Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biodegradable poly(amidoamine)s with uniform degradation fragments via sequence-controlled macromonomers†
M. F.
Ebbesen
 *,
C.
Gerke
,
P.
Hartwig
and
L.
Hartmann
*
*,
C.
Gerke
,
P.
Hartwig
and
L.
Hartmann
*
Heinrich-Heine-University Düsseldorf, Institute of Organic Chemistry and Macromolecular Chemistry, Universitätsstr. 1, 40225 Düsseldorf, Germany. E-mail: ebbesen@hhu.de; laura.hartmann@hhu.de
First published on 24th October 2016
Abstract
A new and general strategy for the synthesis of high molecular weight, sequence-controlled and selectively degradable poly(amidoamine)s is presented that employs solid-phase synthesis for incorporating degradable linkers at predefined positions within macromonomers. Subsequent molecular weight expansion via Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC)-mediated addition polymerization yields polymers up to an average Mn of 21 kDa. Control of the number and position of degradable linkers within the polymer backbone thus translates into complete and highly selective enzymatic fragmentation down to uniform degradation products. Hence, the control and selectivity of fragmentation now accessible with our strategy can further promote the development of degradable polymers within diagnostic and therapeutic applications.
Introduction
The short half-life of many modern therapeutics such as nucleic acids and peptides in addition to the nonspecific distribution and toxicity of low molecular weight drugs has prompted the development of a broad range of polymer therapeutics that utilize water-soluble polymers for solubilizing, shielding and stabilizing the drug cargo in the bloodstream.1,2 In addition, high molecular weight therapeutics can provide size-dependent accumulation to certain tissues but are limited to a molecular weight below the renal filtration threshold (20–45 kDa) for proper excretion of non-biodegradable constructs.3,4Over the last decade, this has stimulated an interesting and rapid development towards high molecular weight and biodegradable polymer systems for more efficacious therapeutics that will safely eliminate from the body to ensure biocompatibility.5,6 Kopeček's group based one strategy on reversible addition–fragmentation chain transfer (RAFT) polymerization of N-(2-hydroxypropyl)methacrylamide (HPMA) using a dialkyne chain transfer agent that together with a bis-azido GFLG tetrapeptide linker and via Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) enabled the formation of an enzymatically degradable multiblock p(HPMA) construct.7 Pauly et al. used Michael addition polymerization for synthesizing terpolymers of ethylene glycol dimethacrylate, ethylenedioxy diethanethiol and one of two different cysteine-terminated tripeptides with either leucine or phenylalanine residues for installing degradability.8 Being powerful methodologies for generating high molecular weight biodegradable polymer therapeutics, positions and sequences of functionalities (e.g. drugs or targeting moieties) incorporated along the polymer backbone are inherently statistically distributed and not necessarily uniform among polymer chains within a single sample. These differences could have important implications for degradation kinetics and in vivo distribution.
Degradable polymer therapeutics are metabolized inside the body and thus a proper characterization of both the intact and degraded polymer must be performed for a meaningful assessment of its ADME- (absorption, distribution, metabolism, excretion) and toxicity profile.9 In this context, polymers that degrade down to uniform fragments might allow a more feasible and complete evaluation of degradation and drug release and thus a more rational design of novel polymer–drug-conjugates.
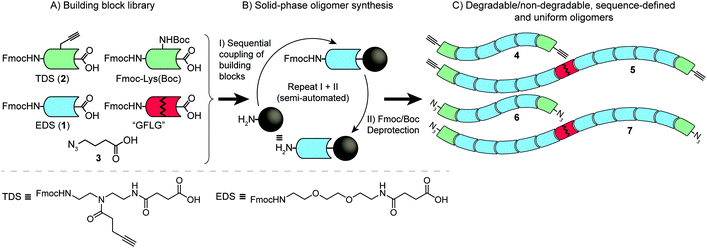
Previously, our group introduced a strategy for the solid phase synthesis of oligo(amidoamine)s with controlled chain length and monomer sequence that utilizes the automated and stepwise addition of specifically designed functional building blocks.10 This introduces synthetic control that allows not only for fine-tuning of oligomer properties such as hydrophilicity and molecular weight but also for sequence-defined insertion of a range of functionalities11,12 such as drugs, ligands for sequence-specific receptor recognition or linkers for degradable oligomer-based structures (Scheme 1).13–15
Polymerization of sequence-defined macromonomers has previously been performed using e.g. CuAAC-,16–18 or thiol–ene/yne polymerization19–21 to obtain sequence-controlled polymers.
Here, we extend our previous sequence-controlled oligo(amidoamine) strategy and demonstrate its application as a tool for the synthesis of a range of poly(amidoamine)s, thereby translating oligomer sequence-control into highly defined polymer structures with selective degradability and improved control of polymer functionality. Furthermore, combining various sequence-defined, degradable and non-degradable α,ω-bis-alkyne/azide oligo(amidoamine)s of different lengths significantly expands the synthetic flexibility with regard to degradable linker density and size of the resulting degradation fragments.
Experimental
Synthesis of building blocks
The preparation of the compound 1-(9H-fluoren-9-yl)-3,14-dioxo-2,7,10-trioxa-4,13-diazaheptadecan-17-oic acid, EDS (1), previously reported by Ponader et al.,11 was carried out using a trityl protection procedure instead of Boc as adapted from a previously published general method for the synthesis of Fmoc-protected building blocks.22 See ESI for the synthesis of EDS (1) and its precursors (Scheme S1 – ESI†) as well as TDS (2) and 4-azidobutanoic acid (3).Solid-phase synthesis (SPS)
Solid-phase reactions (Scheme 1) were performed on a scale of 0.05–0.25 mmol. Coupling of TDS (2) (double coupling, 5 eq.), deprotection of Fmoc-L-Lys(Boc)-OH and final oligomer cleavage were carried out manually in syringe-type polypropylene Chromabond columns of 30 mL with a polypropylene frit and cap. The remaining major part of the solid-phase reactions were carried out using an automated Activotec P11 Peptide Synthesizer. Tentagel S RAM resin (loading 0.23 mmol g−1) was used as solid support and was swollen in DCM for 15 min twice prior to synthesis.Copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC)-mediated oligomer polymerization
The CuAAC mediated polymerization of azide- and alkyne modified oligomers, 4–7, and 1,11-bis-azido-PEG3 (L) (Scheme 2) was performed on a 4 μmol scale. Various reaction parameters were tested (Tables 1 and S1†) with the optimal conditions described here. Oligomers were dissolved in water (50 mmol L−1), mixed according to the desired oligomer composition (at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar eq.) and freeze-dried in glass HPLC vials. Subsequently, solid sodium ascorbate (4 eq.) and a deoxygenated solvent mixture (DMF/water 9/1, purged with Ar for at least 10 min) containing the copper catalyst (CuSO4 or CuBr, 0.4 eq.) was added to an oligomer concentration of 100 mg mL−1 and the vial was capped and mixed thoroughly. Then the vial was shaken (1500 rpm) for 4 h at 25 °C. Hereafter the reaction mixture was diluted with a 1.3 mL aq. solution of sodium diethyldithiocarbamate (10 eq. to the Cu catalyst), stirred 20 min and filtrated through 0.2 um syringe filters to remove the Cu-diethyldithiocarbamate precipitate.24 Complete removal of Cu (to below 0.0005 wt%) was verified by Atomic Absorption Spectroscopy. After extensive dialysis (Vivaspin 2 concentrators, MWCO 2 kDa) against Millipore water and freeze-drying, the resulting polymers were obtained as faint yellow solids in yields between 80% and 90% and analysed by gel permeation chromatography-multi angle light scattering (GPC-MALS) and 1H-NMR (Fig. 2, Tables 1, S1 and Fig. S18, 19 ESI†).
1 molar eq.) and freeze-dried in glass HPLC vials. Subsequently, solid sodium ascorbate (4 eq.) and a deoxygenated solvent mixture (DMF/water 9/1, purged with Ar for at least 10 min) containing the copper catalyst (CuSO4 or CuBr, 0.4 eq.) was added to an oligomer concentration of 100 mg mL−1 and the vial was capped and mixed thoroughly. Then the vial was shaken (1500 rpm) for 4 h at 25 °C. Hereafter the reaction mixture was diluted with a 1.3 mL aq. solution of sodium diethyldithiocarbamate (10 eq. to the Cu catalyst), stirred 20 min and filtrated through 0.2 um syringe filters to remove the Cu-diethyldithiocarbamate precipitate.24 Complete removal of Cu (to below 0.0005 wt%) was verified by Atomic Absorption Spectroscopy. After extensive dialysis (Vivaspin 2 concentrators, MWCO 2 kDa) against Millipore water and freeze-drying, the resulting polymers were obtained as faint yellow solids in yields between 80% and 90% and analysed by gel permeation chromatography-multi angle light scattering (GPC-MALS) and 1H-NMR (Fig. 2, Tables 1, S1 and Fig. S18, 19 ESI†).
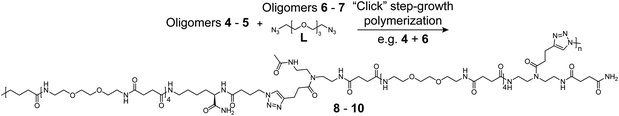 | ||
| Scheme 2 CuAAC mediated polymerization of matching bis-alkyne (4–5) and bis-azide (6–7 + L) oligomer pairs. Polymerization of 4 and 6 are here shown as example. Polymerization of oligomer 4 and the linker 1,11-diazido-PEG3 (L) was performed for initial reaction optimization (see ESI†). | ||
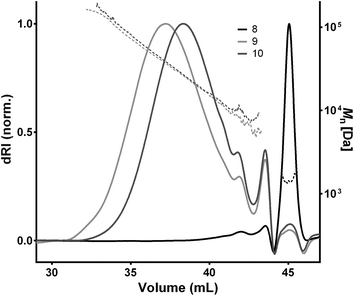 | ||
| Fig. 2 GPC elugrams of polymerizations 8–10 performed in three different reaction solvent compositions (Table 1). The highest molecular weight polymer (9) were observed for solvent compositions DMF/H2O 9/1 with essentially only monomeric species (eluting at 45 mL) shown for DMF only (8). Full lines traces the dRI signal and dashed lines traces the molecular mass. | ||
| # | Bis-C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CH oligomer CH oligomer |
Bis-N3 oligomer | DMSO/DMF/H2O | Catalyst | Rx. [h] |
M
n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a [kDa] a [kDa] |
Đ
M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b b |
X
n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c c |
|---|---|---|---|---|---|---|---|---|
| a Number averaged molecular mass measured using GPC-MALS as described in the ESI. b Molecular mass distribution (Mw/Mn). c Number-average degree of polymerization. | ||||||||
| 8 | 4 (1.51 kDa) | 6 (1.29 kDa) | 0/10/0 | CuSO4 | 4 | 1.7 | 1.6 | 1.2 |
| 9 | — | — | 0/9/1 | — | — | 20.8 | 1.5 | 14.8 |
| 10 | — | — | 0/7/3 | — | — | 16.9 | 1.6 | 12.0 |
| 11 | 5 (3.27 kDa) | 6 (1.29 kDa) | 0/9/1 | CuSO4 | 4 | 17.3 | 2.0 | 7.6 |
| 12 | 5 (3.27 kDa) | 7 (3.04 kDa) | — | — | — | 14.1 | 2.3 | 4.5 |
Enzyme mediated polymer degradation
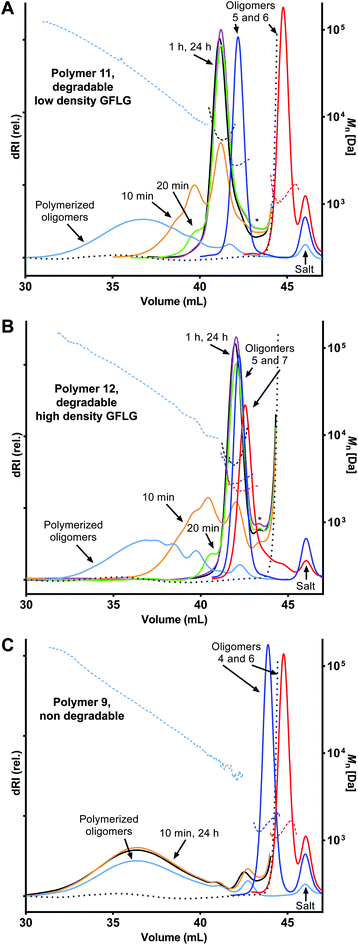
The enzyme-mediated degradation of the polymers (Fig. 3 and 4) was performed following a procedure modified from Luo et al.25 Papain was used as a model enzyme for the degradation. Stock papain enzyme purity was determined by UV at 280 nm from a 1 mg mL−1 enzyme solution in McIlvaine's buffer (50 mM sodium citrate, 100 mM NaH2PO4, 2 mM EDTA, pH 6.0) using the extinction coefficient 1% = 25.26 Equal volumes of glutathione solution (10 mM) and papain stock solution (16 μM) in McIlvaine's buffer were mixed and preincubated for 5 min at 37 °C. Finally, the enzymatic degradation was performed at 37 °C with a polymer concentration of 3 mg mL−1 and papain concentration of 8 μM. Aliquots of 350 μL were taken out at predetermined time points, snap-frozen in filter vials (Mini-UniPrep, 0.2 μm porosity) and stored at −20 °C. GPC analysis was performed after thawing (5 min, 37 °C) and filtration of the individual samples (Fig. 3). MALDI-TOF mass analysis was performed before and 24 h after the enzyme treatment (Fig. S20–22 and Table S2, 3†).Results and discussion
The overall synthetic strategy is based on the solid-phase-mediated introduction of enzymatically degradable sequences within a set of uniform oligomers synthesized from specially designed building blocks (Scheme 1). Subsequent molecular weight expansion through CuAAC-mediated addition polymerization (Scheme 2) leads to the sequence-controlled and selectively degradable precision polymers.Building blocks
Besides being compatible with standard Fmoc solid-phase polyamide synthesis (Scheme 1A), the applied building blocks are individually designed and/or selected towards a particular purpose such as attachment of ligands, drugs or for degradability.11,14 In this study, the CuAAC coupling was selected for the final oligomer addition polymerization (Scheme 2) due to its high coupling efficiency but also due to orthogonality towards the coupling chemistry used with solid phase oligomer synthesis.27,28 Therefore, in addition to a free carboxylic acid and an Fmoc-protected amine group, important features of the applied building blocks were alkyne and azide functionalities. These were introduced via the alkyne-functional building block 1-(fluorenyl)-3,11-dioxo-7-(pent-4-ynoyl)-2-oxa-4,7,10-triazatetradecan-14-oic acid (TDS, 2), previously synthesized in our group,11,22 and via on-resin azido-functionalization of terminal building blocks (Schemes 1 and S2, ESI†). A second building block based on 2,2′(ethylenedioxy)bis(ethylamine) (EDS, 1) was synthesized via a Trt intermediate updating the previous synthetic route via a Boc intermediate and then ubiquitously utilized for conferring hydrophilicity and flexibility to the final polymer backbone (see ESI for synthesis of TDS and EDS (Scheme S1†)).Previously, biodegradability has been installed within sequence-defined oligomers using disulfide containing building blocks as introduced by Hartmann et al.14 The present strategy utilizes a GFLG tetrapeptidyl linker known from work by Kopeček and Duncan29 which is seamlessly introduced into our solid-phase oligo(amidoamine) synthesis via sequential addition of its Fmoc-protected amino acid components to ultimately generate sequence-defined and selectively degradable precision polymers.
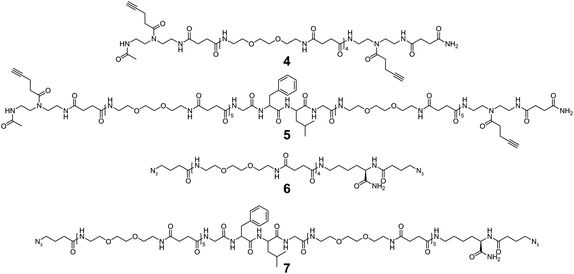
Synthesis of selectively degradable uniform oligomers
Four different oligomer structures varying in length, end group functionality and degradability (Fig. 1) were synthesized using optimized PyBOP-mediated activation chemistry and deprotection protocols that enabled the synthesis of sequence-controlled oligomers in high purity (Scheme 1).22 Coupling of TDS, deprotection of Fmoc-L-Lys(Boc)-OH and final oligomer cleavage were carried out manually in syringe-type polypropylene columns while the remaining major part of the solid-phase reactions could be carried out on an automated peptide synthesizer. After acetylation of the free N-terminal group and cleavage from the resin, oligomer 4 and 5 were precipitated in diethyl ether and lyophilized from aqueous solution. No further purification was performed.The azido-equivalents of oligomer 4 and 5 (6 and 7, Fig. 1) were targeted via functionalizing oligomer terminal residues with 4-azidobutanoic acid (3). This required a selective deprotection of a C-terminal amine group that was initially pursued via orthogonal deprotection of a Fmoc-L-Lys(Aloc)-OH residue but without achieving the deprotected product in the desired purity. A Boc deprotection strategy was therefore employed using Fmoc-L-Lys(Boc)-OH as the initial C-terminal building block. After initial Fmoc deprotection and capping of the lysine α-amine with 3, Boc deprotection was performed following a procedure from Han et al.23 using a cold 4 M solution of HCl in dioxane (see ESI† for further details). This procedure allowed the continued and automated addition of the residual building blocks and after capping the N-terminus with 3, the final oligomer products 6 and 7 could be isolated in a similar way to 4 and 5. Oligomer 7 was cleaved from the resin in 75% crude purity and was therefore purified via preparative RP-HPLC.
Analysis by RP-HPLC/MS (0–50% eluent B in 30 min, UV detection at 214 nm) and ESI-HRMS revealed uniform oligomer products with the expected mass-to-charge (m/z) ratios and purities of 91% (4), 89% (5), 93% (6) and >99% (7), respectively. The slightly higher crude purities of 4 and 6 reflect the lower number of couplings required for these oligomers and the absence of the more hydrophobic Phe and Lys residues, which often lead to more difficult couplings due to aggregation.30 NMR analysis confirmed the desired oligomer structure (see ESI, Fig. S4–17,† for detailed analytical data).
In summary, four uniform, sequence-defined and selectively degradable/non-degradable oligomers (4–7, Fig. 1) with different lengths and end-group functionalities were obtained and next applied for CuAAC-mediated addition polymerization to create selectively degradable polymers.
Polymerization
As the solid-phase synthesis of oligo(amidoamine)s is typically restricted to molecular weights below ∼5 kDa,30 further expansion is necessary for providing certain size-dependent properties such as enhanced biodistribution to the final therapeutic construct.4 CuAAC-mediated addition polymerization is well-known with macromonomers composed especially from atom transfer radical polymerization (ATRP) synthesized polymers31 and polymer–peptide mixtures,25 whereas only a few studies describe all-peptide macromonomers.17,18In this work, four sequence-defined, bis-alkyne and bis-azido terminated oligomers (4–7) were synthesized and applied as homobifunctional macromonomers for CuAAC-mediated addition polymerization (Scheme 2). For initial testing purposes, a macromonomer system comprised of 1,11-bis-azido-PEG3 (L), and an oligomer (4) was used (Table S1†) that was later replaced by oligomer macromonomers 4 and 6 (Table 1, Fig. 2) to obtain the desired products. In general, special care was taken to add a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric mixture of the selected oligomers to optimize polymerization conversion and, moreover, the oligomer/linker concentration was kept as high as possible to increase the reactive group concentration for decreasing the probability for intramolecular ring formation.
1 stoichiometric mixture of the selected oligomers to optimize polymerization conversion and, moreover, the oligomer/linker concentration was kept as high as possible to increase the reactive group concentration for decreasing the probability for intramolecular ring formation.
The influence of the reaction solvent, reaction time and catalyst type was then tested for optimizing the polymerization reaction with regard to the degree of polymerization (Xn) that was measured via GPC analysis. No significant differences were found for reaction times in the range 2 to 24 h (Table S1†). CuBr is a commonly used CuAAC catalyst in DMF due to suitable solubility.25 Here, a direct comparison to CuSO4 catalysis in various DMF/water ratios (Table S1†), however, showed a higher Xn for CuSO4 (Xn = 9.3) compared to CuBr (Xn = 6.7). CuSO4 was therefore used for subsequent polymerizations. A comparison of polymerizations performed under various solvent conditions (Table 1, 8–10 and Table S1,†16–20) illustrated the benefit of fine-tuning reaction solvent parameters with neither water nor DMF alone providing optimal results. Water has been well-recognized for being a good solvent for supporting Cu(I)-acetylides and for the Cu-ligand exchange32 and also seemed a crucial component of the solvent mixture in this study with a Xn of 10.6 for a DMF/water 9/1 mixture and CuSO4 as Cu(I) source. More than 10% water, however, led to decreased Xn, possibly due to alkyne water addition hereby forming an unreactive ketone. Overall, the best reaction parameters were determined to be a CuSO4/NaAsc. mixture (0.4/4 eq.) in a deoxygenated DMF/water 9/1 with 4 h reaction time at 25 °C. The optimized conditions showed a low level of remaining oligomer via GPC analysis (Fig. 2) and 1H-NMR analysis (Fig. S19 – ESI†) showed full consumption of the alkyne proton at 2.36 ppm and confirmed the overall polymer structure.
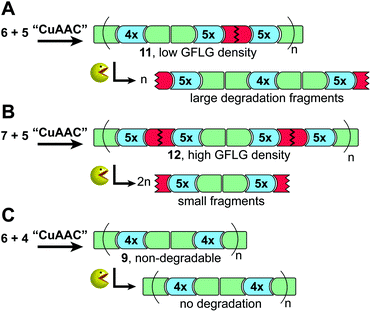
As shown in the literature, the degree of polymerization (Xn) obtained from macromonomer addition polymerization depends on the monomer size due to a lower effective concentration of the reactive end-groups for larger monomers with a typical Xn around 15 for monomers >1 kDa and higher Xn for lower molecular weight monomers.25,33 Despite the larger average monomer size, polymerization of oligomers 4 and 6 showed a higher maximal Xn of 14.8 (9, Table 1, Fig. 2) compared to the test system (L + 4, Table S1 – ESI†) that could probably be ascribed to high oligomer end-group purities. The Xn and ĐM values obtained correlate well with previous examples of CuAAC-mediated macromonomer addition polymerization (before fractionation) with ĐM between ∼1.5 and 2 and maximal Xn values in the range of ∼13–16.17,18,25,31 Refinement of the synthesis by preparative fractionation should lead to polymers of higher Xn with lower ĐM values.25 Subsequently, different sets of degradable and non-degradable oligomers were selected and copolymerized to obtain polymers with pre-determined densities of cleavage sites. Comprised of larger oligomers, these polymers (11–12) displayed a somewhat lower Xn (Table 1) due to the lower reactive group concentration.
In summary, the CuAAC-mediated polymerization of selected combinations of sequence-defined and degradable/non-degradable azide/alkyne terminated macromonomers enabled the synthesis of precision polymers with selective degradation profiles. A set of three selectively degradable/non-degradable polymers were then subjected to enzymatic degradation for demonstrating their controlled degradation down to uniform fragments.
Enzyme-mediated polymer degradation
The enzyme-cleavable tetrapeptide sequence, GFLG, is not a natural substrate for serum enzymes, which ensures stability in serum but prompt degradation when internalized into endocytic vessels containing the cysteine protease cathepsin-B that recognizes and cleaves it.29,34 Enzymatically degradable polymers are, therefore, unique in terms of the high selectivity between enzyme and substrate that allows for spatially (enzyme location) controlled degradation,35,36 but, as shown in this work, potentially also structural (polymer) degradation control by exact positioning of the degradable peptide sequence within the polymer backbone.Polymer degradation was investigated using a low-cost alternative to cathepsin B. Papain is an enzyme in the same group of cysteine proteases and has a substrate specificity similar to cathepsin B.34 The selectively degradable precision polymers comprising GFLG linkers (11, 12) (Fig. 4A and B) and non-degradable polymers without GFLG as control (9) (Fig. 4C) were incubated with 8.0 μM papain under slightly acidic and reducing conditions (pH 6 and 37 °C) up to 24 h leading to rapid onset of polymer degradation with complete degradation within 1 h as determined by GPC analysis (Fig. 3). This was faster than a previous similar study based on degradable multiblock polyHPMA25 which could potentially be attributed to a higher GFLG density and less steric hindrance for the lower Mn structures reported in our study. The presence of low Xn populations (monomer, dimer, etc.) for 11 and 12 entails a higher amount of polymer terminals. Cleavage of terminal GFLG linkers from these polymers thus lead to an observable amount of smaller degradation fragments derived from the polymer terminals (Fig. 3A and B). The presence of these smaller fragments should be significantly reduced in the next generation of materials with optimized and longer polymer chains lowering the relative amount of polymer terminals.
Uniform degradation fragments were obtained with sizes depending on the individual oligomer combinations in terms of size and degradability (Fig. 3 and 4). MALDI-MS analysis was performed (Fig. S20, 21 and Table S2, 3 – ESI†) and confirmed complete polymer degradation down to a very narrow range of fragments corresponding to the average size of the degradable oligomers and any non-degradable oligomers within the polymer (4.2–4.9 kDa and 2.8–3.4 kDa for polymer 11 and 12, respectively, Table S2†). Likewise, MALDI-MS analysis in combination with our sequence-defined polymers also provided a qualitative view on the enzyme peptidase activity towards GFLG. Papain is considered an endopeptidase and cleaves amide bonds within the GFLG sequence except in the sterically crowded site between Phe and Leu residues.37 Interestingly, from our study we found that papain also degraded amide bonds between synthetic EDS building blocks and Gly amino acid residues (see ESI, Fig. S21–22 and Table S3†). This might contribute to further development of enzyme-degradable polymer linkers and opens up further potential e.g. for an all-artificial enzymatically degradable linker synthesized from our present building block library.
No significant changes were observed for the GPC traces between 1 h and 24 h (Fig. 3A and B) that indicates no further degradation of the fragments within this timeframe. The stability of the non-degradable polymers was confirmed in a similar manner (Fig. 3C) that supported the observed degradation to be mediated by the GFLG linker. A low degree of non-enzymatic hydrolysis has previously been reported for the GFLG linker38 and thus further studies on the long-term stability of the degradable and non-degradable polymers are currently ongoing.
The resulting sequence-controlled and selectively degradable precision poly(amidoamines) represent a promising class of materials not previously used within polymer therapeutics. Similar to previous reported biodegradable polymers,25 the design allows efficient enzymatic degradation and with the additional possibilities for controlling the size of polymer degradation fragments, this strategy could ultimately be of great importance e.g. for the development new degradable polymers with a more predictive release and body clearance profile.
Conclusions
The purpose of this study was to introduce a new approach for the synthesis of sequence-defined and biodegradable polymers for further enhancing the spatial and structural controlled fragmentation of polymers. This was demonstrated via the solid-phase-supported synthesis of a set of sequence-defined azide- and alkyne terminated oligomers containing enzymatically degradable linkers at predefined positions. Subsequent CuAAC-mediated addition polymerization expanded the average molecular weight up to 15 times (Mn 21 kDa). Finally, complete and efficient enzymatic degradation down to a narrow range of uniform degradation products validated the approach for the synthesis of selectively degradable precision polymers. The envisioned benefits include highly defined degradation products and a flexible yet more consistent synthesis without inherent heterogeneities in terms of positions and sequences of polymer functionalities that has potential to further promote the use of polymeric therapeutics.Acknowledgements
M. F. E. thanks the Danish Council for Independent Research for support through the grant DFF – 4005-00023. L. H. thanks the Boehringer-Ingelheim-Stiftung for support through the “Perspektivenprogramm Plus-3”. Moreover, the authors are grateful to Assoc. Prof. Nicole Snyder (Davidson College, North Carolina) for fruitful discussions.References
- R. Duncan and M. J. Vicent, Adv. Drug Delivery Rev., 2013, 65, 60–70 CrossRef CAS PubMed.
- M. Ebbesen, K. Bienk, B. Deleuran and K. Howard, Molecular and Cellular Therapies, 2014, 2, 29 CrossRef PubMed.
- D. G. Rudmann, J. T. Alston, J. C. Hanson and S. Heidel, Toxicol. Pathol., 2013, 41, 970–983 CrossRef PubMed.
- J. G. Shiah, M. Dvořák, P. Kopečková, Y. Sun, C. M. Peterson and J. Kopeček, Eur. J. Cancer, 2001, 37, 131–139 CrossRef CAS PubMed.
- A. V. Yurkovetskiy and R. J. Fram, Adv. Drug Delivery Rev., 2009, 61, 1193–1202 CrossRef CAS PubMed.
- J. W. Singer, J. Controlled Release, 2005, 109, 120–126 CrossRef CAS PubMed.
- R. Zhang, K. Luo, J. Yang, M. Sima, Y. Sun, M. M. Janát-Amsbury and J. Kopeček, J. Controlled Release, 2013, 166, 66–74 CrossRef CAS PubMed.
- A. C. Pauly and F. di Lena, Polymer, 2015, 72, 378–381 CrossRef CAS.
- E. Markovsky, H. Baabur-Cohen, A. Eldar-Boock, L. Omer, G. Tiram, S. Ferber, P. Ofek, D. Polyak, A. Scomparin and R. Satchi-Fainaro, J. Controlled Release, 2012, 161, 446–460 CrossRef CAS PubMed.
- L. Hartmann and H. G. Börner, Adv. Mater., 2009, 21, 3425–3431 CrossRef CAS PubMed.
- D. Ponader, F. Wojcik, F. Beceren-Braun, J. Dernedde and L. Hartmann, Biomacromolecules, 2012, 13, 1845–1852 CrossRef CAS PubMed.
- F. Wojcik, A. G. O'Brien, S. Götze, P. H. Seeberger and L. Hartmann, Chem. – Eur. J., 2013, 19, 3090–3098 CrossRef CAS PubMed.
- D. Ponader, P. Maffre, J. Aretz, D. Pussak, N. M. Ninnemann, S. Schmidt, P. H. Seeberger, C. Rademacher, G. U. Nienhaus and L. Hartmann, J. Am. Chem. Soc., 2014, 136, 2008–2016 CrossRef CAS PubMed.
- L. Hartmann, S. Häfele, R. Peschka-Süss, M. Antonietti and H. G. Börner, Macromolecules, 2007, 40, 7771–7776 CrossRef CAS.
- J.-F. Lutz, M. Ouchi, D. R. Liu and M. Sawamoto, Science, 2013, 341, 1238149 CrossRef PubMed.
- M.-A. Berthet, Z. Zarafshani, S. Pfeifer and J.-F. Lutz, Macromolecules, 2010, 43, 44–50 CrossRef CAS.
- T.-B. Yu, J. Z. Bai and Z. Guan, Angew. Chem., Int. Ed., 2009, 48, 1097–1101 CrossRef CAS PubMed.
- S. van der Wal, C. J. Capicciotti, S. Rontogianni, R. N. Ben and R. M. J. Liskamp, MedChemComm, 2014, 5, 1159–1165 RSC.
- F. Driessen, F. E. Du Prez and P. Espeel, ACS Macro Lett., 2015, 4, 616–619 CrossRef CAS.
- W. Xi, S. Pattanayak, C. Wang, B. Fairbanks, T. Gong, J. Wagner, C. J. Kloxin and C. N. Bowman, Angew. Chem., Int. Ed., 2015, 54, 14462–14467 CrossRef CAS PubMed.
- J. Han, Y. Zheng, B. Zhao, S. Li, Y. Zhang and C. Gao, Sci. Rep., 2014, 4, 4387 Search PubMed.
- F. Wojcik, S. Mosca and L. Hartmann, J. Org. Chem., 2012, 77, 4226–4234 CrossRef CAS PubMed.
- G. Han, M. Tamaki and V. J. Hruby, J. Pept. Res., 2001, 58, 338–341 CrossRef CAS PubMed.
- B. J. Strawter, Canada Pat., CA1186076, 1985 Search PubMed.
- K. Luo, J. Yang, P. Kopečková and J. Kopeček, Macromolecules, 2011, 44, 2481–2488 CrossRef CAS PubMed.
- A. N. Glazer and E. L. Smith, J. Biol. Chem., 1961, 236, 2948–2951 CAS.
- W. H. Binder and R. Sachsenhofer, Macromol. Rapid Commun., 2008, 29, 952–981 CrossRef CAS.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS PubMed.
- P. Rejmanová, J. Kopeček, R. Duncan and J. B. Lloyd, Biomaterials, 1985, 6, 45–48 CrossRef.
- W. C. Chan and P. D. White, Fmoc Solid-Phase Peptide Synthesis: A Practical Approach, Oxford University Press, New York, 2000 Search PubMed.
- N. V. Tsarevsky, B. S. Sumerlin and K. Matyjaszewski, Macromolecules, 2005, 38, 3558–3561 CrossRef CAS.
- R. Berg and B. F. Straub, Beilstein J. Org. Chem., 2013, 9, 2715–2750 CrossRef PubMed.
- S. Binauld, F. Boisson, T. Hamaide, J. P. Pascault, E. Drockenmuller and E. Fleury, J. Polym. Sci., Part A: Polym. Chem., 2008, 46, 5506–5517 CrossRef CAS.
- V. Turk, B. Turk and D. Turk, EMBO J., 2001, 20, 4629–4633 CrossRef CAS PubMed.
- Y. Hashimoto, H. Kakegawa, Y. Narita, Y. Hachiya, T. Hayakawa, J. Kos, V. Turk and N. Katunuma, Biochem. Biophys. Res. Commun., 2001, 283, 334–339 CrossRef CAS PubMed.
- H. Appelqvist, P. Wäster, K. Kågedal and K. Öllinger, J. Mol. Cell Biol., 2013, 5, 214–226 CrossRef CAS PubMed.
- M. Azarkan, A. El Moussaoui, D. van Wuytswinkel, G. Dehon and Y. Looze, J. Chromatogr. B: Biomed. Sci. Appl., 2003, 790, 229–238 CrossRef CAS.
- N. Larson, J. Yang, A. Ray, D. L. Cheney, H. Ghandehari and J. Kopeček, Int. J. Pharm., 2013, 454, 435–443 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, synthesis protocols and further analytical data of building blocks, oligomers, polymerizations and polymer degradations. See DOI: 10.1039/c6py01700b |
| This journal is © The Royal Society of Chemistry 2016 |