 2014, 572, 91–98 CrossRef CAS.
- R. Reshmi, G. Sanjay and S. Sugunan, Catal. Commun., 2007, 8, 393–399 CrossRef CAS.
- M. Huckel, H.-J. Wirth and M. T. W. Hearn, J. Biochem. Biophys. Methods, 1996, 31, 165–179 CrossRef CAS PubMed.
- A. Clearfield, G. P. D. Serrette and A. H. Khazi-Syed, Catal. Today, 1994, 20, 295–312 CrossRef CAS.
- Y. Z. Chen, C. B. Ching and R. Xu, Process Biochem., 2009, 44, 1245–1251 CrossRef CAS.
- Y. Masuda, S. Kugimiya and K. Kato, Journal of Asian Ceramic Societies, 2014, 2, 11–19 CrossRef.
- S. Yan, X. Chen, J. Wu and P. Wang, Appl. Microbiol. Biotechnol., 2012, 94, 829–838 CrossRef CAS PubMed.
- A. Homaei and D. Saberi, Process Biochem., 2015, 50, 1394–1399 CrossRef CAS.
- C. Cheng and K. C. Chang, Enzyme Microb. Technol., 2013, 53, 444–451 CrossRef CAS PubMed.
- I. Venditti, C. Palocci, L. Chronopoulou, I. Fratoddi, L. Fontana, M. Diociaiuti and M. Vittoria, J. Chem. Technol. Biotechnol., 2015, 131, 93–101 CAS.
- R. Mas-Ballesté, C. Gómez-Navarro, J. Gómez-Herrero and F. Zamora, Nanoscale, 2011, 3, 20–30 RSC.
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, M. I. Katsnelson, I. V. Grigorieva, S. V. Dubonos and A. A. Firsov, Nature, 2005, 438, 197–200 CrossRef CAS PubMed.
- K. S. K. S. Novoselov, A. K. A. K. Geim, S. V. S. V. Morozov, D. Jiang, Y. Zhang, S. V. V. Dubonos, I. V. V Grigorieva and A. Firsov, Science, 2004, 306, 666–669 CrossRef CAS PubMed.
- Y. Wang, R. Yuan, Y. Chaia, W. Li, Y. Zhuo, Y. Yuan and J. Li, J. Mol. Catal. B: Enzym., 2011, 71, 146–151 CrossRef CAS.
- S. K. Vashist and J. H. T. Luong, Carbon, 2014, 4, 519–550 Search PubMed.
- B. Jiang, K. Yang, L. Zhang, Z. Liang, X. Peng and Y. Zhang, Talanta, 2014, 122, 278–284 CrossRef CAS PubMed.
- A. A. Gokhale, J. Lu and I. Lee, J. Mol. Catal. B: Enzym., 2013, 90, 76–86 CrossRef CAS.
- X. Wang and X. Zhang, Electrochim. Acta, 2013, 112, 774–782 CrossRef CAS.
- C. Shan, H. Yang, J. Song, D. Han, A. Ivaska and L. Niu, Anal. Chem., 2009, 81, 2378–2382 CrossRef CAS PubMed.
- (a) U. Guzik, K. Hupert-Kocurek and D. Wojcieszyńska, Molecules, 2014, 19, 8995–9018 CrossRef PubMed; (b) F. Hu, S. Chen, C. Wang, R. Yuan, Y. Chai, Y. Xiang and C. Wang, J. Mol. Catal. B: Enzym., 2011, 72, 298–304 CrossRef CAS.
- (a) X. Zhu, I. Yuri, X. Gan, I. Suzuki and G. Li, Biosens. Bioelectron., 2007, 22, 1600–1604 CrossRef CAS PubMed; (b) M. Hu, K. P. Giapis and D. Poulikakos, Appl. Phys. Lett., 2011, 98, 211904 CrossRef.
- (a) J. Ge, J. Lei and R. N. Zare, Nat. Nanotechnol., 2012, 7, 428–432 CrossRef CAS PubMed; (b) X. Wu, M. Hou and J. Ge, Catal. Sci. Technol., 2015, 5, 5077–5508 RSC; (c) C. Altinkaynak, S. Tavlasoglu, N. Özdemir and I. Ocsoy, Enzyme Microb. Technol., 2016, 93–94, 105–112 CrossRef CAS PubMed; (d) J.-Y. Lee, J. Han, J. Lee, S. Ji and J.-S. Yeo, Nanoscale Res. Lett., 2015, 10, 1–9 CrossRef PubMed; (e) M. Y. Ge, L. Y. Han, U. Wiedwald, X. B. Xu, C. Wang, K. Kuepper, P. Ziemann and J. Z. Jiang, Nanotechnology, 2010, 21, 425702 CrossRef CAS PubMed.
- X. Wang, J. Shi, Z. Li, S. Zhang, H. Wu, Z. Jiang, C. Yang and C. Tian, ACS Appl. Mater. Interfaces, 2014, 6, 14522–14532 CAS.
- K. Min and Y. J. Yoo, Biotechnol. Bioprocess Eng., 2014, 19, 553–567 CrossRef CAS.
- E. T. Hwang and M. B. Gu, Eng. Life Sci., 2013, 13, 49–61 CrossRef CAS.
- R. E. Abraham, M. L. Verma, C. J. Barrow and M. Puri, Biotechnol. Biofuels, 2014, 7, 90 CrossRef PubMed.
- C. Rafael, F. Terrasan, E. P. Cipolatti, L. T. De, A. Souza, R. O. Henriques, S. Moreno-pérez, W. D. M. Junior, A. O. Chioma and J. M. Guisan, Mycology: Current and Future Developments, ed. E. N. Silva, Benttham e books, 2015, vol. 1, ch. 10, pp. 276–315 Search PubMed.
- Q. Zhang, J. Kang, B. Yang, L. Zhao, Z. Hou and B. Tang, Chin. J. Catal., 2016, 37, 389–397 CrossRef CAS.
- U. V. Sojitra, S. S. Nadar and V. K. Rathod, Food Chem., 2016, 213, 296–305 CrossRef CAS PubMed.
- C. Cheng and K. C. Chang, Enzyme Microb. Technol., 2013, 53, 444–451 CrossRef CAS PubMed.
- H. Liao, D. Chen, L. Yuan, M. Zheng, Y. Zhu and X. Liu, Carbohydr. Polym., 2010, 82, 600–604 CrossRef CAS.
- C. M. T. Santin, R. P. Scherer, N. L. D. Nyari, C. D. Rosa, R. M. Dallago, D. de Oliveira and J. V. Oliveira, Biocatal. Agric. Biotechnol., 2014, 3, 90–94 Search PubMed.
- F. Hasan, A. A. Shah and A. Hameed, Enzyme Microb. Technol., 2006, 39, 235–251 CrossRef CAS.
- X. Zhao, F. Qi, C. Yuan, W. Du and D. Liu, Renewable Sustainable Energy Rev., 2015, 44, 182–197 CrossRef CAS.
- M. Puri, C. J. Barrow and M. L. Verma, Trends Biotechnol., 2013, 31, 215–216 CrossRef CAS PubMed.
- A. MacArio, F. Verri, U. Diaz, A. Corma and G. Giordano, Catal. Today, 2013, 204, 148–155 CrossRef CAS.
- S. Duraiarasan, S. A. Razack, A. Manickam, A. Munusamy, M. B. Syed, M. Y. Ali, G. M. Ahmed and M. S. Mohiuddin, Direct conversion of lipids from marine microalga C. salina to biodiesel with immobilised enzymes using magnetic nanoparticle, 2016, vol. 4 Search PubMed.
- R. Dalla-Vecchia, M. D. G. Nascimento and V. Soldi, Quim. Nova, 2004, 27, 623–630 CrossRef CAS.
- E. Skoronski, T. M. Bonetti, J. J. João and A. Fúrigo Júnior, Cienc. Tecnol. Aliment., 2010, 30, 897–902 CrossRef.
- H. Abbas and L. Comeau, Enzyme Microb. Technol., 2003, 32, 589–595 CrossRef CAS.
- A. Gupta, S. R. Dhakate, M. Pahwa, S. Sinha, S. Chand and R. B. Mathur, Process Biochem., 2013, 48, 124–132 CrossRef CAS.
- M. Guncheva, M. Dimitrov and D. Zhiryakova, Process Biochem., 2011, 46, 2170–2177 CrossRef CAS.
- M. A. Arnold and M. E. Meyerhoff, Anal. Chem., 1984, 56(5), 20–48 CrossRef.
- D. Grieshaber, R. MacKenzie, J. Vörös and E. Reimhult, Sensors, 2008, 8, 1400–1458 CrossRef CAS.
- P. T. Kissinger, Biosens. Bioelectron., 2005, 20, 2512–2516 CrossRef CAS PubMed.
- W. Putzbach and N. J. Ronkainen, Sensors, 2013, 13, 4811–4840 CrossRef CAS PubMed.
- P. Rubens, G. Barrocas, A. Cláudia, S. De Vasconcellos, S. Duque, L. Maria, S. Couto, A. L. Lauria-filgueiras and J. C. Moreira, Cadernos Saúde Coletiva, 2008, 16, 677–700 Search PubMed.
- B. D. Malhotra and A. Chaubey, Sens. Actuators, B, 2003, 91, 117–127 CrossRef CAS.
- C. Dai and S. Choi, Open J. Appl. Biosens., 2013, 2, 83–93 CrossRef.
- B. W. Park, D. Y. Yoon and D. S. Kim, Biosens. Bioelectron., 2010, 26, 1–10 CrossRef CAS PubMed.
- M. Holzinger, A. Le Goff and S. Cosnier, Front. Chem., 2014, 2, 1–10 CAS.
- V. Perumal and U. Hashim, J. Appl. Biomed., 2014, 12, 1–15 CrossRef.
- D. Baratella, M. Magro, G. Sinigaglia, R. Zboril, G. Salviulo and F. Vianello, Biosens. Bioelectron., 2013, 45, 13–18 CrossRef CAS PubMed.
- H. Teymourian, A. Salimi and S. Khezrian, Biosens. Bioelectron., 2013, 49, 1–8 CrossRef CAS PubMed.
- L. Stanciu, Y.-H. Won, M. Ganesana and S. Andreescu, Sensors, 2009, 9, 2976–2999 CrossRef CAS PubMed.
- X. Tan, M. Li, P. Cai, L. Luo and X. Zou, Anal. Biochem., 2005, 337, 111–120 CrossRef CAS PubMed.
- A. M. Salgado, R. O. M. Folly, B. Valdman and F. Valero, Biotechnol. Tech., 1998, 12, 305–307 CrossRef CAS.
- F. Batzias and C. G. Siontorou, J. Environ. Manage., 2007, 82, 221–239 CrossRef CAS PubMed.
- J. Castillo, S. Gáspár, S. Leth, M. Niculescu, A. Mortari, I. Bontidean, V. Soukharev, S. A. Dorneanu, A. D. Ryabov and E. Csöregi, Sens. Actuators, B, 2004, 102, 179–194 CrossRef CAS.
- K. R. Rogers, Biosens. Bioelectron., 1995, 10, 533–541 CrossRef CAS.
- M. Thompson, C. Blaszykowski, S. Sheikh and A. Romaschin, Biosens. Bioelectron., 2014, 67, 3–10 CrossRef PubMed.
- Y. Chen, L. Xiao, Y. Liu, X. Li, J. Zhang and Y. Shu, Microchim. Acta, 2014, 181, 615–621 CrossRef CAS.
- M. Cao, Z. Li, J. Wang, W. Ge, T. Yue, R. Li, V. L. Colvin and W. W. Yu, Trends Food Sci. Technol., 2012, 27, 47–56 CrossRef CAS.
- Y. Lei, W. Chen and A. Mulchandani, Anal. Chim. Acta, 2006, 568, 200–210 CrossRef CAS PubMed.
- R. L. Rich and D. G. Myszka, J. Mol. Recognit., 2011, 24, 892–914 CrossRef CAS PubMed.
- A. A Ansari, M. Alhoshan, M. S. Alsalhi and A. S. Aldwayyan, Biosensors, 2010, 23–47 Search PubMed.
- S. F. D'Souza, Appl. Biochem. Biotechnol., 2001, 96, 225–238 CrossRef.
- L. Yanxiao, Z. Xiao-bo, H. Xiao-wei, S. Ji-yong, Z. Jie-wen, M. Holmes and L. Hao, Biosens. Bioelectron., 2015, 67, 35–41 CrossRef PubMed.
- N. Zehani, S. V. Dzyadevych, R. Kherrat and N. J. Jaffrezic-Renault, Front. Chem., 2014, 2, 1–7 CAS.
- D. Montalvan-Sorrosa, J. L. González-Solis, J. Mas-Oliva and R. Castillo, RSC Adv., 2014, 4, 57329–57336 RSC.
- A. Kaushik, Open J. Appl. Biosens., 2013, 2, 1–11 CrossRef.
- (a) C. Dhand, S. P. Singh, S. K. Arya, M. Datta and B. D. Malhotra, Anal. Chim. Acta, 2007, 602, 244–251 CrossRef CAS PubMed; (b) M. D. Luque de Castro and M. C. Herrera, Biosens. Bioelectron., 2002, 18, 279–294 CrossRef.
- Y. Zhang, G. Guo, F. Zhao, Z. Mo, F. Xiao and B. Zeng, Electroanalysis, 2010, 22, 223–228 CrossRef CAS.
- R. Nenkova, D. Ivanova, J. Vladimirova and T. Godjevargova, Sens. Actuators, B, 2010, 148, 59–65 CrossRef CAS.
- Z. Zhao and H. Jiang, Biosensors, ed. P. A. Serra, INTECH, Croatia, 2010, pp. 1–23 Search PubMed.
- A. Umar, M. M. Rahman, M. Vaseem and Y. B. Hahn, Electrochem. Commun., 2009, 11, 118–121 CrossRef CAS.
- A. Hayat, G. Catanante and J. Marty, Sensors, 2014, 14, 23439–23461 CrossRef PubMed.
- (a) Y. Kuwahara, T. Yamanishi, T. Kamegawa, K. Mori, M. Che and H. Yamashita, Chem. Commun., 2012, 48, 2882–2884 RSC; (b) Y. Kuwahara, T. Yamanishi, T. Kamegawa, K. Mori and H. Yamashita, ChemCatChem, 2013, 5, 2527–2536 CrossRef CAS; (c) L. Zhang, K. Qian, X. Wang, F. Zhang, X. Shi, Y. Jiang, S. Liu, M. Jaroniec and J. Liu, Adv. Sci., 2016 Search PubMed; (d) J. Liu, B. Wang, S. Budi Hartono, T. Liu, P. Kantharidis, A. P. J. Middelberg, G. Q. M. Lu, L. He and S. Z. Qiao, Biomaterials, 2012, 33, 970–978 CrossRef CAS PubMed; (e) T. Liu, L. Qu, K. Qian, J. Liu, Q. Zhang, L. Liu and S. Liu, Chem. Commun., 2016, 52, 1709–1712 RSC.
- (a) A. Popat, S. B. Hartono, F. Stahr, J. Liu, S. Z. Qiao and G. Qing Lu, Nanoscale, 2011, 3, 2801 RSC; (b) A. Popat, B. P. Ross, J. Liu, S. Jambhrunkar, F. Kleitz and S. Z. Qiao, Angew. Chem., Int. Ed., 2012, 51, 12486–12489 CrossRef CAS PubMed; (c) Z. Zhao, J. Tian, Z. Wu, J. Liu, D. Zhao, W. Shen and L. He, J. Mater. Chem. B, 2013, 1, 4719 RSC; (d) Z. Y. Zhao, J. Liu, M. Hahn, S. Qiao, A. P. J. Middelberg and L. He, RSC Adv., 2013, 3, 22008 RSC.
| This journal is © The Royal Society of Chemistry 2016 |