Yeast-derived biosynthesis of silver/silver chloride nanoparticles and their antiproliferative activity against bacteria†
Abstract
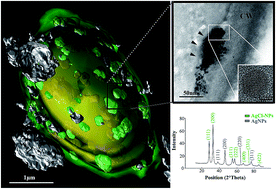
Here, we provide the first evidence of yeast strains assisted Ag/AgCl-NPs production in vitro. The formed nanoparticles were characterized by spectroscopic and electron microscopy approaches. UV-vis supported the biosynthesis. TEM analysis evidenced that the nanoparticles mainly presented a circular shape and their diameters varied mostly being in the range 2 to 10 nm. XRD analysis showed a crystalline structure, with diffraction peaks corresponding to metallic silver and silver chloride nanoparticles, and when analyzed by high-resolution transmission electron microscopy (HRTEM), instead of being round, (111) (octahedral) and (200) (cubic) symmetry facets appeared systematically in one side of the nanoparticles. Analysis of ultra-thin sections by TEM indicated that the domain of the synthesis of Ag/AgCl-NPs was mainly between the cell wall and the plasma membrane. By using 3D reconstruction obtained from focused ion beam scanning electron microscopy (FIB/SEM) the spatial distribution of the domains of nanoparticle synthesis was mapped and nanoaggregates of Ag/AgCl-NPs up 35 nm in diameter were observed. Extracellular synthesis also occurred; in accordance with the fact that conditioned media from yeast isolates were as efficient at producing Ag/AgCl-NPs as live-cell cultures. Exposure of Gram-positive Staphylococcus aureus and Gram-negative Klebsiella pneumoniae cultures to Ag/AgCl-NPs led to a strong growth inhibition as shown by optical density measurements. The Ag/AgCl-NPs described here have characteristics compatible with a strong potential for use in the biotechnology industry, particularly for biomedical applications.

 Please wait while we load your content...
Please wait while we load your content...