Open Access Article
Open Access ArticleAmino acid recognition by fine tuning the association constants: tailored naphthalimides in pillar[5]arene-based indicator displacement assays†
Márton
Bojtár
a,
Adrien
Paudics
b,
Dóra
Hessz
bc,
Miklós
Kubinyi
bc and
István
Bitter
*a
aDepartment of Organic Chemistry and Technology, Budapest University of Technology and Economics, 1521 Budapest, Hungary. E-mail: bitter@oct.bme.hu
bDepartment of Physical Chemistry and Materials Science, Budapest University of Technology and Economics, 1521 Budapest, Hungary
cInstitute of Materials and Environmental Chemistry, Research Center for Natural Sciences, Hungarian Academy of Sciences, 1519 Budapest, P. O. B. 286, Hungary
First published on 5th September 2016
Abstract
Three aminonaphthalimide derivatives were synthesized bearing different anchoring groups in their 4-position in order to adjust the supramolecular interactions with carboxylato-pillar[5]arene (WP5), an anionic, water-soluble host. The modification of the anchor groups resulted in varying association constants embracing three orders of magnitude (Ka from ∼103 to ∼106) in buffered water. Since the fluorophore responded significantly to the electronic environment, large fluorescence quenching was observed with the anionic WP5 host. The naphthalimide indicator–WP5 supramolecular assemblies were used to detect arginine and lysine with complete selectivity over other non-basic α-amino acids by turn-on fluorescence. The same assemblies proved to be highly sensitive fluorescence displacement assays for the detection of cadaverine.
Introduction
The construction of fluorescence indicator displacement (FID) assays with macrocyclic receptors is a prosperous field of supramolecular analytical chemistry.1,2 These systems exploit the principle of competitive binding assays applied extensively in biochemistry: a receptor binds a fluorescent indicator first, creating the sensing assembly, from which the indicator is displaced by the non-fluorescent analyte.3–5 The fluorescence of the system depends on the concentrations of the bound and free forms of the indicator and thereby on the concentration of the analyte.Numerous macrocycle–dye complexes were investigated in the past decade applicable as FID assays for the detection of various analytes.4 The macrocyclic hosts in these complexes were most often cyclodextrins, calixarenes and cucurbiturils.
Pillar[n]arenes6 are a new class of macrocycles, containing n hydroquinone units linked together in p-position with a methylene bridge. In the past years, various applications were discovered mainly by the versatile host–guest chemistry of these compounds.7–17 However, these applications were mainly carried out in organic media using primarily alkyl- and functionalised pillararenes. Water soluble pillararenes containing usually ionic (carboxylato-,18,19 ammonium,20,21 imidazolium22 and recently, phosphoryl23) groups are emerging as popular hosts in aqueous media. Carboxylato-pillar[n]arenes (WPns), first synthesized by Ogoshi in 2010 (ref. 18) (n = 5) are interesting structures due to their hydrophobic, electron-rich cavity and anionic outer rim. These macrocycles are ideal hosts for ammonium-containing and electron-deficient guests.18,24–27 As was demonstrated, WP5 forms strong complexes with basic amino acids and cadaverine, the interactions were, however, monitored only by NMR-studies.28
As a first example for a chemosensor, a pillar[5]arene functionalized with a pyrene fluorophore was synthesized by Stoddart to detect diamines in acetonitrile/water mixtures.29 This was followed by FID assays with WPn hosts. The operations of a quaternary ammonium attached to tetraphenylethene30 and an N-methylacridinium31 fluorescent guest in WP6 and WP5, respectively, were demonstrated as FID assays for paraquat. Recently, we reported the complexation of 4-styryl-N-methylpyridinium iodide and two other stilbazolium dyes with WP5 in water32,33 and proposed a FID system for the colorimetric and fluorescent detection of paraquat with a detection limit of 0.2 μM. To our knowledge, no pillar[5]arene-based indicator displacement assays were constructed for analytes with biological relevance, such as amino acids (AAs).
Detection and quantification of AAs and related compounds is mainly based on chromatographic34 and electrochemical35 methods. Besides, in the past years, there has been a rapid progress in the development of fluorescent and colorimetric chemosensors for the detection of AAs,36 however there is still a need to increase the selectivity and simplicity of these systems. Basic amino acids, arginine (Arg) and lysine (Lys) have N-containing units on the side chain that is vital for the proper function of the living systems. Their fluorescent and/or colorimetric detection is usually achieved by metal complexes,37–40 metal nanoparticles,41–43 cucurbituril- or cucurbituril-analogue-based differential sensing platforms44–46 or by utilizing their nucleophilicity and basicity for bond cleavage.47,48 Biogenic diamines are usually detected by non-selective amine chemosensors49–55 or cucurbituril-based FID systems.4
In this paper, our aim has been to introduce a novel principle to construct FID-based chemosensing systems for amino acids. Using an environment-responsive fluorophore attached to different anchors in order to manipulate the association constants, the fine tuning the displacement process is possible to alter sensing parameters (selectivity, sensitivity). This was demonstrated by supramolecular systems containing 4-amino-1,8-naphthalimide fluorophores with three different anchors as guest, and WP5 as host, the first pillararene-based FID systems to detect basic amino acids. These systems also proved applicable for the detection of biogenic diamines.
Results and discussion
Structures and syntheses
As the fluorophore unit of the guest molecules, the 4-amino-1,8-naphthalimide group was selected, which has the special advantages of convenient derivatization and the easy modulation of its fluorescence via photoinduced electron transfer (PET) process.56 In water, the fluorescence of this fluorophore is quenched due to the stabilization of its ICT state. However, when the substituent attached to the 4-position becomes positively charged, the fluorescence is recovered due to the inhibition of the PET process. This principle is widely used for the fluorescent detection of metal ions.57–59 Since WPns form strong complexes with several cationic guests, such as ammonium, imidazolium and pyridinium compounds, we speculated that some of these groups would be ideal as anchoring moieties.The expected sensing mechanism of our FID systems is illustrated in Scheme 1. The complexation of the naphthalimide derivatives by WP5 leads to a fluorescence quenching, since the negative charges on the rim of the pillararene host compensate the positive charge on the anchors of the guests, restoring the PET effect. These systems will then provide a turn-on fluorescence response to the addition of non-fluorescent analytes as competitive guests.
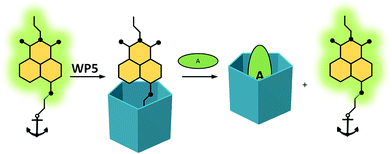 | ||
| Scheme 1 Anchored naphthalimides in WP5-based indicator displacement systems. The green oval represents the analyte. | ||
Guests G1–G3 (Fig. 1) were synthesized using the general methods to yield substituted naphthalimides (see ESI for synthetic details†). WP5 was synthesized as described recently.60G1 has a putrescine anchor and is already mentioned in the literature as an anchored indicator for cucurbituril hosts.61G2 and G3 are new naphthalimide derivatives. G2 contains an imidazolium moiety as anchor and was prepared from 4-(2-bromoethylamino)-1,8-naphthalimide57 and N-methylimidazole. G3 bearing a trimethyl-ammonium end group was synthesized in a similar manner. This latter dye was designed to be a “poorly” complexing indicator since the size of the WP5 cavity allows only a weaker binding of this guest. The compounds were fully characterised using NMR and HRMS techniques. The complexation studies were performed using optical spectroscopic methods and 1H-NMR measurements.
Complexation studies
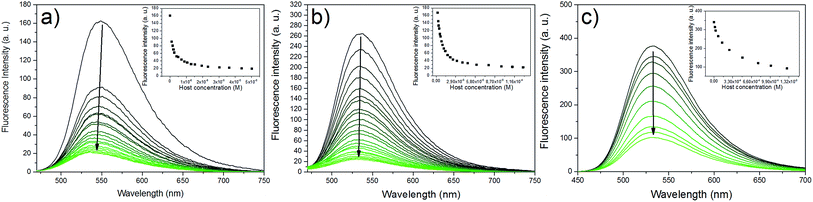
As had been expected, all the three guests fluoresced well in HEPES-buffered aqueous media (pH 7.4) due to the inhibition of the PET process by the primary (G1) and quaternary (G2 and G3) ammonium anchor groups. The results with the spectral properties are summarized in Table 1. The complexation with the anionic WP5 caused a significant reduction of the fluorescence due to the charge-compensating effects of the carboxylate arms of the host (Fig. 2). Since the positive charge was effectively shielded, the fluorescence decreased due to the increased PET.| λ abs (nm) | ε (M−1 cm−1) | λ em (nm) | Φ | |
|---|---|---|---|---|
| G1 | 452 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
550 | 0.12 |
| G1·WP5 | 452 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
550 | 0.02 |
| G2 | 438 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
535 | 0.20 |
| G2·WP5 | 437 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
535 | 0.02 |
| G3 | 434 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
533 | 0.32 |
| G3·WP5 | 434 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
533 | 0.06 |
The Job's plots of the indicator–WP5 systems (ESI Fig. S12–S14†) indicated a stepwise supramolecular reaction: besides the expected 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes, 2
1 complexes, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (guest
1 (guest![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) WP5) complexes were also formed. The stepwise association constants for the two complexes were determined by a least square fitting to the fluorescence spectra of samples with the same indicator and different WP5 concentrations (Table 2, see ESI Section 6 for details†). As at the higher WP5 equivalents, applied in the displacements assays, the dyes were present predominantly in the form of their 1
WP5) complexes were also formed. The stepwise association constants for the two complexes were determined by a least square fitting to the fluorescence spectra of samples with the same indicator and different WP5 concentrations (Table 2, see ESI Section 6 for details†). As at the higher WP5 equivalents, applied in the displacements assays, the dyes were present predominantly in the form of their 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 WP5 complexes, the concentrations of 2
1 WP5 complexes, the concentrations of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes were negligible, we discuss here briefly only the trends in the Ka1 association constants.
1 complexes were negligible, we discuss here briefly only the trends in the Ka1 association constants.
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka1 (Ka1 in M−1) Ka1 (Ka1 in M−1) |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka2 (Ka2 in M−1) Ka2 (Ka2 in M−1) |
|
|---|---|---|
| G1·WP5 | 6.09 | 6.00 |
| G2·WP5 | 5.45 | 5.75 |
| G3·WP5 | 3.68 | — |
The guests showed the expected variations in complex strength, with Ka1 values ranging from 103 to 106 M−1. These results indicate that an efficient supramolecular control could be achieved by the proper selection of the anchoring groups. In the case of G1, a strong 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex was formed (Ka1,G1 = 1.24 × 106 M−1) with WP5, resulting in an 8-fold reduction of the fluorescence intensity. G2 also formed an 1
1 complex was formed (Ka1,G1 = 1.24 × 106 M−1) with WP5, resulting in an 8-fold reduction of the fluorescence intensity. G2 also formed an 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex, with a somewhat lower association constant (Ka1,G2 = 2.83 × 105 M−1) and a similar response in fluorescence. The lowest association constant (Ka1,G3 = 4.75 × 103 M−1) was measured in the case of G3. This demonstrates that the trimethylammonium group functions as a “poorly complexing” anchor.
1 complex, with a somewhat lower association constant (Ka1,G2 = 2.83 × 105 M−1) and a similar response in fluorescence. The lowest association constant (Ka1,G3 = 4.75 × 103 M−1) was measured in the case of G3. This demonstrates that the trimethylammonium group functions as a “poorly complexing” anchor.
The supramolecular interactions in the complexes were further evaluated using 1H-NMR measurements in D2O (Fig. 3). The complexation was fast enough to present one set of signals in each case. In the case of G1, the protons of the anchor moiety displayed unusually large upfield shifts (up to Δδ = −3.4 ppm) making some of the methylene protons appear at negative chemical shifts. The aromatic protons of the fluorophore showed slight downfield shifts with the exception in the 3-position near the anchor unit that exhibited some upfield shift. This can be rationalized by the partial inclusion of the molecule illustrated in Scheme 1: the anchoring unit penetrates the cavity, whereas the naphthalimide moiety remains at the rim of the macrocycle. The protons of the host remained mostly unchanged, which indicates that there was no significant restriction in the movement of the constituent units. Similar effects were observed with G2: large upfield shifts of the N-methylimidazolium protons (up to Δδ = −1.1 ppm) and the spacer methylene units (up to Δδ = −2.4 ppm) accompanied by the broadening of the signals, which is attributable to the shielding effect of the macrocycle. The aromatic protons of G2 exhibited some downfield shifts as well. Due to the positive charge on the G2 guest, some changes could be detected in the signals of the WP5 macrocyclic host. The sterically less favorable guest G3 showed somewhat different changes: we suppose that due to steric hindering, complete inclusion could not be reached, therefore only small changes were detected in the signals of the anchor moiety (i.e. the trimethylammonium group showed Δδ = −0.13 ppm). The different complexation was also suggested by the upfield shifts of the aromatic protons of the naphthalimide unit.
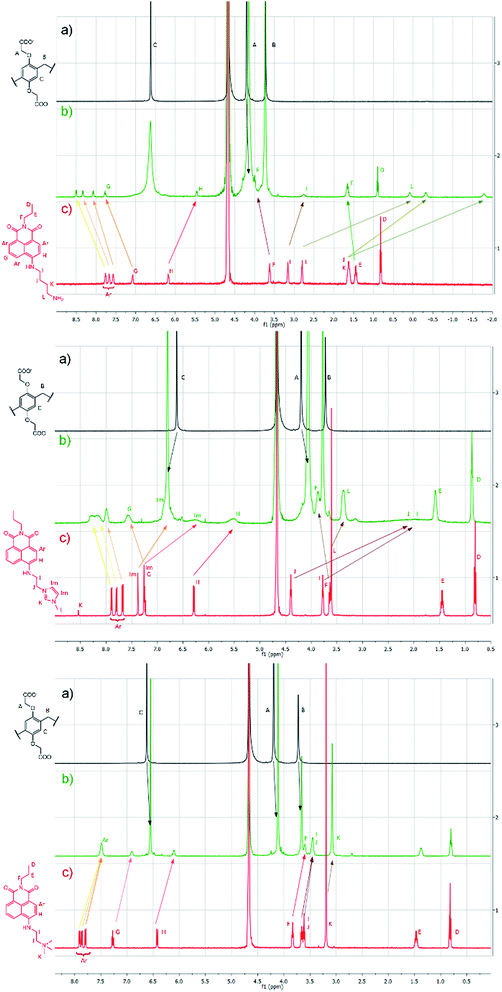 | ||
| Fig. 3 1H-NMR spectra (500 MHz, 25 °C) of the guests G1–G3 (a) WP5 (b) Gx·WP5 (c) Gx in D2O maintaining a constant concentration of 3 mM for all components. | ||
To gain further insight into these distinct host–guest interactions, we calculated the anchor volumes and compared them to the cavity volume of pillar[5]arene. It was confirmed that the ethylammonium ion (anchor for G1) with a volume of 77 Å3, and the N,N′-ethyl-methyl imidazolium cation (anchor for G2) filling a volume of 143 Å3 can be included in the 152 Å3 cavity of the WP5 pillarene host,9 whereas the volume of N,N-ethyl-trimethylammonium moiety (anchor for G3) is 171 Å3, exceeds the above cavity volume of WP5, preventing such type host–guest interaction (see ESI Section 7 for details†).
Fluorescence indicator displacement
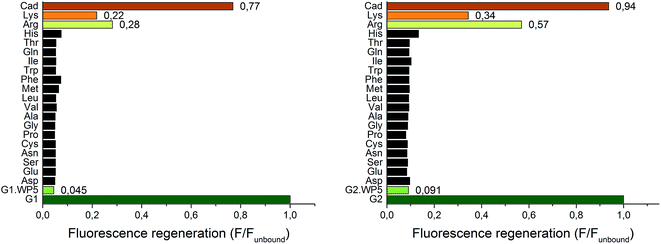
Due to their large signal modulation upon complexation and optimal binding constants with WP5, G1 and G2 were selected as potential indicators for FID systems. Upon addition of various amino acids (3 mM) to the indicator–WP5 complexes, a significant fluorescence enhancement was observed only in the cases of arginine and lysine. The effect could be characterized by the fluorescence regeneration, F/Funbound, which is related to the proportion of the displaced indicators (Fig. 4).The screening experiment showed that the addition of non-basic α-amino acids in high concentration caused negligible changes in the fluorescence, with the exception of histidine, which resulted in a small fluorescence enhancement. Thus, both of our FID systems showed a high selectivity towards the basic amino-acids. The two diamines, putrescine and cadaverine caused an almost complete fluorescence regeneration of the free indicators (only cadaverine shown). Subsequently, titration experiments were performed to evaluate the sensitivity of the two indicator displacement systems for cadaverine, arginine and lysine (Fig. S15 and S16†). A host concentration of 15 μM was selected (15-fold excess related to the indicator guest) to achieve a high level of indicator–WP5 complex, which was ∼95% for the G1·WP5 and ∼80% for the G2·WP5 system as can be calculated from the respective Ka values.
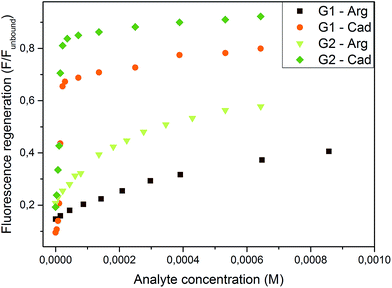
Both systems were found highly sensitive for cadaverine with a large turn-on fluorescence. A smaller, but still significant enhancement was induced by arginine and an even smaller enhancement was reached with lysine. The weaker G2·WP5 complex showed the expected higher sensitivity towards the analytes, as can be seen in Fig. 5. The limit of detection (LOD; for its calculation see ESI Section 8†) was found to be larger by almost one order of magnitude in the case of G2·WP5: for cadaverine, the LOD was found to be 9.44 × 10−7 M and 1.41 × 10−7 M for G1·WP5 and G2·WP5, respectively. In the case of arginine, the LOD value of 1.34 × 10−5 M was obtained for the G1·WP5 assay and 3.27 × 10−6 M for the G2·WP5 assay. This example demonstrates that the sensing properties can be tailored and fine-tuned with varying the anchor unit with the same fluorophore. In addition, due to the different sensitivity of these systems, differential sensing can be achieved based on the anchored naphthalimide indicators and WP5.
The indicator displacement was further evaluated by 1H-NMR-spectroscopy. Following the NMR investigation of the naphthalimide–WP5 mixtures (see Section ‘Complexation studies’), the mixtures of the guests with cadaverine and arginine were tested in the absence of the host (Fig S17–S20†). No significant changes were observed showing the lack of interaction of the indicators with the analytes. In the case of G1, the slight shift of the signals could be attributed to the pH sensitivity of the anchor group. Next, the indicator displacement process was studied by NMR spectroscopy (Fig. S21–S24†). Upon addition of cadaverine in 10-fold excess to the complexes of WP5 with G1 and G2, the displacement of the indicators was clearly visible, as the spectra of the free indicators were regained. However, when arginine was added to the G1·WP5 complex in the same excess, an average signal of the complex and the free G2 was observed as a result of partial displacement. Unfortunately, upon the addition of arginine to G1·WP5 precipitation occurred, it is unlikely, however, that in the latter system the indicator was displaced from the strong complex.
With respect to the acid–base character of components, some aspects of the pH-dependence of the complexation and displacement reactions were also investigated. As could be expected, the complexation of G1 was pH-dependent (ESI Section 10†), however, the association constants for G2·WP5 were almost identical in the 6.8–8.2 pH range. The indicator displacement of G2·WP5 was tested at pH values 6.8, 7.4 and 8.2: evidently, higher fluorescence regeneration values were detected for the basic amino acids at lower pH, whereas, for cadaverine, no correlation was found (Fig. S25†).
As a further test of the G2·WP5 system, the more sensitive FID sensor for basic amino acids, the association constants for the lysine–WP5 and arginine–WP5 supramolecular complexes were determined by a least square fitting to the fluorescence spectra of G2·WP5–lysine/arginine mixtures, with different lysine/arginine concentrations. For the lysine–WP5 complex Ka = 1.12 × 103 M−1 was obtained, for the arginine–WP5 complex Ka = 5.24 × 103 M−1, in good agreement with the values of 1.8 × 103 M−1 for lysine–WP5 and 5.9 × 103 M−1 for arginine–WP5 determined by NMR experiments.28
Conclusions
In conclusion, the synthesized 4-amino-1,8-naphthalimide indicators having different anchoring groups in the 4-position were shown to form complexes with WP5 with varying association constants and large signal modulation. Two complexes were tested as indicator displacement systems for cadaverine, putrescine and α-amino acids. Both systems can detect cadaverine and the basic amino acids with high selectivity and different sensitivity. The FID process was further established using NMR spectroscopy. To our knowledge, this is the first WP5-based FID sensing system for analytes with biological relevance. We believe that the current toolbox of indicator displacement assays could be further expanded with this intriguing host.Acknowledgements
We thank the Hungarian Research Foundation for the financial support of this work (Grant No. K108752). We are grateful to Fanni Bazsó for the mass spectrometric measurements and Zoltán Szakács for the anchor volume calculations.Notes and references
- E. V. Anslyn, J. Org. Chem., 2007, 72, 687–699 CrossRef CAS PubMed.
- L. You, D. Zha and E. V. Anslyn, Chem. Rev., 2015, 115, 7840–7892 CrossRef CAS PubMed.
- B. T. Nguyen and E. V. Anslyn, Coord. Chem. Rev., 2006, 250, 3118–3127 CrossRef CAS.
- R. N. Dsouza, U. Pischel and W. M. Nau, Chem. Rev., 2011, 111, 7941–7980 CrossRef CAS PubMed.
- G. Ghale and W. M. Nau, Acc. Chem. Res., 2014, 47, 2150–2159 CrossRef CAS PubMed.
- T. Ogoshi, S. Kanai, S. Fujinami, T.-a. Yamagishi and Y. Nakamoto, J. Am. Chem. Soc., 2008, 130, 5022–5023 CrossRef CAS PubMed.
- P. J. Cragg and K. Sharma, Chem. Soc. Rev., 2012, 41, 597–607 RSC.
- T. Ogoshi, J. Inclusion Phenom. Macrocyclic Chem., 2012, 72, 247–262 CrossRef CAS.
- M. Xue, Y. Yang, X. Chi, Z. Zhang and F. Huang, Acc. Chem. Res., 2012, 45, 1294–1308 CrossRef CAS PubMed.
- T. Ogoshi and T.-a. Yamagishi, Eur. J. Org. Chem., 2013, 2013, 2961–2975 CrossRef CAS.
- H. Zhang and Y. Zhao, Chem.–Eur. J., 2013, 19, 16862–16879 CrossRef CAS PubMed.
- D. Cao and H. Meier, Asian J. Org. Chem., 2014, 3, 244–262 CrossRef CAS.
- C. Li, Chem. Commun., 2014, 50, 12420–12433 RSC.
- D. Cao and H. Meier, Synthesis, 2015, 26, 1041–1056 CrossRef.
- K. Yang, Y. Pei, J. Wen and Z. Pei, Chem. Commun., 2016, 52, 9316–9326 RSC.
- T. Ogoshi, T. A. Yamagishi and Y. Nakamoto, Chem. Rev., 2016, 116, 7937–8002 CrossRef CAS PubMed.
- Y. Wang, G. Ping and C. Li, Chem. Commun., 2016, 52, 9858–9872 RSC.
- T. Ogoshi, M. Hashizume, T.-a. Yamagishi and Y. Nakamoto, Chem. Commun., 2010, 46, 3708–3710 RSC.
- G. Yu, M. Xue, Z. Zhang, J. Li, C. Han and F. Huang, J. Am. Chem. Soc., 2012, 134, 13248–13251 CrossRef CAS PubMed.
- Y. Ma, X. Ji, F. Xiang, X. Chi, C. Han, J. He, Z. Abliz, W. Chen and F. Huang, Chem. Commun., 2011, 47, 12340–12342 RSC.
- G. C. Yu, J. Zhou, J. Shen, G. P. Tang and F. H. Huang, Chem. Sci., 2016, 7, 4073–4078 RSC.
- Y. Yao, J. Li, J. Dai, X. Chi and M. Xue, RSC Adv., 2014, 4, 9039–9043 RSC.
- W. Cheng, H. Tang, R. Wang, L. Wang, H. Meier and D. Cao, Chem. Commun., 2016, 52, 8075–8078 RSC.
- C. Li, X. Shu, J. Li, S. Chen, K. Han, M. Xu, B. Hu, Y. Yu and X. Jia, J. Org. Chem., 2011, 76, 8458–8465 CrossRef CAS PubMed.
- S. Dasgupta, A. Chowdhury and P. S. Mukherjee, RSC Adv., 2015, 5, 85791–85798 RSC.
- B. Shi, K. Jie, Y. Zhou, J. Zhou, D. Xia and F. Huang, J. Am. Chem. Soc., 2016, 138, 80–83 CrossRef CAS PubMed.
- Y. Sun, F. Guo, T. Zuo, J. Hua and G. Diao, Nat. Commun., 2016, 7, 12042 CrossRef PubMed.
- C. Li, J. Ma, L. Zhao, Y. Zhang, Y. Yu, X. Shu, J. Li and X. Jia, Chem. Commun., 2013, 49, 1924–1926 RSC.
- N. L. Strutt, R. S. Forgan, J. M. Spruell, Y. Y. Botros and J. F. Stoddart, J. Am. Chem. Soc., 2011, 133, 5668–5671 CrossRef CAS PubMed.
- P. Wang, X. Yan and F. Huang, Chem. Commun., 2014, 50, 5017–5019 RSC.
- P. Wang, Y. Yao and M. Xue, Chem. Commun., 2014, 50, 5064–5067 RSC.
- M. Bojtar, Z. Szakacs, D. Hessz, M. Kubinyi and I. Bitter, RSC Adv., 2015, 5, 26504–26508 RSC.
- M. Bojtár, Z. Szakács, D. Hessz, F. L. Bazsó, M. Kállay, M. Kubinyi and I. Bitter, Dyes Pigm., 2016, 133, 415–423 CrossRef.
- J. Le Boucher, C. Charret, C. Coudray-Lucas, J. Giboudeau and L. Cynober, Clin. Chem., 1997, 43, 1421–1428 CAS.
- G. L. Luque, N. F. Ferreyra and G. A. Rivas, Talanta, 2007, 71, 1282–1287 CrossRef CAS PubMed.
- Y. Zhou and J. Yoon, Chem. Soc. Rev., 2012, 41, 52–67 RSC.
- A. Buryak and K. Severin, J. Am. Chem. Soc., 2005, 127, 3700–3701 CrossRef CAS PubMed.
- X. Zhou, X. Jin, D. Li and X. Wu, Chem. Commun., 2011, 47, 3921–3923 RSC.
- L. He, V. L. L. So and J. H. Xin, Sens. Actuators, B, 2014, 192, 496–502 CrossRef CAS.
- J. Cao, L. Ding, Y. Zhang, S. Wang and Y. Fang, J. Photochem. Photobiol., A, 2016, 314, 66–74 CrossRef CAS.
- D. Xiong, M. Chen and H. Li, Chem. Commun., 2008, 880–882, 10.1039/B716270G.
- G. Patel and S. Menon, Chem. Commun., 2009, 3563–3565, 10.1039/B905141D.
- K. A. Rawat and S. K. Kailasa, Microchim. Acta, 2014, 181, 1917–1929 CrossRef CAS.
- D. M. Bailey, A. Hennig, V. D. Uzunova and W. M. Nau, Chem.–Eur. J., 2008, 14, 6069–6077 CrossRef CAS PubMed.
- L. A. Baumes, M. Buaki, J. Jolly, A. Corma and H. Garcia, Tetrahedron Lett., 2011, 52, 1418–1421 CrossRef CAS.
- T. Minami, N. A. Esipenko, B. Zhang, L. Isaacs and P. Anzenbacher, Chem. Commun., 2014, 50, 61–63 RSC.
- X. Lu, W. Wang, Q. Dong, X. Bao, X. Lin, W. Zhang, X. Dong and W. Zhao, Chem. Commun., 2015, 51, 1498–1501 RSC.
- X. Qian, W. Gong, F. Wang, Y. Lin and G. Ning, Tetrahedron Lett., 2015, 56, 2764–2767 CrossRef CAS.
- K. Secor, J. Plante, C. Avetta and T. Glass, J. Mater. Chem., 2005, 15, 4073–4077 RSC.
- G. Lu, J. E. Grossman and J. B. Lambert, J. Org. Chem., 2006, 71, 1769–1776 CrossRef CAS PubMed.
- S. Reinert and G. J. Mohr, Chem. Commun., 2008, 2272–2274, 10.1039/B717796H.
- M.-S. Steiner, R. J. Meier, A. Duerkop and O. S. Wolfbeis, Anal. Chem., 2010, 82, 8402–8405 CrossRef CAS PubMed.
- B. Lee, R. Scopelliti and K. Severin, Chem. Commun., 2011, 47, 9639–9641 RSC.
- J.-F. Lin, J. Kukkola, T. Sipola, D. Raut, A. Samikannu, J.-P. Mikkola, M. Mohl, G. Toth, W.-F. Su, T. Laurila and K. Kordas, J. Mater. Chem. A, 2015, 3, 4687–4694 CAS.
- Y. Hu, X. Ma, Y. Zhang, Y. Che and J. Zhao, ACS Sens., 2016, 1, 22–25 CrossRef CAS.
- R. M. Duke, E. B. Veale, F. M. Pfeffer, P. E. Kruger and T. Gunnlaugsson, Chem. Soc. Rev., 2010, 39, 3936–3953 RSC.
- T. Chen, W. Zhu, Y. Xu, S. Zhang, X. Zhang and X. Qian, Dalton Trans., 2010, 1316–1320 RSC.
- G. R. C. Hamilton, S. K. Sahoo, S. Kamila, N. Singh, N. Kaur, B. W. Hyland and J. F. Callan, Chem. Soc. Rev., 2015, 44, 4415–4432 RSC.
- J. Yin, Y. Hu and J. Yoon, Chem. Soc. Rev., 2015, 44, 4619–4644 RSC.
- R. R. Kothur, J. Hall, B. A. Patel, C. L. Leong, M. G. Boutelle and P. J. Cragg, Chem. Commun., 2014, 50, 852–854 RSC.
- C. P. Carvalho, R. Ferreira, J. P. Da Silva and U. Pischel, Supramol. Chem., 2013, 25, 92–100 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra15003a |
| This journal is © The Royal Society of Chemistry 2016 |