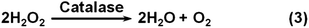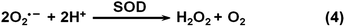Nanozymes in bionanotechnology: from sensing to therapeutics and beyond†
Xiaoyu
Wang
a,
Yihui
Hu
a and
Hui
Wei
*ab
aDepartment of Biomedical Engineering, College of Engineering and Applied Sciences, Collaborative Innovation Center of Chemistry for Life Sciences, Nanjing National Laboratory of Microstructures, Nanjing University, Nanjing 210093, China. E-mail: weihui@nju.edu.cn; Web: http://weilab.nju.edu.cn Fax: +86-25-83594648; Tel: +86-25-83593272
bState Key Laboratory of Electroanalytical Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, Jilin 130022, China
First published on 7th December 2015
Abstract
In the past few decades, researchers have developed lots of artificial enzymes with various materials to mimic the structures and functions of natural enzymes. Recently, nanozymes, nanomaterials with enzyme-like characteristics, are emerging as novel artificial enzymes, and attracting researchers’ enormous interest. Remarkable advances have been made in the area of nanozymes due to their unique properties compared with natural enzymes and classic artificial enzymes. Until now, lots of nanomaterials have been studied to mimic various natural enzymes for wide applications. To highlight the recent progress of nanozymes (especially in bionanotechnology), here we discuss the diverse applications of nanozymes, which range from sensing, imaging, and therapeutics, to logic gates, pollutant removal, water treatment, etc. Finally, we address the current challenges facing nanozyme research as well as possible directions to fulfill their great potential in future.
1. Introduction
Nanozymes are nanomaterials with enzyme-like characteristics.1 As an emerging research area in the field of artificial enzymes, nanozymes have attracted researchers’ enormous interest due to their unique properties compared with natural enzymes and classic artificial enzymes.1–4 Compared with natural enzymes, nanozymes are advantageous in several aspects, such as low cost, ease of mass production, robustness to harsh environments, high stability, long-term storage, and size/composition dependent activity.1 Besides, nanozymes have also shown unique properties compared with other artificial enzymes in terms of their size- (shape-, structure-, composition-) dependent catalytic activities, integrated (multi-)functions besides catalysis, large surface area for further modification and bioconjugation, smart response to external stimuli, self-assembly capability, etc.1 Until now, lots of nanomaterials have been explored to mimic various natural enzymes, such as catalase, oxidase, peroxidase, superoxide dismutase (SOD), esterase, nuclease, phosphatase, protease and ferroxidase (Fig. 1).5–26 These nanozymes have been extensively investigated for diverse applications in bionanotechnology, ranging from biosensing and bioimaging to tissue engineering, therapeutics and beyond. In 2013, we have made a comprehensive review on nanozymes.1 Since then, substantial progress has been achieved in the field. To highlight the recent exciting progress, the present review discusses the key reports of nanozymes, especially their applications in bionanotechnology. A few early studies are also included to provide a historic view of nanozyme research. Several well established mechanisms about nanozyme catalytic reactions are also covered. The current challenges facing nanozyme research as well as possible directions to fulfill their great potential are discussed in the final section.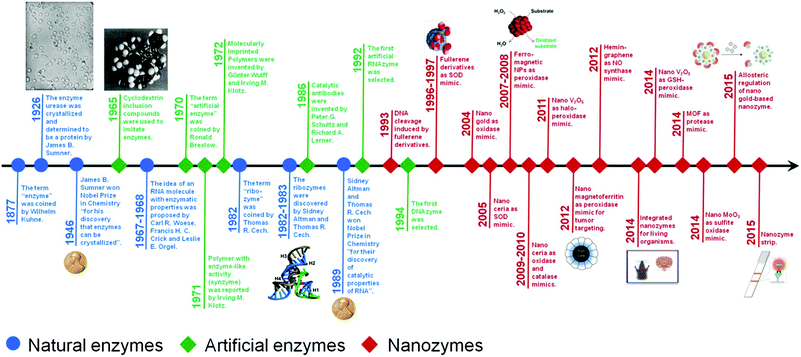 | ||
| Fig. 1 A brief timeline for the development of artificial enzymes (natural enzymes are also listed for comparison) (see Table S1, ESI† for related references). Adapted with permission from ref. 1. Copyright (2013) Royal Society of Chemistry. | ||
We do not attempt to cover all the related publications on nanozymes due to space limitations. Readers are referred to numerous critical reviews and the references therein for more information.1–4,27–36
2. Nanozymes for sensing and imaging
Since the seminal reports by Yan's and Wang's groups,11,12 nanozymes have been widely used for detecting a variety of important targets, such as bioactive small molecules, metal ions, nucleic acids, cancer cells and even bacteria.2.1 Nanozymes for H2O2 sensing
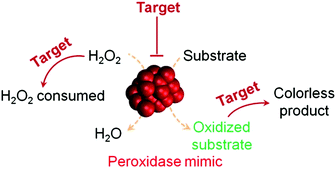
H2O2 detection is of great interest owing to its important roles in biology, medicine, food industry and environmental protection.37,38 H2O2 detection is usually achieved by using nanomaterials’ peroxidase mimicking activities, in which the H2O2-mediated oxidation of a substrate is catalyzed by a peroxidase mimic (Scheme 1 and Fig. 2). By monitoring the production of the oxidized substrate (i.e., A in Scheme 1), H2O2 can be determined.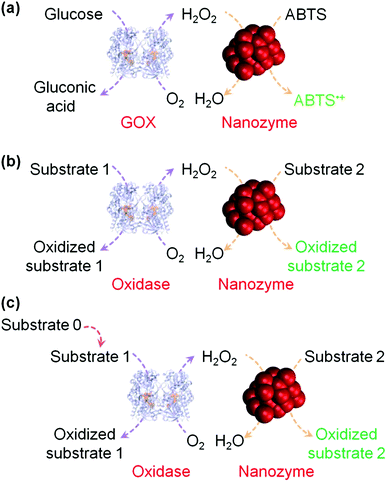 | ||
| Fig. 2 (a) Nanozyme as peroxidase mimic for colorimetric sensing of H2O2, and glucose when combined with glucose oxidase. (b) The sensing format in (a) could be extended to other targets (substrate 1 here) when combined with a suitable oxidase. (c) Target of interest as substrate 0 could be determined if it could be converted into an oxidase substrate. Numerous transduction signals can be adopted for sensing (such as colorimetric, fluorometric, chemiluminescent, and SERS signals when the corresponding substrates are used; and electrochemical signals when a nanozyme is immobilized on an electrode). Adapted with permission from ref. 1. Copyright (2013) Royal Society of Chemistry. | ||
Wei and Wang have developed a colorimetric method for H2O2 detection.12 They used Fe3O4 magnetic nanoparticles (MNPs) as the peroxidase mimic and ABTS as the substrate for signalling (Fig. 2a). The presence of H2O2 generated green coloured ABTS˙+, which could be quantified by absorption spectra or even visualized by the naked eye. Since then, considerable amount of studies have been devoted to H2O2 detection by exploring various nanomaterials’ peroxidase mimicking activities (also see Table S2, ESI†).39–86 Besides colorimetric detection with peroxidase substrates (such as ABTS, 4-AAP, DPD, OPD, and TMB), H2O2 has also been determined via fluorescent, electrochemical and surface enhanced Raman scattering (SERS) methods.42,52,56,58,64,68,72,73,77,79,82,84
Recently, Zhang and co-workers developed 3D Fe- and N-doped carbon nanostructures to mimic peroxidase.52 Due to the presence of highly active Fe–N and doped-N species as well as the large surface area, the nanostructures exhibited good peroxidase mimicking activity. They further constructed a fluorescent sensor for H2O2 detection with the developed nanozyme, which had a linear range from 100 nM to 100 μM and a detection limit of 68 nM. In another study, Fe7S8 nanowires with intrinsic peroxidase-like activity have been studied for fluorescent detection of H2O2.59
Fang et al. used Fe3O4/reduced graphene oxide (rGO) nanocomposites as the peroxidase mimic to prepare the modified glassy carbon electrode for electrochemical sensing of H2O2 (Fig. 3).67 The developed electrochemical sensor showed good selectivity toward H2O2 detection over several metal ions (Na+, K+, Ag+, Mg2+, and Cu2+) and bioactive small molecules (glutathione (GSH), glucose, ascorbic acid, L-cysteine, and uric acid). Interestingly, with the developed electrochemical biosensor, they were able to monitor CdTe quantum dot-stimulated extracellular H2O2 release from living HeLa cells. This study may be useful for understanding the biological effects of nanomaterials.
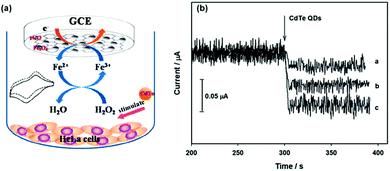 | ||
| Fig. 3 Electrochemical monitoring of H2O2 release from living cells stimulated by CdTe quantum dots with a nanozyme-modified electrode. Reprinted with permission from ref. 67. Copyright (2014) Royal Society of Chemistry. | ||
When a target of interest is used as the substrate for the peroxidase, it can be measured with nanozymes as peroxidase mimics. 5-Hydroxyindole-3-acetic acid (5-HIAA), an indoleamine metabolite, is an important diagnostic biomarker for carcinoid tumours. To selectively oxidize 5-HIAA with hemin as a peroxidase mimic, magnetic molecularly imprinted catalytic polymers were fabricated.87 The products of 5-HIAA oxidation were then separated and detected with HPLC to quantitatively measure 5-HIAA.
2.2 Nanozymes for glucose (and other oxidase substrate) sensing
As shown in Fig. 2, when an oxidase is combined with a peroxidase mimic, the corresponding oxidase substrate can be determined. By combining glucose oxidase with Fe3O4 MNPs as the peroxidase mimic, a sensitive and selective colorimetric approach to glucose detection was reported by Wei et al. (Fig. 4).12 | ||
| Fig. 4 Colorimetric detection of glucose by combining glucose oxidase with Fe3O4 MNPs as peroxidase mimic. Reprinted with permission from ref. 12. Copyright (2008) American Chemical Society. | ||
Until now, varieties of nanozymes with peroxidase activities have been developed and used for glucose detection when they were combined with glucose oxidase (also see Table S3, ESI†).39–44,46–52,59–65,69–71,78,88–121 For example, NiTe thorny nanostructures have been synthesized and used for sensitive and selective glucose detection by Wan et al.105 Some of the nanozymes have already been successfully used for determining glucose in drinks and biological samples (such as blood and urine).89,94,104,106,108,112
To further promote cascade reactions, glucose oxidase and a nanozyme could be assembled together as integrated nanocomposites.118,119 Qu et al. recently reported an inspiring method for glucose detection.104 They replaced both glucose oxidase and peroxidase with the nanomaterial mimics (i.e., gold nanoparticles (AuNPs) as the glucose oxidase mimic and V2O5 nanowires as the peroxidase mimic) (Fig. 5). The two nanozymes were assembled together via polymerized dopamine for cascade reactions. By eliminating the use of both natural enzymes, the current system displayed higher robustness but at much lower cost.
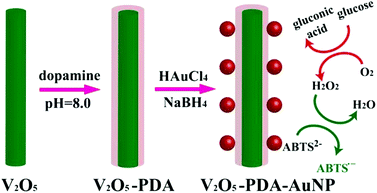 | ||
| Fig. 5 Colorimetric detection of glucose by assembled AuNPs onto V2O5 nanowires as glucose oxidase and peroxidase mimics, respectively. Reprinted with permission from ref. 104. Copyright (2014) John Wiley and Sons. | ||
As indicated by Fig. 2b, when other oxidases are used, their corresponding substrates as targets of interest can be detected. Choline, D-alanine, lactate, uric acid, and xanthine have been determined with the strategy shown in Fig. 2b.64,74,122–124 For example, when NaYF4:Yb,Er nanoparticles as a peroxidase mimic was combined with uricase, uric acid has been determined.124 The proposed method had good selectivity against several biomolecules, such as creatinine, ascorbic acid, glucose, cholesterol, triglyceride, and urea. Moreover, the uric acid in a clinical human serum sample was successfully measured via the developed method, which matched well with the established method.
Several studies also demonstrated that the targets of interest as substrate 0 could be determined if they could be converted into oxidase substrates (Fig. 2c).123,125–127 Using acetylcholinesterase, choline oxidase and Au/Ag nanoparticles as the peroxidase mimic, Wang et al. reported the sensitive detection of acetylcholine.125 With Wang's method, a detection limit of 0.21 nM was obtained. Based on the inhibition of acetylcholinesterase's activity with organophosphates, Liang and co-workers later developed a rapid and sensitive strategy for detecting organophosphorus pesticides and nerve agents (such as Sarin).126
2.3 Sensing targets of interest by modulating peroxidase mimics (or oxidase mimics) catalyzed reactions
As shown in Fig. 6, by modulating the peroxidase mimic catalyzed reactions, the targets of interest can be detected.62,128–138 So far, these targets have already covered a wide range of molecules, such as ascorbic acid, biothiols (e.g., cysteine), catechol, dopamine, melamine, phosphate, trypsin, etc.62,128–138By using C-rich DNA as the template, Sun et al. synthesized bimetallic AuxPty and studied their peroxidase mimicking activities.134 Their results revealed that the nanozymes’ activities could be tuned by manipulating the ratios of x![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) y. After demonstrating the high peroxidase mimicking activity of the Au2Pt1 nanozyme, they then used it for detection of biothiols (i.e., cysteine and homocysteine), which was based on the thiol-induced inhibition of the nanozyme's activities. The proposed strategy showed good selectivity against non-thiol-containing amino acids. Moreover, with Sun's sensing strategy, cysteine spiked in human serum was successfully detected with good recovery. Lin et al. suggested that the presence of cysteine not only inhibited the nanozyme's activities but also reduced the oxidized substrate (such as oxidized TMB).135 Due to such a synergetic effect, a detection limit of 1.2 nM was obtained for cysteine detection. Ascorbic acid, one of the physiologically important neurochemicals,139 was detected on the basis of its inhibition effect on CuNPs@C nanocomposites’ peroxidase-like activity.131 Several ions such as phosphate and uranyl ions have been detected based on their inhibition effects on peroxidase-like nanozymes.136,137
y. After demonstrating the high peroxidase mimicking activity of the Au2Pt1 nanozyme, they then used it for detection of biothiols (i.e., cysteine and homocysteine), which was based on the thiol-induced inhibition of the nanozyme's activities. The proposed strategy showed good selectivity against non-thiol-containing amino acids. Moreover, with Sun's sensing strategy, cysteine spiked in human serum was successfully detected with good recovery. Lin et al. suggested that the presence of cysteine not only inhibited the nanozyme's activities but also reduced the oxidized substrate (such as oxidized TMB).135 Due to such a synergetic effect, a detection limit of 1.2 nM was obtained for cysteine detection. Ascorbic acid, one of the physiologically important neurochemicals,139 was detected on the basis of its inhibition effect on CuNPs@C nanocomposites’ peroxidase-like activity.131 Several ions such as phosphate and uranyl ions have been detected based on their inhibition effects on peroxidase-like nanozymes.136,137
By reacting with and consuming H2O2, a number of bioactive small molecules (such as ascorbic acid, catechol, dopamine, GSH) have been detected.62,130–133 For example, catechol in the Yellow river was determined using Fe3O4 MNPs as the peroxidase mimic.132
An innovative concept for the chemiluminescence detection of pesticides was reported.140 The Fe3O4 MNPs as peroxidase mimics catalyzed the generation of chemiluminescence from luminol, which could be quenched by ethanol. However, the quenching was reversed when pesticides were bound onto Fe3O4 MNPs. By “turning on” the chemiluminescence of luminol, nonredox active pesticides were sensitively detected. More interestingly, Fe3O4 MNPs with different surface ligands exhibited unique chemiluminescence patterns towards different pesticides. Thus, simultaneous detection of pesticides could be achieved via the developed method.
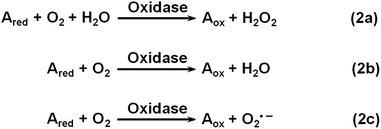
Oxidase uses oxygen to oxidize its substrates (Scheme 2). Similar to natural oxidase, nanomaterial-based oxidase mimics have been used for sensing the corresponding substrates.141–144 Hayat et al. evaluated the oxidase mimicking activity of nanoceria and developed a colorimetric assay for dopamine and catechol.143 By carefully choosing the nanoceria type, buffer composition, pH, etc., they demonstrated good selectivity for dopamine and catechol detection with respect to several interferences.
Sulfite is an important additive in food and wine. It can tune the oxidase mimicking activity of CoFe2O4 nanoparticles. Zhang et al. discovered that sulfite at low concentration inhibited the oxidase mimicking activity of CoFe2O4 nanoparticles while a high concentration of sulfite exhibited enhancement effects.141 On the basis of this phenomenon, they reported a chemiluminescent assay for sulfite determination in white wines. Sulfite in food was also measured by exploring its inhibition effect on the oxidase mimicking activity of Co3O4 nanoparticles.142
2.4 Nanozymes for nucleic acid sensing
Several strategies have been developed for nucleic acid detection using nanozymes.104,145–155 The reported protocols can be roughly classified into two types. One type of assay uses nanozymes as alternative tags to classic dyes (or enzymes) to label the nucleic acid probe strand while the other type employs nucleic acids to tailor the nanozyme’ activities.For example, graphene-supported ferric porphyrin, acting as the peroxidase mimic, was bioconjugated onto streptavidin for electrochemically detecting the target DNA (Fig. 7a).146 The hairpin DNA was immobilized onto a AuNP-modified electrode via Au–S interaction. In the presence of target DNA, the hairpin structure was opened and the pre-conjugated biotin was thus available for binding to the streptavidin–nanozyme conjugates. With the developed electrochemical sensor, as low as 22 aM of target DNA was detected. Thiramanas and co-workers reported the detection of bacterial DNA with Fe3O4 MNPs as peroxidase mimics via a sandwich assay.147 The developed assay was used for monitoring bacteria in drinking and tap water.
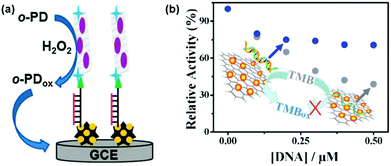 | ||
| Fig. 7 Nucleic acid detection with nanozymes. (a) Electrochemical detection of DNA with graphene-supported ferric porphyrin as a peroxidase mimic. (b) DNA detection based on modulating the peroxidase mimicking activity of AuNP/graphene hybrids with ssDNA and dsDNA. (a) Reprinted with permission from ref. 146. Copyright (2013) Royal Society of Chemistry. (b) Reprinted with permission from ref. 156. Copyright (2012) Royal Society of Chemistry. | ||
Single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) exhibit different affinities towards several nanozymes. By modulating the peroxidase mimicking activity of AuNP/graphene hybrids with ssDNA and dsDNA, Liu et al. reported a colorimetric method for DNA detection (Fig. 7b).156
Despite the current progress, no RNA detection with nanozymes has been reported. Future efforts should also be focused on RNA detection, which is critical for disease diagnostics, etc.
Interestingly, while some reports showed that DNA could enhance the activity of several nanozymes,157,158 others demonstrated that DNA could also inhibit the activity of certain nanozymes.156,159,160 Liu et al. suggested that the enhanced activity of Fe3O4- and nanoceria-based peroxidase mimics by DNA was attributed to (i) the electrostatic interactions between the positively charged substrate TMB and the negatively charged phosphate backbone and (ii) the aromatic stacking between the DNA bases and TMB.158 On the other hand, the much stronger binding between ssDNA and the gold nanoparticle-based nanozyme could completely prevent the interaction between the nanozyme and the substrate (even for positively-charged TMB), therefore inhibition effects were observed.156,159 However, since the enhancing/inhibiting effects of DNA on the nanozyme’ activity depend on many factors (such as the nature of nanozymes, the substrates, the DNA sequence, structures, etc.), more systematic studies are needed to understand the mechanisms involved.161
2.5 Nanozymes for aptasensors
Aptamers are selected short nucleic acid sequences, which are capable of recognizing specific targets and thus have been widely used for constructing various aptasensors.162–177 Recently, nanozymes have been employed to develop numerous aptasensors for bioactive small molecules, proteins and metal ions.154,157,159,160,178–183By making use of the AuNPs’ peroxidase mimicking activities and an S-18 aptamer's high affinity and specificity to acetamiprid, a colorimetric assay for rapid pesticide monitoring was demonstrated.159 The aptamer inhibited the nanozyme's activities due to its binding-induced surface passivation. Acetamiprid present in samples would interact with its aptamer and thus prevent the aptamer's binding onto AuNPs, which in turn recovered the AuNPs’ catalytic activities. With Weerathunge's approach, as low as 0.1 ppm of acetamiprid was detected within 10 minutes, which met the requirement of the United States Environmental Protection Agency. The same group also showed their approach was applicable to a wide range of targets, such as antibiotics (kanamycin).178
Using the corresponding aptamers, several studies reported the detection of proteins, such as thrombin and lysozyme.179–182 For example, sensitive and selective detection of thrombin was achieved in a recent study by Xu and co-workers.180 The aptasensor for thrombin employed a multiple amplification strategy, in which peroxidase-like MnO2 nanoflowers, PtNPs, toluidine blue, and hemin/G-quadruplex were co-assembled together onto multi-walled carbon nanotubes (CNTs). It was demonstrated that the designed multiple-catalysts were superior to other catalyst assemblies. The detection of thrombin in 10-fold-diluted human blood serum was also demonstrated. Using DNA stabilized At/Pt nanoclusters as the peroxidase mimic, Zheng et al. reported a colorimetric aptasensor for thrombin detection.181
Based on the highly specific interaction between Hg2+ and T-rich DNA (i.e., T–Hg2+–T base pairing), Kim et al. reported a colorimetric approach for Hg2+ detection using the Fe3O4 MNP based peroxidase mimic.160 The binding of T-rich DNA onto Fe3O4 MNPs inhibited the peroxidase mimicking activity. The presence of Hg2+ would recover the Fe3O4 MNPs’ catalytic activity by Hg2+-mediated release of T-rich DNA via T–Hg2+–T coordination. In another study, it has been shown that G-rich DNA could enhance the AuNPs’ peroxidase mimicking activity.157 Since the G-rich DNA could specifically interact with K+ by forming a G-quadruplex structure, a signal-off approach to K+ detection was proposed.
In future, other functional nucleic acid-based sensing systems (such as DNAzyme-based sensors) should also be investigated, which will broaden the nanozyme research.164,184
2.6 Nanozymes for metal ion sensing
Quite a few studies have been devoted to metal ion sensing with nanozymes.154,157,160,185–196 In an early report, a sensitive and selective sensor for Cu2+ was developed by using Cu2+-based click chemistry.185Via the click chemistry, CNTs and magnetic silica nanoparticles were assembled together, leading to synergistic enhancement for CNTs’ peroxidase mimicking activity. As low as 1 μM of Cu2+ was detected with the sensor.As discussed above, Hg2+ has been detected based on T–Hg2+–T coordination.160 Other strategies were also studied for developing Hg2+ sensors.186,190,193 For example, Li et al. prepared 2 nm PtNPs with BSA as the template and studied their peroxidase mimicking activity.193 Interestingly, they found that Hg2+ specifically inhibited the nanozyme's activity due to Hg2+–Pt0 metallophilic interactions. They therefore developed a sensitive and selective sensor for Hg2+ detection. The sensor had a linear range of 0–120 nM and a detection limit of 7.2 nM. It has been applied for monitoring Hg2+ in drinking water. On the basis of the Ag+ inhibition effect on CoFe2O4 nanoparticles’ peroxidase mimicking activity, a chemiluminescent sensor for Ag+ has been reported recently.192
2.7 Nanozymes for immunoassay
Enormous efforts have been made to develop immunoassays with nanozymes.11,50,53,197–216Several formats have been employed for immunoassays using nanozymes as signalling elements. For example, Gao et al. reported an antigen-down immunoassay format and a capture-detection sandwich immunoassay format.11 Later, many research groups adopted the classic sandwich immunoassay format for detection using peroxidase and oxidase mimicking nanozymes.50,197,201,204,206,210,212 For example, Kim et al. prepared a highly active peroxidase mimic by encapsulating Fe3O4 MNPs and PtNPs within porous carbon.206 They then used the nanozyme as the signalling element for labelling antibodies. With their nanozyme-labelled detection antibody, as low as 1.5 ng mL−1 of HER2 (human epidermal growth factor receptor 2) was detected within 3 minutes via a sandwich assay. Moreover, the diarrhea causing rotavirus was also successfully detected with the developed immunoassay. Gao and co-workers fabricated urchin-like Au@Pt core shell nanostructures as a highly efficient peroxidase mimic.210 They then developed a sandwich assay for prostate specific antigen (PSA) detection. Using their nanozyme-labelled anti-PSA antibody as the detection antibody, as low as 2.9 pg mL−1 of PSA was determined. More excitingly, PSA concentrations in the clinical serum specimen were analyzed, which exhibited very good correlation with the referenced method. These studies suggested that nanozymes could be used as alternatives to conventional natural enzyme-based immunoassays in future.
Recently, several studies demonstrated that nanozymes can be used for point-of-care (POC) (Fig. 8).207,211,213,216 To combat highly infectious diseases such as the Ebola virus, it is critical to perform rapid diagnosis, especially rapid local detection with portable devices. To meet this challenge, Duan and co-workers recently reported a nanozyme-based strip for Ebola virus detection, which exhibited 100 times more sensitivity compared to the AuNP-based strip (Fig. 8a).211 They showed that as low as 240 pfu mL−1 of pseudo-EBOV could be detected within 30 minutes. As shown in Fig. 8b, Kim et al. developed a similar strip for hCG detection.213 With the help of a smart phone (or other portable devices), the concentration of hCG could be readily quantified and communicated with patients/physicians and other clinical workers.
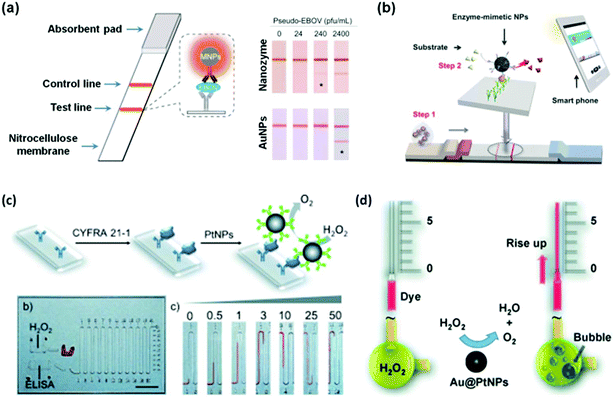 | ||
| Fig. 8 Nanozymes for immunoassay. Nanozyme-based strips for Ebola detection using Fe3O4 MNPs as peroxidase mimic (a) and for hCG detection using hierarchically structured PtNPs as the peroxidase mimic (b). PtNPs (c) and Au@PtNPs (d) as catalase mimics for the immunoassay on a volumetric bar-chart chip. (a) Reprinted with permission from ref. 211. Copyright (2015) Elsevier. (b) Reprinted with permission from ref. 213. Copyright (2015) John Wiley and Sons. (c) Reprinted with permission from ref. 207. Copyright (2014) John Wiley and Sons. (d) Reprinted with permission from ref. 216. Copyright (2014) John Wiley and Sons. | ||
Catalase catalyzes the decomposition of H2O2 into H2O and O2 gas (Scheme 3). Though many nanomaterials have been used to mimic catalase, only a few studies have been focused on their applications.1,207,216 Recently, Song et al. showed that the O2 gas from catalase mimic-catalyzed H2O2 decomposition could be used for diagnosis (Fig. 8c).207 They used PtNPs as the catalase mimic, which were then used to label the detection antibody for signalling. To efficiently and conveniently measure the produced O2 gas, they developed a microfluidic platform called the volumetric bar-chart chip, which could easily gauge the volume of O2 gas with coloured solution in the chip channel. With such a device, they have detected the cancer biomarker cytokeratin 19 fragment (CYFRA 21-1) in serum and HER2 expressed on three breast cancer cell surfaces. By pre-encapsulating the catalase mimic (i.e., Au@PtNPs) within an aptamer hydrogel, Zhu and co-workers devised a similar bar-chart chip for cocaine detection (Fig. 8d).216
2.8 Nanozymes for detection of cells and bacteria
Cells (usually cancer cells) and bacteria have been detected with nanozymes.53,198,202,205,209,217–220Cancer cells over-express characteristic receptors as cancer biomarkers. Therefore cancer cells can be detected with the corresponding antibodies or ligands, which recognize the receptors specifically. When an antibody or a ligand is conjugated with nanozymes, the conjugates can be employed for cancer cell detection. For example, Asati et al. employed antibody conjugated ceria nanoparticles for detection of folate receptor over-expressed lung carcinoma cells (A-549) and EpCAM over-expressed MCF-7 cells.217 Recently, several studies also showed that folate receptor over-expressed cancer cells could be detected with various folate-modified nanozymes.53,198,205,209 In Wang and co-workers’ study, they discovered that chitosan stabilized silver halide (AgX) nanoparticles exhibited interesting peroxidase mimicking activities (Fig. 9a).205 In the absence of H2O2, the AgX nanoparticles could still oxidize the substrate (such as TMB) if stimulated by light irradiation. Using AgI nanoparticles as the nanozyme, folate receptor over-expressed MDA-MB-231 cells were detected under light irradiation.
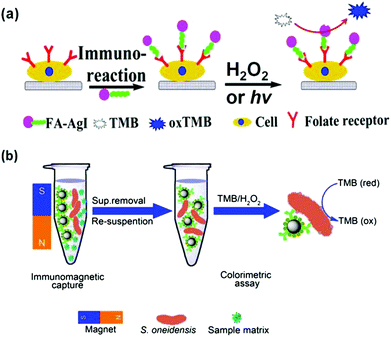 | ||
| Fig. 9 Nanozymes for cancer cell (a) and bacterial detection (b). (a) Reprinted with permission from ref. 205. Copyright (2014) American Chemical Society. (b) Reprinted with permission from ref. 202. Copyright (2014) Nature Publishing Group. | ||
Wen reported the colorimetric detection of Shewanella oneidensis, a facultative anaerobic bacterium, on the basis of immunomagnetic capture of the bacteria and bacterial intrinsic peroxidase mimicking activities for signalling (Fig. 9b).202 The developed method exhibited good selectivity toward S. oneidensis and has been used for identifying spiked S. oneidensis in river water.
2.9 Nanozymes for imaging
Fan et al. reported the imaging tumour tissue by staining them with magnetoferritin nanoparticles as a peroxidase mimic (Fig. 10).221 Due to the presence of over-expressed transferrin receptor 1, the tumour tissues were specifically stained with the magnetoferritin nanoparticles, which were coated with recombinant human heavy-chain ferritin. For the 474 patient specimens examined, the nanozyme-based imaging technique successfully distinguished cancer samples from normal ones with a sensitivity of 98% and a specificity of 95%. Recently, Cai and co-workers also reported the staining and imaging tumour tissues with the magnetoferritin nanoparticles, which confirmed Fan's discovery.208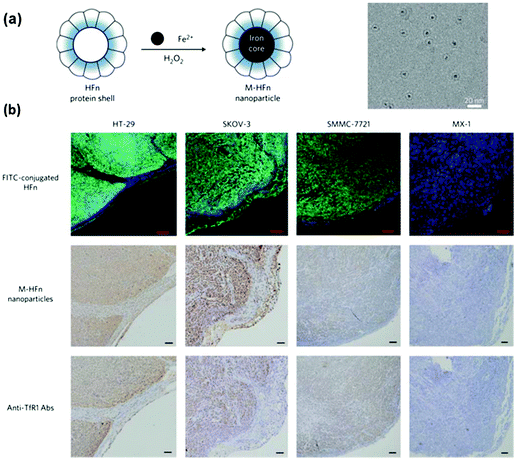 | ||
| Fig. 10 Magnetoferritin nanoparticles as peroxidase mimic for tumour tissue staining and imaging. (a) Preparation of magnetoferritin nanoparticles. (b) Magnetoferritin nanoparticle staining of tumour tissues. Reprinted with permission from ref. 221. Copyright (2012) Nature Publishing Group. | ||
3. Nanozymes for therapeutics
By mainly eliminating reactive oxygen species (ROS) and/or reactive nitrogen species (RNS), nanozymes have been exploited for potential therapeutics.13–17,25,209,222–237 The nanozymes’ ROS scavenging capabilities mainly originated from their SOD mimicking activities, which convert superoxide into H2O2 (Scheme 4). In this section, the therapeutic effects of nanozymes are discussed, which cover anti-aging effects, anti-inflammatory effects, anti-oxidation effects, neuroprotection, promotion of stem cell growth, etc.3.1 Neuroprotection
Fullerene derivatives are among the first nanozymes used for therapeutic studies.14,15 Dugan et al. pioneered the use of carboxyfullerenes as SOD mimics to protect neural cells from free radical damage.14,15 In a later study, Dugan's group not only investigated the SOD mimicking activity of C60[C(COOH)2]3 and the associated catalytic mechanism but also studied the nanozyme's therapeutic effect on SOD2 knockout mice (Fig. 11).222 For the Sod2−/− mice, which cannot express mitochondrial manganese superoxide dismutase (MnSOD), their life span has been increased by 300% after either utero or postnatal treatment with the C60[C(COOH)2]3 nanozyme. Since the nanozyme has been detected within mitochondria, it was proposed that the nanozyme may functionally replace MnSOD.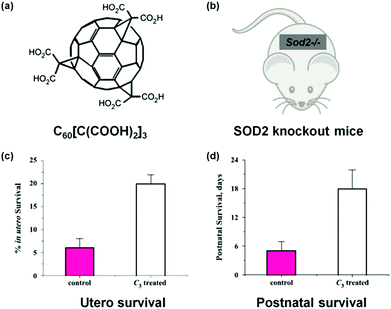 | ||
| Fig. 11 Treatment of SOD2 knockout mice with C60[C(COOH)2]3. (a) Structure of C60[C(COOH)2]3. (b) SOD2 knockout mice. Utero (c) and postnatal (d) survival of SOD2 knockout mice after treatment with C60[C(COOH)2]3. Adapted with permission from ref. 222. Copyright (2004) Elsevier. | ||
Ceria nanoparticles could also mimic SOD and exhibit interesting neuroprotective activity.17,227,230 Chen and co-workers demonstrated that ceria nanoparticles could prevent retinal neuron cells from ROS damage, which was induced by intracellular hydrogen peroxide.17 Remarkably, their animal studies also demonstrated that the ceria nanoparticle-based nanozyme exhibited protective activity towards light-induced rat retina photoreceptor cell degeneration.
ROS, generated and accumulated during ischemia, plays a critical role in ischemic stroke and associated neural injuries and diseases.238 By eliminating the ischemic ROS, it would be possible to protect the brain against ischemic stroke. For the first time, Kim et al. demonstrated that PEGylated ceria nanoparticles indeed exhibited excellent protective activity against ischemic stroke.227 The animal studies showed that ceria nanoparticles of optimal dosage reduced ischemic brain damage (Fig. 12).
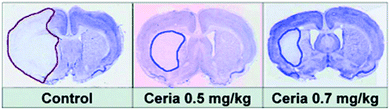 | ||
| Fig. 12 Ceria nanoparticles protect against ischemic stroke. Reprinted with permission from ref. 227. Copyright (2012) John Wiley and Sons. | ||
RNS and amyloid beta (Aβ) peptides are involved in Alzheimer's disease (AD). However, effective therapy towards AD with suitable antioxidants is still lacking. To address this challenge, Dowding and co-workers studied the protective roles of nanoceria against RNS and Aβ-induced mitochondrial fragmentation and neuronal cell death.230 The electron microscopic studies showed that the nanoceria were internalized by neurons and localized at the mitochondrial outer membrane and inner leaflet of the plasma membrane, where most of RNS were produced.230 Due to the switchable oxidation states between Ce3+ and Ce4+, ceria nanoparticles could scavenge RNS and thus protect the neurons against degeneration. The results demonstrated that ceria nanoparticles protected neurons from nitrosative-associated mitochondrial fragmentation, DRP1 S616 hyperphosphorylation and neuronal cell death.
3.2 Anti-aging
It is known that redox reactions participate in the aging process. For example, the detoxification of ROS is involved in regulating aging. Therefore, the ROS scavenging nanozymes may help to detoxify the ROS and thus prevent aging-related diseases. To this end, Quick et al. showed that the C60[C(COOH)2]3-based SOD mimic exhibited interesting anti-aging effects and improved wild-type mice's cognition ability.224 Detailed studies revealed that the nanozyme decreased the age-associated mitochondrial superoxide production and thus improved the mitochondrial biological function, which in turn rescued age-related cognitive impairment due to its localization within the mitochondria.2243.3 Anti-inflammatory
ROS can induce inflammatory responses, which may lead to endothelial dysfunction and tissue injury.239 By scavenging ROS with SOD or its mimics, it can protect cells and tissues from ROS-induced inflammation. Several nanozymes have exhibited interesting anti-inflammatory effects due to their ROS scavenging properties.232,240 Hirst et al. studied the anti-inflammatory properties of ceria nanoparticles.240 They demonstrated that the biocompatible ceria nanoparticles inhibited the production of ROS and radical nitric oxide (the two key inflammatory mediators) in J774A.1 murine macrophage cells. In a recent report, Son and co-workers integrated ceria nanoparticles as anti-inflammatory agents onto a bioresorbable electronic stent, which was a therapeutic device for endovascular diseases (Fig. 13).232 Using human umbilical vein endothelial cells as a model, they demonstrated that the nanozyme was able to improve the cell viability under oxidative stress. Moreover, they showed that the nanozyme also exhibited excellent anti-inflammatory effects in animal models after implantation of the device in the canine common carotid artery. The nanozyme's anti-inflammatory properties were attributed to their ROS scavenging effect. This study offers a new direction for nanozyme research.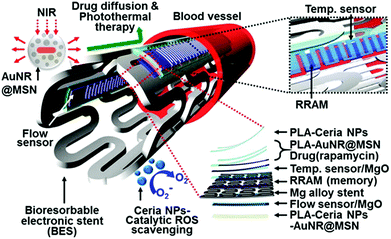 | ||
| Fig. 13 Ceria nanoparticles as anti-inflammatory agents in a bioresorbable electronic stent for endovascular diseases. Reprinted with permission from ref. 232. Copyright (2015) American Chemical Society. | ||
3.4 Anti-oxidation
The anti-oxidation effects of several nanozymes have been investigated.13,234,241 Early studies demonstrated that the nanoceria possessed interesting anti-oxidation properties.241 On the basis of the self-regenerating antioxidant mechanism, nanoceria protected cardiac progenitor cells from H2O2-induced cytotoxicity for one week.241Su et al. examined the protective effects of PVP-stabilized iridium nanoparticles against H2O2-induced oxidative damage to A549 lung cancer cells.234 They showed that IrNPs significantly reduced intracellular ROS levels and thus enhanced cell viability in a dose-dependent manner. Vernekar and co-workers studied the cytoprotective effect of vanadia (V2O5) nanowires (Fig. 14).13 They found that the vanadia nanowires exhibited interesting glutathione peroxidase (GPx) mimicking activity (Fig. 14a). Moreover, by combining the nanozyme with glutathione reductase (GR), the GSH was recycled. Using the genetically encoded H2O2-specific probe HyPer, they showed that the nanozyme indeed exhibited ROS scavenging properties against both extrinsic H2O2 and intrinsic cellular peroxide (induced by CuSO4) in HEK293T cells (Fig. 14b). The cytoprotective effects of the nanozyme were further confirmed by staining the cells with H2O2-specific dye Amplex Red and ROS-sensitive fluorescent dye DCFDA-H2 (Fig. 14c). In a following study, it was demonstrated that nanoceria could also mimic GPx and protect the cells via anti-oxidative effects.233
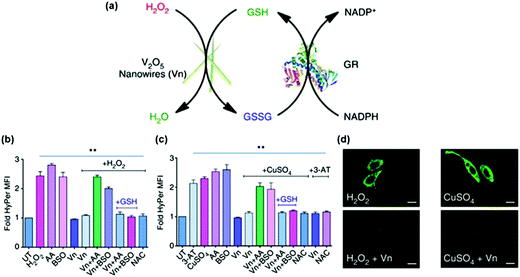 | ||
| Fig. 14 Vanadia nanowires as anti-oxidation nanozymes for cytoprotection. (a) Schematic depicting the glutathione peroxidase (GPx)-like antioxidant activity of vanadia nanowires (Vn) and GSH recycling by glutathione reductase (GR). Extrinsic H2O2 (b) and intrinsic cellular peroxide (induced by CuSO4) (c) scavenging activities of Vn measured using genetically encoded H2O2-specific probe HyPer in HEK293T cells. Cells were either left untreated or pretreated with various agents. (d) HeLa cells were treated with Vn before the treatment with H2O2 or CuSO4 and then stained with 15 μM DCFDA-H2 dye. Reprinted with permission from ref. 13. Copyright (2014) Nature Publishing Group. | ||
3.5 Anti-biofouling
Nanozymes have also been used for anti-biofouling.231,242,243 Tremel's group discovered that vanadia nanowires could mimic haloperoxidase.244 They further investigated the vanadia nanowires’ anti-biofouling properties in a subsequent report.242 They carried out a long-term (60 days) in situ study of the nanozyme's activity by fixing the testing samples to a boat hull. Remarkably, because of its excellent anti-biofouling activity, the nanozyme-coated stainless steel plates significantly prevented marine biofouling when compared with the uncoated stainless steel plate. Due to its high efficiency and low toxicity to marine biota, the nanozyme may be used as an alternative to conventional anti-biofouling agents.Gao et al. recently studied the Fe3O4 MNP-enhanced oxidative cleavage of biofilm components (such as nucleic acids, proteins, oligosaccharides, and bacteria).231 They used the Fe3O4 MNPs as a peroxidase mimic, which efficiently eliminated biofilms and thus prevented their formation in the presence of H2O2. Due to the nanozyme's unique penetrating properties into the protective, organic matrix, the developed Fe3O4 MNP–H2O2 system may provide a new approach to biofilm elimination in oral health, etc.
3.6 Other therapeutic applications
Nanozymes have also other interesting therapeutic potential, such as in promoting stem cell growth for tissue engineering and regulating iron homeostasis.25,226,228,229,237,245–247In Kito's report, they investigated the effects of Fe3O4 MNPs-containing liposomes on the proliferation of induced pluripotent stem (iPS) cells (Fig. 15).228 The proliferated iPS cells would form the cell sheets, which were then used for angiogenic cell therapy. It was found that the extracellular matrix (ECM) alone did not form a truly sheet-like structure and thus could not promote angiogenesis effectively. Surprisingly, when ECM was combined with Fe3O4 MNPs-containing liposomes for iPS cell proliferation, the formed iPS cell sheets significantly accelerated revascularization of the ischemic hindlimbs after implanting the sheets in nude mice. The Fe3O4 MNP-enhanced angiogenesis was attributed to the peroxidase-like anti-oxidative activities. The presence of Fe3O4 MNPs also improved the sheets’ mechanical properties, providing sufficient strength for handling.228 This study provides an interesting approach to tissue engineering for reparative angiogenesis.
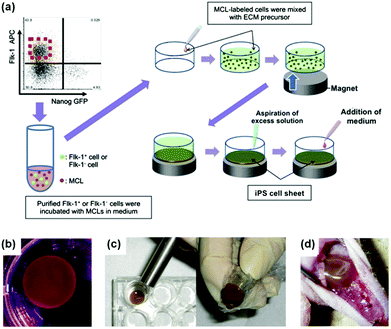 | ||
| Fig. 15 Fe3O4 MNP-facilitated formation of iPS cell sheets for reparative angiogenesis. (a) Procedure for construction of iPS cell-derived cell sheet. (b) An alnico magnet was positioned on the surface of the culture medium. The Flk-1 cell sheet floated up to the surface of the culture medium without disruption. (c) The magnetized Flk-1 cell sheet attached to an Alnico magnet covered with polyvinylidene difluoride membrane via a magnetic force. (d) Flk-1 cell sheets were placed on the adductor muscles of nude mice using the Alnico magnet. Reprinted with permission from ref. 228. Copyright (2013) Nature Publishing Group. | ||
Li et al. prepared PtNPs within L-chain apoferritin cages, which showed ferroxidase mimicking activity.25 They further showed that the nanozyme could regulate iron homeostasis, providing great promise for therapeutic applications. Molybdenum trioxide nanoparticles exhibited sulfite oxidase mimicking activity.229 The cellular study showed that the nanozyme could act as an alternative to natural sulfite oxidase, suggesting the potential use for treating sulfite oxidase-defect diseases.
4. Other applications
Nanozymes have also found other exciting applications, such as in constructing logic gates, pollutant removal and water treatment, etc.23,65,187,189,247–2554.1 Logic gates
Several logic gates have been constructed with various nanozymes.187,189,248,249As shown in Fig. 16, when the ceria nanoparticles were combined with natural enzymes, label-free, resettable, and colorimetric logic gates have been fabricated.248 For ceria nanoparticles, they were colourless when Ce3+ was dominant on the nanoparticles’ surface. However, they would turn into yellow colour when the surface Ce3+ was oxidized to Ce4+ with H2O2. Moreover, by heating, the yellow coloured ceria nanoparticles would return to colourless (Fig. 16a). Lin et al. exploited the unique colour change of the ceria nanoparticles as a signalling readout to construct various logic gates. For example, when the ceria nanoparticles were combined with β-galactosidase and glucose oxidase, an “AND” logic gate was built, and “OR” and “INHIBIT” logic gates were also obtained (Fig. 16c). Due to the thermally responsive colour changing properties, the ceria nanoparticles could be transformed into colourless ones, which in turn reset the logic gates. The potential application of the logic gates for multiplex detection was also studied.
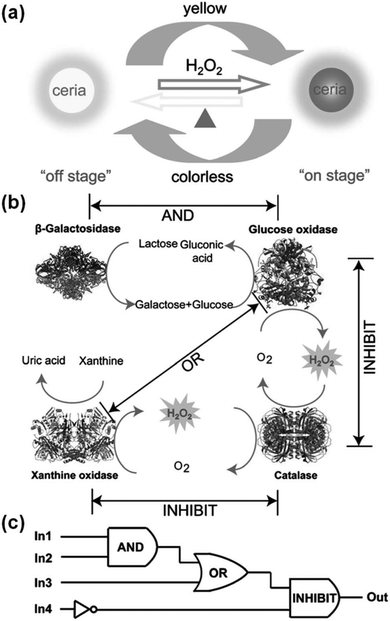 | ||
| Fig. 16 Ceria nanoparticles for colorimetric logic gates. (a) Illustration of a thermally responsive switch based on CeO2 nanoparticles. (b) The operation of logic gates based on biocatalytic reactions. (c) Logic circuitry for the integrated logic system. In1 = β-galactosidase, In2 = glucose oxidase, In3 = xanthine oxidase, In4 = catalase. Reprinted with permission from ref. 248. Copyright (2012) John Wiley and Sons. | ||
Lien and co-workers reported several logic gates on the basis of tuning the AuNPs’ multiple enzyme-like activities with metal ions.187,189 For example, both Be3+ and Hg2+ enhanced AuNPs’ catalase mimicking activity, respectively. Thus, an “OR” logic gate was developed using Be3+ and Hg2+ as inputs and the AuNPs’ catalase activity as the output (Fig. 17a).187 To enhance the AuNPs’ oxidase mimicking activity, both Pt4+ and Hg2+ were necessary. On the basis of this phenomenon, an “AND” logic gate was fabricated (Fig. 17b). Similarly, an “INHIBIT” logic gate was developed by tuning AuNPs’ peroxidase mimicking activity with Pb2+ and Hg2+ (Fig. 17c). Pb2+ enhanced the peroxidase mimicking activity while Hg2+ exhibited an inhibition effect. The presence of both Pb2+ and Hg2+, however, also inhibited the mimicking activity. An “XOR” logic gate was also reported by tuning the AuNPs’ peroxidase mimicking activity with Hg2+ and Bi3+ (Fig. 17d).
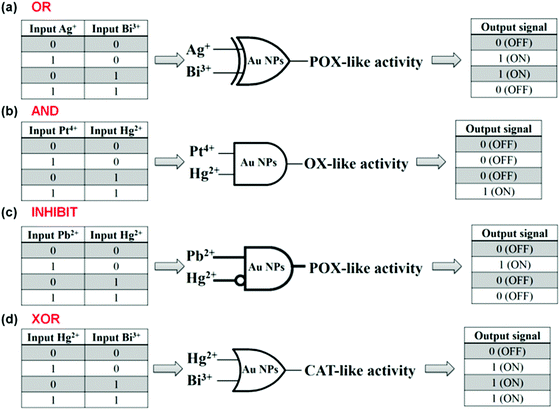 | ||
| Fig. 17 Logical regulation of the enzyme-like activity of AuNPs by using metal ions. “OR” (a), “AND” (b), “INHIBIT” (c), and “XOR” (d) logic gates on the basis of tuning AuNPs’ enzyme mimicking activities with various metal ions. Reprinted with permission from ref. 187. Copyright (2013) Royal Society of Chemistry. | ||
4.2 Pollutant removal and water treatment
Yan's group reported the removal of phenolic pollutants from aqueous solutions using the Fe3O4 MNP-based peroxidase mimic.250 After that, numerous studies have been devoted to pollutant removal and water treatment with nanozymes.23,65,250–252In a recent report, Janos et al. investigated the phosphatase mimicking behaviour of the CeO2/γ-Fe2O3 nanocomposites.23 Due to the adsorption and phosphatase mimetic properties, the nanozyme has been used to decompose organophosphorus pesticide parathion methyl as well as the chemical warfare agents Soman and VX at ambient temperature. Since the nanozyme still retained the magnetic properties, it could be recycled and used in various decontamination strategies.23
5. Mechanisms
Though relatively few studies have been focused on elucidating the mechanisms of the nanozyme based catalysis, several mechanisms have been established.32,222 In this section, the catalytic mechanisms of the fullerene-based SOD mimic and the nanoceria-based SOD and catalase mimics are discussed, which should be helpful for understanding the mechanisms of other nanozymes in the future.5.1 C60[C(COOH)2]3 based SOD mimic
The detailed mechanism of C60[C(COOH)2]3 based SOD mimic has been proposed by Ali and co-workers.222 By combining the experimental results (such as electron paramagnetic resonance (EPR) measurements and kinetics studies) with semiempirical quantum-mechanical calculations, they proposed the mechanism shown in Fig. 18.222 Their study suggested that the electron-deficient regions on the C60[C(COOH)2]3 nanozyme attracted the substrate anions (i.e., O2˙−) towards the surface of the nanozyme electrostatically and then directed the substrate for further dismutation. The whole process was facilitated by the protons from the carboxyl groups of the nanozyme and/or surrounding water molecules around the nanozyme.222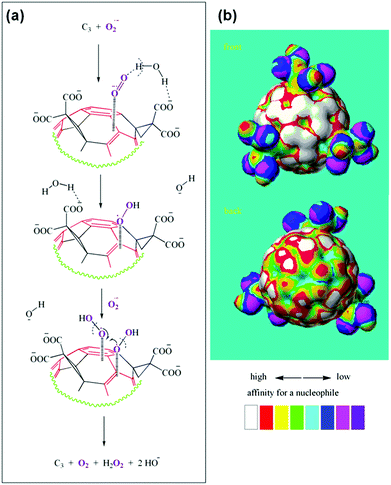 | ||
| Fig. 18 Proposed mechanism of C60[C(COOH)2]3 based SOD mimic. Reprinted with permission from ref. 222. Copyright (2004) Elsevier. | ||
5.2 Nanoceria based SOD mimic
The SOD mimetic activity of nanoceria has been validated by the competitive assay against cytochrome c.256 Based on EPR measurements, kinetics studies as well as other observations, Self's group proposed a dismutation mechanism of nanoceria-based SOD mimics, which is similar to the mechanism of Fe- and Mn-SOD.256 As shown in Fig. 19, an alternative mechanism was proposed by Celardo et al., which showed the redox cycle between Ce3+ and Ce4+.32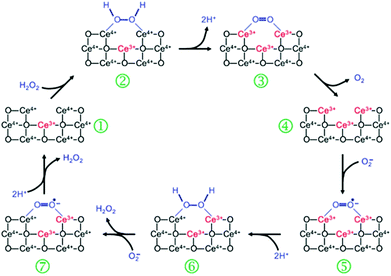 | ||
| Fig. 19 Proposed mechanism of nanoceria based SOD mimic. Reprinted with permission from ref. 32. Copyright (2011) Royal Society of Chemistry. | ||
5.3 Nanoceria based catalase mimic
Nanoceria has also shown catalase mimetic activity. Fig. 20 shows a possible mechanism for the reaction cycle similar to the one of natural catalase.32 The overall reaction involved in Fig. 20 can be expressed by eqn (3) in Scheme 3.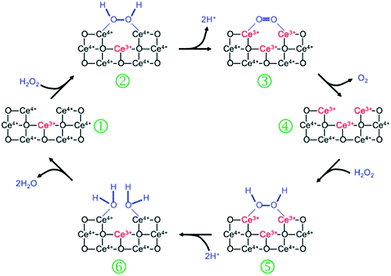 | ||
| Fig. 20 Proposed mechanism of nanoceria based catalase mimic. Reprinted with permission from ref. 32. Copyright (2011) Royal Society of Chemistry. | ||
6. Summary and outlook
This review highlights the recent progress of nanozymes, especially their applications in bionanotechnology. Using selected examples, the broad applications in various areas (such as sensing, imaging, therapeutics, logic gates, pollutant removal and water treatment) were discussed.As evidenced by the above-mentioned examples, the research field of nanozymes has grown substantially. To fulfill their great potential however, there are still numerous challenges remaining to be tackled.1
First, though many nanomaterials have been exploited to mimic various natural enzymes,81,101,257–267 currently the redox enzyme mimics are still dominant in the developed nanozymes. Therefore, new strategies are needed to design and prepare other types of nanozymes. Recent progress in theoretical and computational protein design has enabled the design of new functional proteins, including enzymes.268,269 Such an approach should also be applicable to nanozyme design by combining experiments with theoretical and/or computational strategies.
Second, nanozymes are superior to natural enzymes in their enhanced stability, recyclability, low-cost, etc.105,270 Their catalytic activity and selectivity, however, still cannot compete with natural enzymes. By further understanding their mechanisms, high-performance nanozymes could be rationally designed in future.271–274 For example, by taking advantage of the synergistic effect, nanozymes with improved activities have been prepared.275
Third, natural enzymes usually work together within a confined environment. To truly mimic natural enzymes, cascade reactions catalyzed by nanozyme assemblies (or assemblies of nanozyme and natural enzyme) should be further exploited.238,276,277
Fourth, like natural enzymes, nanozyme’ activities can be regulated.47,78,158,278–285 For example, by tailoring the nanozymes’ composition, their activities could be tuned.78,278 pH and even light have been investigated for modulating the nanozymes’ activities.280,281,283,284 Despite the current progress, more approaches and factors are still needed to intentionally modulate nanozymes’ catalytic properties. Among them, biomolecules will play critical roles in manipulating nanozymes’ catalytic behaviours.47,158,286–290
Fifth, for biomedical applications, a broader range of targets should be considered. More importantly, translational studies should be carried out whenever possible in future. For example, when the nanozyme-based test strips are combined with available telemed devices, they may help us address the need for point-of-care and even precision medicine. For the therapeutic nanozymes, their effective windows should be carefully optimized to avoid potential toxicity.
Sixth, for biomedical applications, the toxicity of nanozymes should be systematically studied. Though previous reports have shown that several nanozymes have been employed for therapeutic studies in animal models, their translation into clinical applications remains a great challenge. Currently, a few FDA approved superparamagnetic iron oxide nanoparticles, such as Resovist (Ferucarbotran), have exhibited peroxidase-like activity and been used for promoting stem cell proliferation.237 However, the toxicity of other promising nanozymes should be tested for clinical trials.
Finally, chiral catalysis using nanozyme has been reported recently.255 Here we speculate that nanozymes may be even involved in the origin of life since there must have been some catalysts to carry out the catalysis before biocatalysts evolved.
Abbreviations
| 4-AAP | 4-Minoantipyrine |
| ABTS | 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) |
| CNTs | Carbon nanotubes |
| DPD | N,N-Diethyl-p-phenylenediamine |
| DRP1 | Dynamin-related protein 1 |
| ECM | Extracellular matrix |
| EpCAM | Epithelial cell adhesion molecule |
| EPR | Electron paramagnetic resonance |
| FDA | Food and drug administration |
| GPx | Glutathione peroxidase |
| GSH | Glutathione |
| hCG | Human chorionic gonadotropin |
| HER2 | Human epidermal growth factor receptor 2 |
| 5-HIAA | 5-Hydroxyindole-3-acetic acid |
| HPLC | High-performance liquid chromatography |
| iPS | Induced pluripotent stem |
| LOD | Limit of detection |
| MNPs | Magnetic nanoparticles |
| MS | Mass spectrometry |
| NPs | Nanoparticles |
| OPD | o-Phenylenediamine |
| POC | Point-of-care |
| PSA | Prostate specific antigen |
| PVP | Polyvinylpyrrolidone |
| rGO | Reduced graphene oxide |
| SERS | Surface enhanced Raman scattering |
| SOD | Superoxide dismutase |
| TMB | 3,3′,5,5′-Tetramethylbenzidine |
Acknowledgements
We thank the National Natural Science Foundation of China (no. 21405081), the Natural Science Foundation of Jiangsu Province (no. BK20130561), the 973 Program (no. 2015CB659400), the 985 Program of Nanjing University, the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Shuangchuang Program of Jiangsu Province, the Six Talents Summit Program of Jiangsu Province, Fundamental Research Funds for the Central Universities, Open Funds of the State Key Laboratory of Electroanalytical Chemistry (SKLEAC201501), and the Thousand Talents Program for Young Researchers for financial support.Notes and references
- H. Wei and E. K. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- L. Z. Gao and X. Y. Yan, Prog. Biochem. Biophys., 2013, 40, 892–902 CAS.
- L. Pasquato, P. Pengo and P. Scrimin, Supramol. Chem., 2005, 17, 163–171 CrossRef CAS.
- L. Z. Gao and X. Y. Yan, in Nanomaterials: emerging characteristics and biomedical applications, ed. X. Y. Yan, Science Press, Beijing, 2014 Search PubMed.
- T. Pirmohamed, J. M. Dowding, S. Singh, B. Wasserman, E. Heckert, A. S. Karakoti, J. E. S. King, S. Seal and W. T. Self, Chem. Commun., 2010, 46, 2736–2738 RSC.
- S. Singh, T. Dosani, A. S. Karakoti, A. Kumar, S. Seal and W. T. Self, Biomaterials, 2011, 32, 6745–6753 CrossRef CAS PubMed.
- W. W. He, Y. Liu, J. S. Yuan, J. J. Yin, X. C. Wu, X. N. Hu, K. Zhang, J. B. Liu, C. Y. Chen, Y. L. Ji and Y. T. Guo, Biomaterials, 2011, 32, 1139–1147 CrossRef CAS PubMed.
- Z. W. Chen, J. J. Yin, Y. T. Zhou, Y. Zhang, L. Song, M. J. Song, S. L. Hu and N. Gu, ACS Nano, 2012, 6, 4001–4012 CrossRef CAS PubMed.
- M. Comotti, C. Della Pina, R. Matarrese and M. Rossi, Angew. Chem., Int. Ed., 2004, 43, 5812–5815 CrossRef CAS PubMed.
- A. Asati, S. Santra, C. Kaittanis, S. Nath and J. M. Perez, Angew. Chem., Int. Ed., 2009, 48, 2308–2312 CrossRef CAS PubMed.
- L. Z. Gao, J. Zhuang, L. Nie, J. B. Zhang, Y. Zhang, N. Gu, T. H. Wang, J. Feng, D. L. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- H. Wei and E. Wang, Anal. Chem., 2008, 80, 2250–2254 CrossRef CAS PubMed.
- A. A. Vernekar, D. Sinha, S. Srivastava, P. U. Paramasivam, P. D'Silva and G. Mugesh, Nat. Commun., 2014, 5, 5301 CrossRef CAS PubMed.
- L. L. Dugan, J. K. Gabrielsen, S. P. Yu, T. S. Lin and D. W. Choi, Neurobiol. Dis., 1996, 3, 129–135 CrossRef CAS PubMed.
- L. L. Dugan, D. M. Turetsky, C. Du, D. Lobner, M. Wheeler, C. R. Almli, C. K. F. Shen, T. Y. Luh, D. W. Choi and T. S. Lin, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 9434–9439 CrossRef CAS.
- R. W. Tarnuzzer, J. Colon, S. Patil and S. Seal, Nano Lett., 2005, 5, 2573–2577 CrossRef CAS PubMed.
- J. P. Chen, S. Patil, S. Seal and J. F. McGinnis, Nat. Nanotechnol., 2006, 1, 142–150 CrossRef CAS PubMed.
- H. Tokuyama, S. Yamago, E. Nakamura, T. Shiraki and Y. Sugiura, J. Am. Chem. Soc., 1993, 115, 7918–7919 CrossRef CAS.
- A. S. Boutorine, H. Tokuyama, M. Takasugi, H. Isobe, E. Nakamura and C. Helene, Angew. Chem., Int. Ed. Engl., 1995, 33, 2462–2465 CrossRef.
- S. Bosi, T. Da Ros, G. Spalluto and M. Prato, Eur. J. Med. Chem., 2003, 38, 913–923 CrossRef CAS PubMed.
- L. Pasquato, F. Rancan, P. Scrimin, F. Mancin and C. Frigeri, Chem. Commun., 2000, 2253–2254 RSC.
- F. Manea, F. B. Houillon, L. Pasquato and P. Scrimin, Angew. Chem., Int. Ed., 2004, 43, 6165–6169 CrossRef CAS PubMed.
- P. Janos, P. Kuran, V. Pilarova, J. Trogl, M. Stastny, O. Pelant, J. Henych, S. Bakardjieva, O. Zivotsky, M. Kormunda, K. Mazanec and M. Skoumal, Chem. Eng. J., 2015, 262, 747–755 CrossRef CAS.
- B. Li, D. M. Chen, J. Q. Wang, Z. Y. Yan, L. Jiang, D. L. Duan, J. He, Z. R. Luo, J. P. Zhang and F. G. Yuan, Sci. Rep., 2014, 4, 6759 CrossRef PubMed.
- L. Li, L. Zhang, U. Carmona and M. Knez, Chem. Commun., 2014, 50, 8021–8023 RSC.
- X. Li, F. Wen, B. Creran, Y. Jeong, X. Zhang and V. M. Rotello, Small, 2012, 8, 3589–3592 CrossRef CAS PubMed.
- E. Nakamura and H. Isobe, Acc. Chem. Res., 2003, 36, 807–815 CrossRef CAS PubMed.
- W. He, W. Wamer, Q. Xia, J.-j. Yin and P. P. Fu, J. Environ. Sci. Health Environ. Part C: Environ Carcinog. Ecotoxicol. Rev., 2014, 32, 186–211 CAS.
- L. J. Prins, Acc. Chem. Res., 2015, 48, 1920–1928 CrossRef CAS PubMed.
- Y. H. Lin, J. S. Ren and X. G. Qu, Adv. Mater., 2014, 26, 4200–4217 CrossRef CAS PubMed.
- J. X. Xie, X. D. Zhang, H. Wang, H. Z. Zheng and Y. M. Huang, TrAC, Trends Anal. Chem., 2012, 39, 114–129 CrossRef CAS.
- I. Celardo, J. Z. Pedersen, E. Traversa and L. Ghibelli, Nanoscale, 2011, 3, 1411–1420 RSC.
- L. Zheng, J. Zhao, X. Niu and Y. Yang, Mater. Rev., 2015, 29, 115 CAS.
- Y. H. Lin, J. S. Ren and X. G. Qu, Acc. Chem. Res., 2014, 47, 1097–1105 CrossRef CAS PubMed.
- A. Karakoti, S. Singh, J. M. Dowding, S. Seal and W. T. Self, Chem. Soc. Rev., 2010, 39, 4422–4432 RSC.
- C. Xu and X. G. Qu, NPG Asia Mater., 2014, 6, e90 CrossRef CAS.
- C. L. Guo, Y. H. Song, H. Wei, P. C. Li, L. Wang, L. L. Sun, Y. J. Sun and Z. Li, Anal. Bioanal. Chem., 2007, 389, 527–532 CrossRef CAS PubMed.
- Z. Liu, B. Zhao, Y. Shi, C. Guo, H. Yang and Z. Li, Talanta, 2010, 81, 1650–1654 CrossRef CAS PubMed.
- K. Zhao, W. Gu, S. S. Zheng, C. L. Zhang and Y. Z. Xian, Talanta, 2015, 141, 47–52 CrossRef CAS PubMed.
- Y. H. Wang, B. Zhou, S. Wu, K. M. Wang and X. X. He, Talanta, 2015, 134, 712–717 CrossRef CAS PubMed.
- F. M. Qiao, L. J. Chen, X. N. Li, L. F. Li and S. Y. Ai, Sens. Actuators, B, 2014, 193, 255–262 CrossRef CAS.
- A. K. Dutta, S. K. Maji, A. Mondal, B. Karmakar, P. Biswas and B. Adhikary, Sens. Actuators, B, 2012, 173, 724–731 CrossRef CAS.
- Y. F. Zhang, C. L. Xu and B. X. Li, RSC Adv., 2013, 3, 6044–6050 RSC.
- W. S. Yang, J. H. Hao, Z. Zhang, B. P. Lu, B. L. Zhang and J. L. Tang, RSC Adv., 2014, 4, 35500–35504 RSC.
- P. C. Pandey, D. Panday and G. Pandey, RSC Adv., 2014, 4, 60563–60572 RSC.
- A. Dalui, B. Pradhan, U. Thupakula, A. H. Khan, G. S. Kumar, T. Ghosh, B. Satpati and S. Acharya, Nanoscale, 2015, 7, 9062–9074 RSC.
- L. J. Chen, K. F. Sun, P. P. Li, X. Z. Fan, J. C. Sun and S. Y. Ai, Nanoscale, 2013, 5, 10982–10988 RSC.
- Q. Chang and H. Q. Tang, Microchim. Acta, 2014, 181, 527–534 CrossRef CAS.
- Q. Y. Liu, L. Y. Zhang, H. Li, Q. Y. Jia, Y. L. Jiang, Y. T. Yang and R. R. Zhu, Mater. Sci. Eng., C, 2015, 55, 193–200 CrossRef CAS PubMed.
- W. Zhang, Y. Zhang, Y. H. Chen, S. Y. Li, N. Gu, S. L. Hu, Y. Sun, X. Chen and Q. Li, J. Nanosci. Nanotechnol., 2013, 13, 60–67 CrossRef CAS PubMed.
- D. Wu, X. Deng, X. M. Huang, K. Wang and Q. Y. Liu, J. Nanosci. Nanotechnol., 2013, 13, 6611–6616 CrossRef PubMed.
- R. Z. Zhang, S. J. He, C. M. Zhang and W. Chen, J. Mater. Chem. B, 2015, 3, 4146–4154 Search PubMed.
- C. L. Sun, X. L. Chen, J. Xu, M. J. Wei, J. J. Wang, X. G. Mi, X. H. Wang, Y. Wu and Y. Liu, J. Mater. Chem. A, 2013, 1, 4699–4705 CAS.
- T. H. Han, M. M. Khan, J. Lee and M. H. Cho, J. Ind. Eng. Chem., 2014, 20, 2003–2009 CrossRef CAS.
- L. Zhan, Y. Zhang, Q. L. Zeng, Z. D. Liu and C. Z. Huang, J. Colloid Interface Sci., 2014, 426, 293–299 CrossRef CAS PubMed.
- X. Yang, L. N. Wang, G. Z. Zhou, N. Sui, Y. X. Gu and J. Wan, J. Cluster Sci., 2015, 26, 789–798 CrossRef CAS.
- Y. Gao, Z. Wei, F. Li, Z. M. Yang, Y. M. Chen, M. Zrinyi and Y. Osada, Green Chem., 2014, 16, 1255–1261 RSC.
- S. H. Ma, X. J. Wang and X. J. Han, Chin. J. Anal. Chem., 2013, 41, 1719–1723 CrossRef CAS.
- W. T. Yao, H. Z. Zhu, W. G. Li, H. B. Yao, Y. C. Wu and S. H. Yu, ChemPlusChem, 2013, 78, 723–727 CrossRef CAS.
- T. Wang, Y. C. Fu, L. Y. Chai, L. Chao, L. J. Bu, Y. Meng, C. Chen, M. Ma, Q. J. Xie and S. Z. Yao, Chem. – Eur. J., 2014, 20, 2623–2630 CrossRef CAS PubMed.
- F. X. Qin, S. Y. Jia, F. F. Wang, S. H. Wu, J. Song and Y. Liu, Catal. Sci. Technol., 2013, 3, 2761–2768 CAS.
- A. X. Zheng, Z. X. Cong, J. R. Wang, J. Li, H. H. Yang and G. N. Chen, Biosens. Bioelectron., 2013, 49, 519–524 CrossRef CAS PubMed.
- Q. Y. Liu, Y. T. Yang, H. Li, R. R. Zhu, Q. Shao, S. G. Yang and J. J. Xu, Biosens. Bioelectron., 2015, 64, 147–153 CrossRef CAS PubMed.
- A. L. Hu, Y. H. Liu, H. H. Deng, G. L. Hong, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Biosens. Bioelectron., 2014, 61, 374–378 CrossRef CAS PubMed.
- Y. Pan, N. Li, J. S. Mu, R. H. Zhou, Y. Xu, D. Z. Cui, Y. Wang and M. Zhao, Appl. Microbiol. Biotechnol., 2015, 99, 703–715 CrossRef CAS PubMed.
- J. F. Guan, J. Peng and X. Y. Jin, Anal. Methods, 2015, 7, 5454–5461 RSC.
- H. T. Fang, Y. L. Pan, W. Q. Shan, M. L. Guo, Z. Nie, Y. Huang and S. Z. Yao, Anal. Methods, 2014, 6, 6073–6081 RSC.
- A. D. Ryabov, R. Ceron-Camacho, O. Saavedra-Diaz, M. A. Denardo, A. Ghosh, R. Le Lagadec and T. J. Collins, Anal. Chem., 2012, 84, 9096–9100 CAS.
- L. P. Lin, X. H. Song, Y. Y. Chen, M. C. Rong, T. T. Zhao, Y. R. Wang, Y. Q. Jiang and X. Chen, Anal. Chim. Acta, 2015, 869, 89–95 CrossRef CAS PubMed.
- F. M. Qiao, Z. Z. Wang, K. Xu and S. Y. Ai, Analyst, 2015, 140, 6684–6691 RSC.
- Q. Chen, J. Chen, C. J. Gao, M. L. Zhang, J. Y. Chen and H. D. Qiu, Analyst, 2015, 140, 2857–2863 RSC.
- Y. Shi, P. Su, Y. Y. Wang and Y. Yang, Talanta, 2014, 130, 259–264 CrossRef CAS PubMed.
- F. T. Zhang, X. Long, D. W. Zhang, Y. L. Sun, Y. L. Zhou, Y. R. Ma, L. M. Qi and X. X. Zhang, Sens. Actuators, B, 2014, 192, 150–156 CrossRef CAS.
- F. M. Qian, J. M. Wang, S. Y. Ai and L. F. Li, Sens. Actuators, 2015, 216, 418–427 CrossRef.
- F. Pogacean, C. Socaci, S. Pruneanu, A. R. Biris, M. Coros, L. Magerusan, G. Katona, R. Turcu and G. Borodi, Sens. Actuators, B, 2015, 213, 474–483 CrossRef CAS.
- X. M. Chen, B. Y. Su, Z. X. Cai, X. Chen and M. Oyama, Sens. Actuators, B, 2014, 201, 286–292 CrossRef CAS.
- P. C. Pandey, R. Singh and Y. Pandey, RSC Adv., 2015, 5, 49671–49679 RSC.
- J. Q. Tian, Q. Liu, A. M. Asiri, A. H. Qusti, A. O. Al-Youbi and X. P. Sun, Nanoscale, 2013, 5, 11604–11609 RSC.
- P. Gayathri and A. S. Kumar, Chem. – Eur. J., 2013, 19, 17103–17112 CrossRef CAS PubMed.
- L. H. Ai, L. L. Li, C. H. Zhang, J. Fu and J. Jiang, Chem. – Eur. J., 2013, 19, 15105–15108 CrossRef CAS PubMed.
- Y. Tao, E. G. Ju, J. S. Ren and X. G. Qu, Chem. Commun., 2014, 50, 3030–3032 RSC.
- K. S. McKeating, S. Sloan-Dennison, D. Graham and K. Faulds, Analyst, 2013, 138, 6347–6353 RSC.
- T. B. Zhang, Y. C. Lu and G. S. Luo, ACS Appl. Mater. Interfaces, 2014, 6, 14433–14438 CAS.
- J. A. R. Guivar, E. G. R. Fernandes and V. Zucolotto, Talanta, 2015, 141, 307–314 CrossRef CAS PubMed.
- J. Zhuang, J. B. Zhang, L. Z. Gao, Y. Zhang, N. Gu, J. Feng, D. L. Yang and X. Y. Yan, Mater. Lett., 2008, 62, 3972–3974 CrossRef CAS.
- S. Chen, X. Hai, X. W. Chen and J. H. Wang, Anal. Chem., 2014, 86, 6689–6694 CrossRef CAS PubMed.
- D. Antuna-Jimenez, M. C. Blanco-Lopez, A. J. Miranda-Ordieres and M. J. Lobo-Castanon, Polymer, 2014, 55, 1113–1119 CrossRef CAS.
- Y. Shi, J. Huang, J. N. Wang, P. Su and Y. Yang, Talanta, 2015, 143, 457–463 CrossRef CAS PubMed.
- A. K. Dutta, S. Das, S. Samanta, P. K. Samanta, B. Adhikary and P. Biswas, Talanta, 2013, 107, 361–367 CrossRef CAS PubMed.
- Q. Li, G. G. Tang, X. W. Xiong, Y. L. Cao, L. L. Chen, F. G. Xu and H. L. Tan, Sens. Actuators, B, 2015, 215, 86–92 CrossRef CAS.
- A. K. Dutta, S. K. Maji, P. Biswas and B. Adhikary, Sens. Actuators, B, 2013, 177, 676–683 CrossRef CAS.
- W. F. Dong, X. D. Liu, W. B. Shi and Y. M. Huang, RSC Adv., 2015, 5, 17451–17457 RSC.
- J. Huang, Q. Chang, G. Jiang, Y. Qiu and H. Tang, Phys. Test. Chem. Anal. B Chem. Anal., 2014, 50, 417–420 CAS.
- T. R. Lin, L. S. Zhong, L. Q. Guo, F. F. Fu and G. N. Chen, Nanoscale, 2014, 6, 11856–11862 RSC.
- L. Han, L. X. Zeng, M. D. Wei, C. M. Li and A. H. Liu, Nanoscale, 2015, 7, 11678–11685 RSC.
- Q. Y. Liu, H. Li, Q. R. Zhao, R. R. Zhu, Y. T. Yang, Q. Y. Jia, B. Bian and L. H. Zhuo, Mater. Sci. Eng., C, 2014, 41, 142–151 CrossRef CAS PubMed.
- Q. Y. Liu, Q. Y. Jia, R. R. Zhu, Q. Shao, D. M. Wang, P. Cui and J. C. Ge, Mater. Sci. Eng., C, 2014, 42, 177–184 CrossRef CAS PubMed.
- Y. X. Chen, H. H. Zhang, H. G. Xue, X. Y. Hu, G. X. Wang and C. Y. Wang, Mater. Sci. Eng., C, 2014, 35, 420–425 CrossRef CAS PubMed.
- B. Liu, Z. Sun, P.-J. J. Huang and J. Liu, J. Am. Chem. Soc., 2015, 137, 1290–1295 CrossRef CAS PubMed.
- H. Wang, S. Li, Y. M. Si, Z. Z. Sun, S. Y. Li and Y. H. Lin, J. Mater. Chem. B, 2014, 2, 4442–4448 RSC.
- N. J. Lang, B. W. Liu and J. W. Liu, J. Colloid Interface Sci., 2014, 428, 78–83 CrossRef CAS PubMed.
- L. Li, C. Zeng, L. Ai and J. Jiang, J. Alloys Compd., 2015, 639, 470–477 CrossRef CAS.
- T. Tian, L. H. Ai, X. M. Liu, L. L. Li, J. Li and J. Jiang, Ind. Eng. Chem. Res., 2015, 54, 1171–1178 CrossRef CAS.
- K. G. Qu, P. Shi, J. S. Ren and X. G. Qu, Chem. – Eur. J., 2014, 20, 7501–7506 CrossRef CAS PubMed.
- L. J. Wan, J. H. Liu and X. J. Huang, Chem. Commun., 2014, 50, 13589–13591 RSC.
- Y. J. Chen, H. Y. Cao, W. B. Shi, H. Liu and Y. M. Huang, Chem. Commun., 2013, 49, 5013–5015 RSC.
- Z. C. Xing, J. Q. Tian, A. M. Asiri, A. H. Qusti, A. O. Al-Youbi and X. P. Sun, Biosens. Bioelectron., 2014, 52, 452–457 CrossRef CAS PubMed.
- L. Su, W. J. Qin, H. G. Zhang, Z. U. Rahman, C. L. Ren, S. D. Ma and X. G. Chen, Biosens. Bioelectron., 2015, 63, 384–391 CrossRef CAS PubMed.
- Y. Liu, M. Yuan, L. J. Qiao and R. Guo, Biosens. Bioelectron., 2014, 52, 391–396 CrossRef CAS PubMed.
- T. R. Lin, L. S. Zhong, J. Wang, L. Q. Guo, H. Y. Wu, Q. Q. Guo, F. F. Fu and G. N. Chen, Biosens. Bioelectron., 2014, 59, 89–93 CrossRef CAS PubMed.
- T. R. Lin, L. S. Zhong, Z. P. Song, L. Q. Guo, H. Y. Wu, Q. Q. Guo, Y. Chen, F. F. Fu and G. N. Chen, Biosens. Bioelectron., 2014, 62, 302–307 CrossRef CAS PubMed.
- R. M. Li, M. M. Zhen, M. R. Guan, D. Q. Chen, G. Q. Zhang, J. C. Ge, P. Gong, C. R. Wang and C. Y. Shu, Biosens. Bioelectron., 2013, 47, 502–507 CrossRef CAS PubMed.
- K. Mitra, A. B. Ghosh, A. Sarkar, N. Saha and A. K. Dutta, Biochem. Biophys. Res. Commun., 2014, 451, 30–35 CrossRef CAS PubMed.
- J. X. Xie, H. Y. Cao, H. Jiang, Y. J. Chen, W. B. Shi, H. Z. Zheng and Y. M. Huang, Anal. Chim. Acta, 2013, 796, 92–100 CrossRef CAS PubMed.
- S. Zhu, X.-E. Zhao, J. You, G. Xu and H. Wang, Analyst, 2015, 140, 6398–6403 RSC.
- Y. L. Liu, X. J. Zhao, X. X. Yang and Y. F. Li, Analyst, 2013, 138, 4526–4531 RSC.
- Y. Guo, W. W. Li, M. Y. Zheng and Y. Huang, Acta Chim. Sin., 2014, 72, 713–719 CrossRef CAS.
- R. Qu, L. L. Shen, A. T. Qu, R. L. Wang, Y. L. An and L. Q. Shi, ACS Appl. Mater. Interfaces, 2015, 7, 16694–16705 CAS.
- C. Lu, X. J. Liu, Y. F. Li, F. Yu, L. H. Tang, Y. J. Hu and Y. B. Yine, ACS Appl. Mater. Interfaces, 2015, 7, 15395–15402 CAS.
- L. Han, C. C. Li, T. Zhang, Q. L. Lang and A. H. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 14463–14470 CAS.
- L. Su, J. Feng, X. M. Zhou, C. L. Ren, H. H. Li and X. G. Chen, Anal. Chem., 2012, 84, 5753–5758 CrossRef CAS PubMed.
- W. Haider, A. Hayat, Y. Raza, A. A. Chaudhry, R. Ihtesham Ur and J. L. Marty, RSC Adv., 2015, 5, 24853–24858 RSC.
- S. B. He, G. W. Wu, H. H. Deng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Biosens. Bioelectron., 2014, 62, 331–336 CrossRef CAS PubMed.
- Y. R. Tang, Y. Zhang, R. Liu, Y. Y. Su and Y. Lu, Chin. J. Anal. Chem., 2013, 41, 330–336 CrossRef CAS.
- C. I. Wang, W. T. Chen and H. T. Chang, Anal. Chem., 2012, 84, 9706–9712 CrossRef CAS PubMed.
- M. M. Liang, K. L. Fan, Y. Pan, H. Jiang, F. Wang, D. L. Yang, D. Lu, J. Feng, J. J. Zhao, L. Yang and X. Y. Yan, Anal. Chem., 2013, 85, 308–312 CrossRef CAS PubMed.
- J. Qian, X. W. Yang, L. Jiang, C. D. Zhu, H. P. Mao and K. Wang, Sens. Actuators, B, 2014, 201, 160–166 CrossRef CAS.
- M. Shamsipur, A. Safavi and Z. Mohammadpour, Sens. Actuators, B, 2014, 199, 463–469 CrossRef CAS.
- P. J. Ni, H. C. Dai, Y. L. Wang, Y. J. Sun, Y. Shi, J. T. Hu and Z. Li, Biosens. Bioelectron., 2014, 60, 286–291 CrossRef CAS PubMed.
- X. Y. Niu, Y. Y. Xu, Y. L. Dong, L. Y. Qi, S. D. Qi, H. L. Chen and X. G. Chen, J. Alloys Compd., 2014, 587, 74–81 CrossRef CAS.
- H. L. Tan, C. J. Ma, L. Gao, Q. Li, Y. H. Song, F. G. Xu, T. Wang and L. Wang, Chem. – Eur. J., 2014, 20, 16377–16383 CrossRef CAS PubMed.
- Y. Ma, C. Yu and X. Chen, J. Anal. Sci., 2014, 30, 709–712 CAS.
- J. R. Li, G. N. Zhang, L. H. Wang, A. G. Shen and J. M. Hu, Talanta, 2015, 140, 204–211 CrossRef CAS PubMed.
- Y. Sun, J. Wang, W. Li, J. Zhang, Y. Zhang and Y. Fu, Biosens. Bioelectron., 2015, 74, 1038–1046 CrossRef CAS PubMed.
- X. Q. Lin, H. H. Deng, G. W. Wu, H. P. Peng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Analyst, 2015, 140, 5251–5256 RSC.
- D. Y. Zhang, Z. Chen, H. Omar, L. Deng and N. M. Khashab, ACS Appl. Mater. Interfaces, 2015, 7, 4589–4594 CAS.
- C. X. Chen, L. X. Lu, Y. Zheng, D. Zhao, F. Yang and X. R. Yang, Anal. Methods, 2015, 7, 161–167 RSC.
- G. L. Wang, L. Y. Jin, Y. M. Dong, X. M. Wu and Z. J. Li, Biosens. Bioelectron., 2015, 64, 523–529 CrossRef CAS PubMed.
- H. Cheng, X. Wang and H. Wei, Anal. Chem., 2015, 87, 8889–8895 CrossRef CAS PubMed.
- G. J. Guan, L. Yang, Q. S. Mei, K. Zhang, Z. P. Zhang and M. Y. Han, Anal. Chem., 2012, 84, 9492–9497 CrossRef CAS PubMed.
- X. D. Zhang, S. H. He, Z. H. Chen and Y. M. Huang, J. Agric. Food Chem., 2013, 61, 840–847 CrossRef CAS PubMed.
- W. J. Qin, L. Su, C. Yang, Y. H. Ma, H. J. Zhang and X. G. Chen, J. Agric. Food Chem., 2014, 62, 5827–5834 CrossRef CAS PubMed.
- A. Hayat, J. Cunningham, G. Bulbul and S. Andreescu, Anal. Chim. Acta, 2015, 885, 140–147 CrossRef CAS PubMed.
- Y. H. Xiong, S. H. Chen, F. G. Ye, L. J. Su, C. Zhang, S. F. Shen and S. L. Zhao, Chem. Commun., 2015, 51, 4635–4638 RSC.
- Y. J. Guo, L. Deng, J. Li, S. J. Guo, E. K. Wang and S. J. Dong, ACS Nano, 2011, 5, 1282–1290 CrossRef CAS PubMed.
- Q. B. Wang, J. P. Lei, S. Y. Deng, L. Zhang and H. X. Ju, Chem. Commun., 2013, 49, 916–918 RSC.
- R. Thiramanas, K. Jangpatarapongsa, P. Tangboriboonrat and D. Polpanich, Anal. Chem., 2013, 85, 5996–6002 CrossRef CAS PubMed.
- M. I. Kim, K. S. Park and H. G. Park, Chem. Commun., 2014, 50, 9577–9580 RSC.
- G. F. Wang, L. Chen, X. P. He, Y. H. Zhu and X. J. Zhang, Analyst, 2014, 139, 3895–3900 RSC.
- Z. F. Wang, X. Yang, J. Feng, Y. J. Tang, Y. Y. Jiang and N. Y. He, Analyst, 2014, 139, 6088–6091 RSC.
- S. Y. Deng, P. X. Yuan, X. B. Ji, D. Shan and X. J. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 543–552 CAS.
- G. L. Wang, K. L. Liu, J. X. Shu, T. T. Gu, X. M. Wu, Y. M. Dong and Z. J. Li, Biosens. Bioelectron., 2015, 69, 106–112 CrossRef CAS PubMed.
- G. Y. Zhang, S. Y. Deng, W. R. Cai, S. Cosnier, X. J. Zhang and D. Shan, Anal. Chem., 2015, 87, 9093–9100 CrossRef CAS PubMed.
- G. L. Wang, L. Y. Jin, X. M. Wu, Y. M. Dong and Z. J. Li, Anal. Chim. Acta, 2015, 871, 1–8 CrossRef CAS PubMed.
- Y. Song, Y. Wang and L. Qin, J. Am. Chem. Soc., 2013, 135, 16785–16788 CrossRef CAS PubMed.
- M. Liu, H. M. Zhao, S. Chen, H. T. Yu and X. Quan, Chem. Commun., 2012, 48, 7055–7057 RSC.
- B. Zou, Y. Liu, J. Wang and C. Huang, Sci. Sin. Chim., 2014, 44, 1641–1646 CrossRef CAS.
- B. W. Liu and J. W. Liu, Nanoscale, 2015, 7, 13831–13835 RSC.
- P. Weerathunge, R. Ramanathan, R. Shukla, T. K. Sharma and V. Bansal, Anal. Chem., 2014, 86, 11937–11941 CrossRef CAS PubMed.
- Y. S. Kim and J. Jurng, Sens. Actuators, B, 2013, 176, 253–257 CrossRef CAS.
- R. Pautler, E. Y. Kelly, P. J. J. Huang, J. Cao, B. W. Liu and J. W. Liu, ACS Appl. Mater. Interfaces, 2013, 5, 6820–6825 CAS.
- L. C. Bock, L. C. Griffin, J. A. Latham, E. H. Vermaas and J. J. Toole, Nature, 1992, 355, 564–566 CrossRef CAS PubMed.
- T. Hermann and D. J. Patel, Science, 2000, 287, 820–825 CrossRef CAS PubMed.
- J. W. Liu, Z. H. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- H. Wei, B. L. Li, J. Li, E. K. Wang and S. J. Dong, Chem. Commun., 2007, 3735–3737 RSC.
- Y. Wang, H. Wei, B. Li, W. Ren, S. Guo, S. Dong and E. Wang, Chem. Commun., 2007, 5220–5222 RSC.
- B. L. Li, H. Wei and S. J. Dong, Chem. Commun., 2007, 73–75 RSC.
- Y. Du, B. L. Li, H. Wei, Y. L. Wang and E. K. Wang, Anal. Chem., 2008, 80, 5110–5117 CrossRef CAS PubMed.
- B. L. Li, Y. L. Wang, H. Wei and S. J. Dong, Biosens. Bioelectron., 2008, 23, 965–970 CrossRef CAS PubMed.
- L. Y. Fang, Z. Z. Lv, H. Wei and E. Wang, Anal. Chim. Acta, 2008, 628, 80–86 CrossRef CAS.
- B. L. Li, Y. Du, H. Wei and S. J. Dong, Chem. Commun., 2007, 3780–3782 RSC.
- L. B. Zhang, H. Wei, J. Li, T. Li, D. Li, Y. H. Li and E. K. Wang, Biosens. Bioelectron., 2010, 25, 1897–1901 CrossRef CAS PubMed.
- J. G. Bai, H. Wei, B. L. Li, L. H. Song, L. Y. Fang, Z. Z. Lv, W. H. Zhou and E. K. Wang, Chem. – Asian J., 2008, 3, 1935–1941 CrossRef CAS PubMed.
- L. Y. Fang, Z. Z. Lu, H. Wei and E. K. Wang, Biosens. Bioelectron., 2008, 23, 1645–1651 CrossRef CAS PubMed.
- B. Wang, C. L. Guo, G. J. Chen, B. Park and B. Q. Xu, Chem. Commun., 2012, 48, 1644–1646 RSC.
- B. Wang, C. L. Guo, M. M. Zhang, B. Park and B. Q. Xu, J. Phys. Chem. B, 2012, 116, 5316–5322 CrossRef CAS PubMed.
- S. Y. Zhang, Y. B. Ding and H. Wei, Molecules, 2014, 19, 11933–11987 CrossRef PubMed.
- T. K. Sharma, R. Ramanathan, P. Weerathunge, M. Mohammadtaheri, H. K. Daima, R. Shukla and V. Bansal, Chem. Commun., 2014, 50, 15856–15859 RSC.
- P. Jing, W. J. Xu, H. Y. Yi, Y. M. Wu, L. J. Bai and R. Yuan, Analyst, 2014, 139, 1756–1761 RSC.
- W. J. Xu, S. Y. Xue, H. Y. Yi, P. Jing, Y. Q. Chai and R. Yuan, Chem. Commun., 2015, 51, 1472–1474 RSC.
- C. Zheng, A. X. Zheng, B. Liu, X. L. Zhang, Y. He, J. Li, H. H. Yang and G. N. Chen, Chem. Commun., 2014, 50, 13103–13106 RSC.
- L. Chen, L. Sha, Y. W. Qiu, G. F. Wang, H. Jiang and X. J. Zhang, Nanoscale, 2015, 7, 3300–3308 RSC.
- S. M. Taghdisi, N. M. Danesh, P. Lavaee, A. S. Emrani, M. Ramezani and K. Abnous, RSC Adv., 2015, 5, 43508–43514 RSC.
- H. Wei, B. L. Li, J. Li, S. J. Dong and E. K. Wang, Nanotechnology, 2008, 19, 095501 CrossRef PubMed.
- Y. Song, K. Qu, C. Xu, J. Ren and X. Qu, Chem. Commun., 2010, 46, 6572–6574 RSC.
- R. Zhu, Y. Zhou, X. L. Wang, L. P. Liang, Y. J. Long, Q. L. Wang, H. J. Zhang, X. X. Huang and H. Z. Zheng, Talanta, 2013, 117, 127–132 CrossRef CAS PubMed.
- C.-W. Lien, Y.-C. Chen, H.-T. Chang and C.-C. Huang, Nanoscale, 2013, 5, 8227–8234 RSC.
- Y. S. Wu, F. F. Huang and Y. W. Lin, ACS Appl. Mater. Interfaces, 2013, 5, 1503–1509 CAS.
- C.-W. Lien, Y.-T. Tseng, C.-C. Huang and H.-T. Chang, Anal. Chem., 2014, 86, 2065–2072 CrossRef CAS PubMed.
- Z. Mohammadpour, A. Safavi and M. Shamsipur, Chem. Eng. J., 2014, 255, 1–7 CrossRef CAS.
- Y. Fu, H. X. Zhang, S. D. Dai, X. Zhi, J. L. Zhang and W. Li, Analyst, 2015, 140, 6676–6683 RSC.
- H. Abdolmohammad-Zadeh and E. Rahimpour, Sens. Actuators, B, 2015, 209, 496–504 CrossRef CAS.
- W. Li, B. Chen, H. Zhang, Y. Sun, J. Wang, J. Zhang and Y. Fu, Biosens. Bioelectron., 2015, 66, 251–258 CrossRef CAS PubMed.
- H. G. Yang, Y. H. Xiong, P. Zhang, L. J. Su and F. G. Ye, Anal. Methods, 2015, 7, 4596–4601 RSC.
- W. Li, H. X. Zhang, J. L. Zhang and Y. Fu, Anal. Methods, 2015, 7, 4464–4471 RSC.
- G. W. Wu, S. B. He, H. P. Peng, H. H. Deng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Anal. Chem., 2014, 86, 10955–10960 CrossRef CAS PubMed.
- Z. Q. Gao, M. D. Xu, L. Hou, G. N. Chen and D. P. Tang, Anal. Chem., 2013, 85, 6945–6952 CrossRef CAS PubMed.
- Y. C. Lee, M. I. Kim, M. A. Woo, H. G. Park and J. I. Han, Biosens. Bioelectron., 2013, 42, 373–378 CrossRef CAS PubMed.
- Z. F. Wang, S. Zheng, J. Cai, P. Wang, J. Feng, X. Yang, L. M. Zhang, M. Ji, F. G. Wu, N. Y. He and N. Wan, Anal. Chem., 2013, 85, 11602–11609 CrossRef CAS PubMed.
- J. L. Dong, L. N. Song, J. J. Yin, W. W. He, Y. H. Wu, N. Gu and Y. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 1959–1970 CAS.
- Z. H. Yang, Y. Q. Chai, R. Yuan, Y. Zhuo, Y. Li, J. Han and N. Liao, Sens. Actuators, B, 2014, 193, 461–466 CrossRef CAS.
- J. L. Wen, S. G. Zhou and J. H. Chen, Sci. Rep., 2014, 4, 5191 CAS.
- X. N. Hu, A. Saran, S. Hou, T. Wen, Y. L. Ji, W. Q. Liu, H. Zhang and X. C. Wu, Chin. Sci. Bull., 2014, 59, 2588–2596 CrossRef CAS.
- L. Zhan, C. M. Li, W. B. Wu and C. Z. Huang, Chem. Commun., 2014, 50, 11526–11528 RSC.
- G. L. Wang, X. F. Xu, L. Qiu, Y. M. Dong, Z. J. Li and C. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 6434–6442 CAS.
- M. I. Kim, Y. Ye, M. A. Woo, J. Lee and H. G. Park, Adv. Healthcare Mater., 2014, 3, 36–41 CrossRef CAS PubMed.
- Y. Song, X. Xia, X. Wu, P. Wang and L. Qin, Angew. Chem., Int. Ed., 2014, 53, 12451–12455 CAS.
- Y. Cai, C. Q. Cao, X. Q. He, C. Y. Yang, L. X. Tian, R. X. Zhu and Y. X. Pan, Int. J. Nanomed., 2015, 10, 2619–2634 CAS.
- S. K. Maji, A. K. Mandal, K. T. Nguyen, P. Borah and Y. L. Zhao, ACS Appl. Mater. Interfaces, 2015, 7, 9807–9816 CAS.
- Z. Gao, M. Xu, M. Lu, G. Chen and D. Tang, Biosens. Bioelectron., 2015, 70, 194–201 CrossRef CAS PubMed.
- D. Duan, K. Fan, D. Zhang, S. Tan, M. Liang, Y. Liu, J. Zhang, P. Zhang, W. Liu, X. Qiu, G. P. Kobinger, G. F. Gao and X. Yan, Biosens. Bioelectron., 2015, 74, 134–141 CrossRef CAS PubMed.
- Z. Tian, J. Li, Z. Zhang, W. Gao, X. Zhou and Y. Qu, Biomaterials, 2015, 59, 116–124 CrossRef CAS PubMed.
- M. Kim, M. S. Kim, S. H. Kweon, S. Jeong, M. H. Kang, M. I. Kim, J. Lee and J. Doh, Adv. Healthcare Mater., 2015, 4, 1311–1316 CrossRef CAS PubMed.
- J. Shu, Z. Qiu, Q. Wei, J. Zhuang and D. Tang, Sci. Rep., 2015, 5, 15113 CrossRef CAS PubMed.
- Y. Li, J. Xuan, Y. Song, P. Wang and L. Qin, Lab Chip, 2015, 15, 3300–3306 RSC.
- Z. Zhu, Z. Guan, S. Jia, Z. Lei, S. Lin, H. Zhang, Y. Ma, Z.-Q. Tian and C. J. Yang, Angew. Chem., Int. Ed., 2014, 53, 12503–12507 CAS.
- A. Asati, C. Kaittanis, S. Santra and J. M. Perez, Anal. Chem., 2011, 83, 2547–2553 CrossRef CAS PubMed.
- Y. Tao, Y. H. Lin, Z. Z. Huang, J. S. Ren and X. G. Qu, Adv. Mater., 2013, 25, 2594–2599 CrossRef CAS PubMed.
- Z. J. Zhang, Y. J. Guan, M. Li, A. D. Zhao, J. S. Ren and X. G. Qu, Chem. Sci., 2015, 6, 2822–2826 RSC.
- L. N. Zhang, H. H. Deng, F. L. Lin, X. W. Xu, S. H. Weng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Anal. Chem., 2014, 86, 2711–2718 CrossRef CAS PubMed.
- K. Fan, C. Cao, Y. Pan, D. Lu, D. Yang, J. Feng, L. Song, M. Liang and X. Yan, Nat. Nanotechnol., 2012, 7, 459–464 CrossRef CAS PubMed.
- S. S. Ali, J. I. Hardt, K. L. Quick, J. S. Kim-Han, B. F. Erlanger, T. T. Huang, C. J. Epstein and L. L. Dugan, Free Radicals Biol. Med., 2004, 37, 1191–1202 CrossRef CAS PubMed.
- S. S. Ali, J. I. Hardt and L. L. Dugan, Nanomed. Nanotechnol. Biol. Med., 2008, 4, 283–294 CrossRef CAS PubMed.
- K. L. Quick, S. S. Ali, R. Arch, C. Xiong, D. Wozniak and L. L. Dugan, Neurobiol. Aging, 2008, 29, 117–128 CrossRef CAS PubMed.
- D. M. Huang, J. K. Hsiao, Y. C. Chen, L. Y. Chien, M. Yao, Y. K. Chen, B. S. Ko, S. C. Hsu, L. A. Tai, H. Y. Cheng, S. W. Wang, C. S. Yang and Y. C. Chen, Biomaterials, 2009, 30, 3645–3651 CrossRef CAS PubMed.
- C. Mandoli, F. Pagliari, S. Pagliari, G. Forte, P. Di Nardo, S. Licoccia and E. Traversa, Adv. Funct. Mater., 2010, 20, 1617–1624 CrossRef CAS.
- C. K. Kim, T. Kim, I.-Y. Choi, M. Soh, D. Kim, Y.-J. Kim, H. Jang, H.-S. Yang, J. Y. Kim, H.-K. Park, S. P. Park, S. Park, T. Yu, B.-W. Yoon, S.-H. Lee and T. Hyeon, Angew. Chem., Int. Ed., 2012, 51, 11039–11043 CrossRef CAS PubMed.
- T. Kito, R. Shibata, M. Ishii, H. Suzuki, T. Himeno, Y. Kataoka, Y. Yamamura, T. Yamamoto, N. Nishio, S. Ito, Y. Numaguchi, T. Tanigawa, J. K. Yamashita, N. Ouchi, H. Honda, K. Isobe and T. Murohara, Sci. Rep., 2013, 3, 1418 Search PubMed.
- R. Ragg, F. Natalio, M. N. Tahir, H. Janssen, A. Kashyap, D. Strand, S. Strand and W. Tremel, ACS Nano, 2014, 8, 5182–5189 CrossRef CAS PubMed.
- J. M. Dowding, W. Song, K. Bossy, A. Karakoti, A. Kumar, A. Kim, B. Bossy, S. Seal, M. H. Ellisman, G. Perkins, W. T. Self and E. Bossy-Wetzel, Cell Death Differ., 2014, 21, 1622–1632 CrossRef CAS PubMed.
- L. Z. Gao, K. M. Giglio, J. L. Nelson, H. Sondermann and A. J. Travis, Nanoscale, 2014, 6, 2588–2593 RSC.
- D. Son, J. Lee, D. J. Lee, R. Ghaffari, S. Yun, S. J. Kim, J. E. Lee, H. R. Cho, S. Yoon, S. Yang, S. Lee, S. Qiao, D. Ling, S. Shin, J.-K. Song, J. Kim, T. Kim, H. Lee, J. Kim, M. Soh, N. Lee, C. S. Hwang, S. Nam, N. Lu, T. Hyeon, S. H. Choi and D.-H. Kim, ACS Nano, 2015, 9, 5937–5946 CrossRef CAS PubMed.
- M. J. Akhtar, M. Ahamed, H. A. Alhadlaq, M. A. M. Khan and S. A. Alrokayan, J. Colloid Interface Sci., 2015, 453, 21–27 CrossRef CAS PubMed.
- H. Su, D. D. Liu, M. Zhao, W. L. Hu, S. S. Xue, Q. Cao, X. Y. Le, L. N. Ji and Z. W. Mao, ACS Appl. Mater. Interfaces, 2015, 7, 8233–8242 CAS.
- M. J. Akhtar, M. Ahamed, H. A. Alhadlaq, A. Alshamsan, M. A. M. Khan and S. A. Alrokayan, J. Colloid Interface Sci., 2015, 457, 370–377 CrossRef CAS PubMed.
- A. B. Shcherbakov, N. M. Zholobak, A. E. Baranchikov, A. V. Ryabova and V. K. Ivanov, Mater. Sci. Eng., C, 2015, 50, 151–159 CrossRef CAS PubMed.
- D.-M. Huang, J.-K. Hsiao, Y.-C. Chen, L.-Y. Chien, M. Yao, Y.-K. Chen, B.-S. Ko, S.-C. Hsu, L.-A. Tai, H.-Y. Cheng, S.-W. Wang, C.-S. Yang and Y.-C. Chen, Biomaterials, 2009, 30, 3645–3651 CrossRef CAS PubMed.
- H. J. Cheng and H. Wei, Gordon Research Conference: Bioanalytical Sensors, Newport, RI, USA, June 22–27, 2014.
- M. Mittal, M. R. Siddiqui, K. Tran, S. P. Reddy and A. B. Malik, Antioxid. Redox Signaling, 2014, 20, 1126–1167 CrossRef CAS PubMed.
- S. M. Hirst, A. S. Karakoti, R. D. Tyler, N. Sriranganathan, S. Seal and C. M. Reilly, Small, 2009, 5, 2848–2856 CrossRef CAS PubMed.
- F. Pagliari, C. Mandoli, G. Forte, E. Magnani, S. Pagliari, G. Nardone, S. Licoccia, M. Minieri, P. Di Nardo and E. Traversa, ACS Nano, 2012, 6, 3767–3775 CrossRef CAS PubMed.
- F. Natalio, R. Andre, A. F. Hartog, B. Stoll, K. P. Jochum, R. Wever and W. Tremel, Nat. Nanotechnol., 2012, 7, 530–535 CrossRef CAS PubMed.
- T. Xue, B. Peng, M. Xue, X. Zhong, C.-Y. Chiu, S. Yang, Y. Qu, L. Ruan, S. Jiang, S. Dubin, R. B. Kaner, J. I. Zink, M. E. Meyerhoff, X. Duan and Y. Huang, Nat. Commun., 2014, 5, 3200 Search PubMed.
- R. Andre, F. Natalio, M. Humanes, J. Leppin, K. Heinze, R. Wever, H. C. Schroeder, W. E. G. Mueller and W. Tremel, Adv. Funct. Mater., 2011, 21, 501–509 CrossRef CAS.
- J. Zhuang, K. L. Fan, L. Z. Gao, D. Lu, J. Feng, D. L. Yang, N. Gu, Y. Zhang, M. M. Liang and X. Y. Yan, Mol. Pharm., 2012, 9, 1983–1989 CrossRef CAS PubMed.
- F. Xiong, H. Wang, Y. D. Feng, Y. M. Li, X. Q. Hua, X. Y. Pang, S. Zhang, L. Song, Y. Zhang and N. Gu, Sci. Rep., 2015, 5, 8579 CrossRef CAS PubMed.
- Z. Liu, X. Liu, Y. Du, J. Ren and X. Qu, ACS Nano, 2015, 9, 10335–10346 CrossRef CAS PubMed.
- Y. Lin, C. Xu, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2012, 51, 12579–12583 CrossRef CAS PubMed.
- B. Lin, Q. Q. Sun, K. Liu, D. Q. Lu, Y. Fu, Z. A. Xu and W. Zhang, Langmuir, 2014, 30, 2144–2151 CrossRef CAS PubMed.
- J. Zhang, J. Zhuang, L. Gao, Y. Zhang, N. Gu, J. Feng, D. Yang, J. Zhu and X. Yan, Chemosphere, 2008, 73, 1524–1528 CrossRef CAS PubMed.
- Q. Cai, S. K. Lu, F. Liao, Y. Q. Li, S. Z. Ma and M. W. Shao, Nanoscale, 2014, 6, 8117–8123 RSC.
- X. C. Wu, Y. Zhang, T. Han, H. X. Wu, S. W. Guo and J. Y. Zhang, RSC Adv., 2014, 4, 3299–3305 RSC.
- H. Y. Chang, J. S. Cang, P. Roy, H. T. Chang, Y. C. Huang and C. C. Huang, ACS Appl. Mater. Interfaces, 2014, 6, 8305–8312 CAS.
- Z. Liu, X. Liu, X. Ran, E. Ju, J. Ren and X. Qu, Biomaterials, 2015, 69, 56–64 CrossRef CAS PubMed.
- P. F. Zhan, Z. G. Wang, N. Li and B. Q. Ding, ACS Catal., 2015, 5, 1489–1498 CrossRef CAS.
- C. Korsvik, S. Patil, S. Seal and W. T. Self, Chem. Commun., 2007, 1056–1058 RSC.
- Z. Y. Yang, S. L. Luo, H. Li, S. W. Dong, J. He, H. Jiang, R. Li and X. C. Yang, RSC Adv., 2014, 4, 59965–59969 RSC.
- Y. Fu, X. Y. Zhao, J. L. Zhang and W. Li, J. Phys. Chem. C, 2014, 118, 18116–18125 CAS.
- Y. Liu, H. Wu, Y. Chong, W. G. Wamer, Q. Xia, L. Cai, Z. Nie, P. P. Fu and J.-J. Yin, ACS Appl. Mater. Interfaces, 2015, 7, 19709–19717 CAS.
- Y. L. Guo, X. Y. Liu, C. D. Yang, X. D. Wang, D. Wang, A. Iqbal, W. S. Liu and W. W. Qin, ChemCatChem, 2015, 7, 2467–2474 CrossRef CAS.
- W. Shi, X. Zhang, S. He, J. Li and Y. Huang, Sci. Sin. Chim., 2013, 43, 1591–1598 CrossRef CAS.
- J. Xu, J. Wu, C. Zong, H. X. Ju and F. Yan, Anal. Chem., 2013, 85, 3374–3379 CrossRef CAS PubMed.
- Z. Y. Can, A. Uzer, K. Turkekul, E. Ercag and R. Apak, Anal. Chem., 2015, 87, 9589–9594 CrossRef CAS PubMed.
- B. Reuillard, S. Gentil, M. Carriere, A. Le Goff and S. Cosnier, Chem. Sci., 2015, 6, 5139–5143 RSC.
- Y. Tao, E. G. Ju, J. S. Ren and X. G. Qu, Adv. Mater., 2015, 27, 1097–1104 CrossRef CAS PubMed.
- R. Cai, D. Yang, S. Peng, X. Chen, Y. Huang, Y. Liu, W. Hou, S. Yang, Z. Liu and W. Tan, J. Am. Chem. Soc., 2015, 137, 13957–13963 CrossRef CAS PubMed.
- Y. Liu, D. L. Purich, C. Wu, Y. Wu, T. Chen, C. Cui, L. Zhang, S. Cansiz, W. Hou, Y. Wang, S. Yang and W. Tan, J. Am. Chem. Soc., 2015, 137, 14952–14958 CrossRef CAS PubMed.
- Y. Lu, N. Yeung, N. Sieracki and N. M. Marshall, Nature, 2009, 460, 855–862 CrossRef CAS PubMed.
- I. Samish, C. M. MacDermaid, J. M. Perez-Aguilar and J. G. Saven, Annu. Rev. Phys. Chem., 2011, 62, 129–149 CrossRef CAS PubMed.
- J. X. Xie, X. D. Zhang, H. Jiang, S. Wang, H. Liu and Y. M. Huang, RSC Adv., 2014, 4, 26046–26049 RSC.
- X. Ai, Y. Wang, X. D. Hou, L. Yang, C. B. Zheng and L. Wu, Analyst, 2013, 138, 3494–3501 RSC.
- S. Saraf, C. J. Neal, S. Das, S. Barkam, R. McCormack and S. Seal, ACS Appl. Mater. Interfaces, 2014, 6, 5472–5482 CAS.
- H. Sun, A. Zhao, N. Gao, K. Li, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2015, 54, 7176–7180 CrossRef CAS PubMed.
- Y. Li, X. He, J.-J. Yin, Y. Ma, P. Zhang, J. Li, Y. Ding, J. Zhang, Y. Zhao, Z. Chai and Z. Zhang, Angew. Chem., Int. Ed., 2015, 54, 1832–1835 CrossRef CAS PubMed.
- X. F. Lu, X. J. Bian, Z. C. Li, D. M. Chao and C. Wang, Sci. Rep., 2013, 3, 2955 Search PubMed.
- Y. H. Lin, L. Wu, Y. Y. Huang, J. S. Ren and X. G. Qu, Chem. Sci., 2015, 6, 1272–1276 RSC.
- Y. Y. Huang, X. Ran, Y. H. Lin, J. S. Ren and X. G. Qu, Chem. Commun., 2015, 51, 4386–4389 RSC.
- X. N. Hu, A. Saran, S. Hou, T. Wen, Y. L. Ji, W. Q. Liu, H. Zhang, W. W. He, J. J. Yin and X. C. Wu, RSC Adv., 2013, 3, 6095–6105 RSC.
- U. Carmona, L. B. Zhang, L. Li, W. Munchgesang, E. Pippel and M. Knez, Chem. Commun., 2014, 50, 701–703 RSC.
- Y. W. Fan, W. B. Shi, X. D. Zhang and Y. M. Huang, J. Mater. Chem. A, 2014, 2, 2482–2486 CAS.
- L. L. Li, L. H. Ai, C. H. Zhang and J. Jiang, Nanoscale, 2014, 6, 4627–4634 RSC.
- Y. H. Lin, A. D. Zhao, Y. Tao, J. S. Ren and X. G. Qu, J. Am. Chem. Soc., 2013, 135, 4207–4210 CrossRef CAS PubMed.
- J. Li, W. Liu, X. Wu and X. Gao, Biomaterials, 2015, 48, 37–44 CrossRef CAS PubMed.
- W. P. R. Deleu, G. Rivero, R. F. A. Teixeira, F. E. Du Prez and D. E. De Vos, Chem. Mater., 2015, 27, 5495–5502 CrossRef CAS.
- G. Y. Tonga, Y. D. Jeong, B. Duncan, T. Mizuhara, R. Mout, R. Das, S. T. Kim, Y. C. Yeh, B. Yan, S. Hou and V. M. Rotello, Nat. Chem., 2015, 7, 597–603 CrossRef CAS PubMed.
- H. Wei, Z. D. Wang, J. Zhang, S. House, Y. G. Gao, L. M. Yang, H. Robinson, L. H. Tan, H. Xing, C. J. Hou, I. M. Robertson, J. M. Zuo and Y. Lu, Nat. Nanotechnol., 2011, 6, 93–97 CrossRef CAS PubMed.
- H. Wei and Y. Lu, Chem. – Asian J., 2012, 7, 680–683 CrossRef CAS PubMed.
- H. Wei, S. House, J. J. X. Wu, J. Zhang, Z. D. Wang, Y. He, E. J. Gao, Y. G. Gao, H. Robinson, W. Li, J. M. Zuo, I. M. Robertson and Y. Lu, Nano Res., 2013, 6, 627–634 CrossRef CAS.
- Y. B. Ding, L. L. Shi and H. Wei, J. Mater. Chem. B, 2014, 2, 8268–8291 RSC.
- Y. H. Hu, W. J. Guo and H. Wei, Isr. J. Chem., 2015, 55, 682–697 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary tables. See DOI: 10.1039/c5qi00240k |
| This journal is © the Partner Organisations 2016 |