Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis and activity of a novel inhibitor of nonsense-mediated mRNA decay†
Victoria J. B.
Gotham
ab,
Melanie C.
Hobbs
a,
Ryan
Burgin
a,
David
Turton
b,
Carl
Smythe
*b and
Iain
Coldham
*a
aDepartment of Chemistry, University of Sheffield, Sheffield, S3 7HF, UK. E-mail: i.coldham@sheffield.ac.uk
bDepartment of Biomedical Science, University of Sheffield, Sheffield, S10 2TN, UK
First published on 24th December 2015
Abstract
During efforts to prepare the known compound NMDI1, a new tetracyclic compound, called VG1, was prepared in six steps. This compound was found to have good activity as an inhibitor of nonsense-mediated mRNA decay.
A significant proportion of genetic diseases, as well as many cancers, are associated with a particular type of mutation which gives rise to a premature termination codon (PTC) before the genuine stop codon.1 PTC-associated diseases include cases of Duchenne muscular dystrophy, cystic fibrosis, β-thalassaemia, retinitis pigmentosa, von Willebrand disease, Robinow syndrome, haemophilia, spinal muscular atrophy and Rett syndrome. mRNAs containing PTCs are detected and degraded by a process known as nonsense-mediated mRNA decay (NMD),2–4 which reduces the expression of the corresponding gene to a low level. It is the subsequent lack of protein that causes the symptoms of PTC-associated diseases. The PTCs that trigger NMD can occur due to single base pair mutations, reading frame shifts due to deletions or insertions, or mutations in splice sites or splicing regulatory sequences that give rise to aberrant splicing.2–4 At the mRNA level, PTCs can arise from errors in transcription, transcription initiation upstream of the correct site coupled with the location of a termination codon in the region before the proper start site, alternative splicing, or programmed frameshifts during translation.2–4
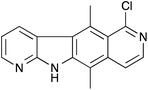
One potential approach to the treatment of diseases caused by PTC mutations is the inhibition of NMD. Therefore there is a need to discover and develop drugs that inhibit NMD selectively. In addition, NMD inhibitors can be useful tools for studying the NMD mechanism and related pathways that utilise some of the same factors. In 2007, Lejeune and co-workers5 found that a tetracyclic compound called NMDI1 (Fig. 1) was able to inhibit both nucleus-associated and cytoplasmic NMD. The compound appears to be a specific inhibitor of NMD: it does not affect the splicing of pre-mRNA reporter transcripts, the amount of pre-mRNA, translation efficiency or the miRNA decay pathway.5 It is non-cytotoxic, even at high concentrations, and does not induce the formation of stress granules.5
The synthesis of NMDI1 was reported by Bisagni and co-workers.6,7 The overall yield was poor (0.5% over 11 steps) and we had difficulties on attempted repetition of the published procedure (see ESI†). Keeling and co-workers have recently reported a synthesis of NMDI1 by a modified route (total 13 steps) in which the final 7 steps were identical to those used by Bisagni and co-workers; however, they report no experimental details.8
There is clearly a need for a NMD inhibitor whose synthesis is more efficient and reproducible. We have synthesised in only 6 steps a close structural analogue of NMDI1, called VG1, by using a different route from that described above.6–8 We found that VG1 inhibits NMD equally well as NMDI1 and is therefore a suitable candidate for use as a tool to investigate NMD and related mechanisms, and possibly for development as a drug for diseases caused by PTC mutations.
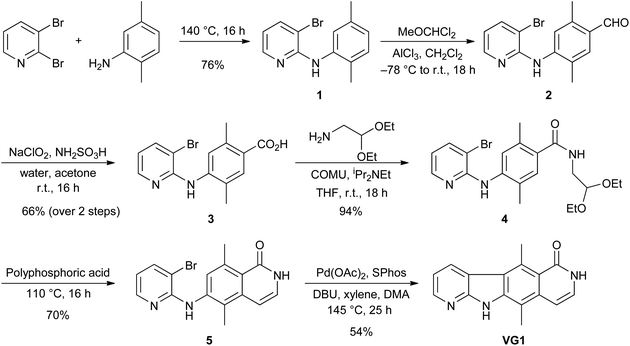
VG1 was synthesised from commercially available 2,3-dibromopyridine and 2,5-dimethylaniline (Scheme 1). These were heated at 140 °C for 16 h to give bromide 1 in 76% yield. An aldehyde was then installed para to the amino group. The use of POCl3 and DMF (Vilsmeier reaction)9,10 failed to give the desired aldehyde, but the use of AlCl3 and MeOCHCl2 (Rieche formylation)11,12 was successful and gave aldehyde 2. This compound was not purified but was treated directly with NaClO2 and NH2SO3H as a chlorine scavenger,13–15 to give the carboxylic acid 3 in 66% yield from bromide 1. The acid was converted to amide 4 in 94% yield by using the commercially available aminoacetaldehyde diethylacetal and the coupling reagent COMU.16 Deprotection of the acetal with polyphosphoric acid was followed by in situ cyclisation and dehydration17 to give compound 5 in 70% yield. An intramolecular palladium-catalysed cross-coupling reaction18 was used to convert bromide 5 into tetracycle VG1 in 54% yield.
Attempted conversion of the lactam 5 to the chloropyridine was unsuccessful by using POCl3, PhP(O)Cl2, PCl3, or PCl5 in POCl3. Chlorination of VG1 was also attempted but the only isolable products obtained, after treatment of either compound 5 or VG1 with PCl5 in POCl3, were N-chloro derivatives (see ESI†). Chlorination on the nitrogen atom of an amide has literature precedent.19–21 Unfortunately, this meant that we were not able to prepare NMDI1 using this synthesis. However, we had prepared a close analogue in VG1 and decided to test the activity of this compound.
To test the ability of VG1 to inhibit NMD we utilised a well-established assay for the measurement of NMD within cells.22 HeLa cells were transfected with either one of two reporter plasmids, pmCMV-Gl (NORM) or pmCMV-Gl (TER),22–24 referred to as NORM and TER. The NORM plasmid contains a hybrid human/mouse β-globin gene. This was constructed by Maquat and Kinniburgh23 by substituting sequences in the human β-globin gene extending from the BamH1 site in exon II with analogous sequences from the mouse β-globin gene. The resulting hybrid gene contains exon I, intron I and most of exon II of the human β-globin gene, and 18 base pairs of exon II, intron II and exon III of the mouse β-globin gene. The TER plasmid contains the same chimaeric gene but with a PTC mutation (CAG to TAG) in codon 39, located in exon II. The cloning of NORM and TER sequences into the mRNA producing plasmid pCMV, and the characterisation of resultant mRNA has been previously described.22 TER and NORM transcripts are subject to splicing, and TER, but not NORM, spliced mRNA is subject to NMD.22Fig. 2 shows a plasmid map and the structure of the hybrid genes. In order to control for transfection efficiency, cells were also transfected with the reference plasmid phCMV-MUP (referred to as MUP), which encodes mouse major urinary protein.25 After 24 h, the transfection medium was removed from cells and replaced with culture medium containing either DMSO or NMDI1 or VG1 at a final concentration of 5 μM made up from a 5 mM stock solution in DMSO. Each compound was added both to cells transfected with NORM and cells transfected with TER. After 20 h, total RNA was extracted and reverse transcribed. The amount of NORM, TER and MUP cDNA was determined by qPCR, where the amount of cDNA serves as a proxy measurement for mRNA as has been described previously.27,28 To ensure that the qPCR reaction products reflected the appropriately spliced mature mRNA, the cDNA generated from RNA extracted from both NORM and TER transfected cells was amplified by standard PCR using identical primers and DNA products which correspond to their cognate mRNAs were analysed for size by agarose gel electrophoresis. These showed that PCR products correspond to correctly spliced mRNA (Fig. S2 in ESI†).
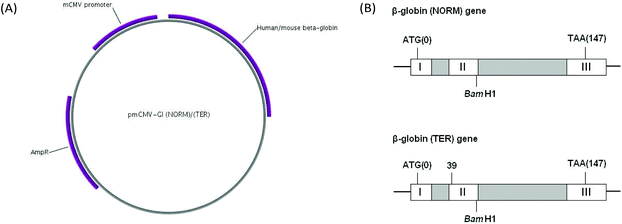 | ||
| Fig. 2 pmCMV-Gl (NORM) and pmCMV-Gl (TER) plasmids. (A) Plasmid map. The plasmids contain a mouse cytomegalovirus (mCMV) promoter, a hybrid human/mouse β-globin gene and a gene for ampicillin resistance. The map was created using the PlasMapper program.26 (B) Structure of the hybrid human/mouse β-globin gene. Grey boxes are introns. Start (ATG) and stop (TAA) codons are shown. The BamH1 site marks the boundary between sequences from the human and mouse genes. The TER plasmid is identical to NORM, except that a PTC mutation (CAG to TAG), has been introduced in codon 39 located in exon II. | ||
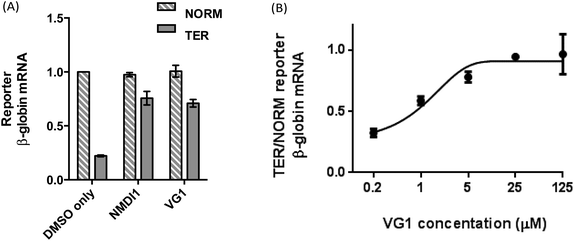
The inferred amounts of NORM and TER mRNA, each expressed as a ratio of the respective inferred amount of MUP mRNA in each sample, were compared in order to determine the NMD efficiency. As expected, for cells treated with DMSO only, the amount of TER mRNA was significantly lower than the amount of NORM mRNA (0.22 times the amount of NORM mRNA), indicating that TER mRNA was efficiently degraded by NMD (Fig. 3A, first and second bars from left). When cells were treated with VG1, the amount of TER mRNA increased to 0.70 times the amount of NORM mRNA, showing that NMD was inhibited (Fig. 3A, fifth and sixth bars from left). VG1 inhibited NMD to the same extent as NMDI1 (Fig. 3A, third and fourth bars from left).
The effect of the dose of VG1 on NMD inhibition was then investigated. Cells were transfected with the NMD reporter plasmids, as described above. After 24 h, the transfection medium was removed from cells and replaced with culture medium containing either DMSO or VG1 at a final concentration of 0.2, 1, 5, 25 or 125 μM made up from a stock solution in DMSO. After 20 h, total RNA was extracted and reverse transcribed. The amount of NORM, TER and MUP cDNA was determined by qPCR and the inferred amounts of NORM and TER mRNA, each expressed as a ratio of the respective inferred amount of MUP mRNA in each sample, were compared in order to determine the NMD efficiency. To ensure that the PCR primers used generated a reaction product corresponding to spliced RNA and not unspliced pre-mRNA, the amplified PCR products from RNA extracted from cells transfected with either NORM, or TER, were examined by DNA agarose gel electrophoresis (Fig. S2†). In each case, these showed the presence of a DNA fragment of 150 bp corresponding to correctly spliced mRNA. In qPCR analyses, the amount of TER mRNA relative to NORM mRNA, and therefore NMD inhibition, increased as VG1 concentration increased, reaching a maximum at 25 μM, at which concentration the amount of TER mRNA was 0.94 times the amount of NORM mRNA (Fig. 3B). There was no further NMD inhibition at 125 μM. Thus high levels of inhibition of NMD were achieved by treating cells with 5–25 μM VG1, assuming that if NMD were fully inhibited, the amount of TER mRNA would be equal to the amount of NORM mRNA.
NMDI1 has been reported to have little effect on cell viability.5 When cells were seeded as normal (104 cells per cm2) and treated with 5 μM VG1 for 24 h, the rate of proliferation appeared similar to cells treated with DMSO alone (Fig. S1A in ESI†) and there was little evidence of increased pyknosis or karyorrhexis (indicators of necrosis or late-stage apoptosis) suggesting that, as with NMDI1, effective doses on this timescale are not toxic for cell survival. The effect of VG1 dose on cells after 20 h exposure was also investigated (Fig. S1B†). As before, at concentrations up to 5 μM, there was no obvious effect on cell number or morphology. When VG1 was added to cell media at 25 μM and cells were incubated with compound for 20 h, there was some indication of altered cell morphology in a minority of cells, suggestive of some toxicity at higher concentrations and possible induction of apoptosis (Fig. S1B†). Further analyses of the effects of longer exposure at 25 μM or the use of higher concentrations were not possible due to insolubility of VG1 in cell media at high concentrations. Exploration of the induction of apoptosis at an intermediate concentration of VG1 (10 μM, Fig. S1C†) was evaluated by immunoblotting total cell lysates for the appearance of the cell marker of apoptotic induction, activated (cleaved) Caspase 3.29 These data (Fig. S1C†) indicated that, at this concentration and time scale, there was no evidence of elevated levels of apoptotic cell death under these conditions. Taken together our data suggest that, as with NMDI1, effective doses of VG1 over a 24 h time scale are not toxic for cell survival.
Inhibiting NMD is a promising approach towards treating diseases caused by PTC mutations as, in many cases, the truncated protein that would result if the PTC-containing mRNA were to be translated could be at least partially functional and could relieve some of the symptoms of the disease. Obviously, the function of the truncated protein would depend on the location of the PTC within the mRNA. As an example of the potential of this approach, when cultured fibroblasts from a patient with Ullrich's disease (which is caused by a PTC in the gene for the extracellular matrix protein collagen VI α2) were treated with inhibitors of the NMD factor SMG1, there was an increase in the expression of truncated collagen VI α2 which partially restored the function of the extracellular matrix.30 However, SMG1 inhibitors are unlikely to be able to be used clinically as they are toxic and are not specific for SMG1. Other small molecules that have been used to inhibit NMD have not yet reached clinical trials of NMD inhibition.31–36
One approach for treating PTC-associated diseases involves the use of a class of compounds called ribosome readthrough promoters.37,38 These compounds induce the ribosome to “read through” a PTC, allowing a full-length protein to be produced from the transcript. However, the efficacy of readthrough promoters is limited by a paucity of PTC-containing mRNAs available for readthrough, as most are degraded rapidly by NMD. Inhibiting NMD, in combination with readthrough therapy, should mean that fewer PTC-containing mRNAs are degraded and therefore more are available for readthrough. This approach has been used by Lejeune and co-workers,33 who found that the effect of 5 μM NMD inhibitor amlexanox combined with 25 μM readthrough promoter PTC124 was greater than that of either drug on its own in increasing iodide efflux in 6CFSMEo cystic fibrosis cells. Keeling and co-workers have shown that NMDI1 was able to enhance the therapeutic effect of at least a subset of readthrough promoters called aminoglycosides in IduaW392X mice (which have a premature stop codon in the gene encoding α-L-iduronidase).8
In this paper, we have developed a synthesis of an analogue of NMDI1 and have found that our compound, VG1, inhibited NMD to the same extent as NMDI1. Our synthesis requires only 6 steps and proceeds in 18% overall yield. Lejeune and co-workers presented evidence that NMDI1 inhibits the interaction between two key NMD factors, UPF1 and SMG5.5 UPF1 forms part of a complex that recognises the PTC-containing mRNA after the first round of translation terminates at the PTC. The protein undergoes cycles of phosphorylation and de-phosphorylation, which allow it to take part in multiple rounds of NMD. SMG5 is one of the proteins that mediates the de-phosphorylation of UPF1, and inhibition of the UPF1-SMG5 interaction may cause UPF1 to become trapped in a hyper-phosphorylated state. Given the similarity between the structures of VG1 and NMDI1, it is likely that the compounds inhibit NMD by the same mechanism, and therefore, that VG1 also inhibits the interaction between UPF1 and SMG5, although this still needs to be confirmed. It is conceivable that NMDI1 is converted into VG1 in the cell and that the active form of the compounds is the same. The compound VG1 passed the filter on an online PAINS (pan assay interference compounds) false positive remover tool,39 which helps to validate VG1 as a genuine NMD inhibitor. Work is required to establish the mechanism of action of VG1. Whatever the outcome, VG1 will be a useful tool for developing an understanding of both the mechanism of NMD and of other related mRNA decay pathways.
Acknowledgements
We thank the EPSRC and the University of Sheffield for funding.Notes and references
- P. A. Frischmeyer and H. C. Dietz, Hum. Mol. Genet., 1999, 8, 1893 CrossRef CAS PubMed.
- O. Isken and L. E. Maquat, Genes Dev., 2007, 21, 1833 CrossRef CAS PubMed.
- P. Nicholson, H. Yepiskoposyan, S. Metze, R. Z. Orozco, N. Kleinschmidt and O. Mühlemann, Cell. Mol. Life Sci., 2010, 67, 677 CrossRef CAS PubMed.
- C. Schweingruber, S. C. Rufener, D. Zünd, A. Yamashita and O. Mühlemann, Biochim. Biophys. Acta, 2013, 1829, 612 CrossRef CAS PubMed.
- S. Durand, N. Cougot, F. Mahuteau-Betzer, C. H. Nguyen, D. S. Grierson, E. Bertrand, J. Tazi and F. Lejeune, J. Cell Biol., 2007, 178, 1145 CrossRef CAS PubMed.
- C. Rivalle, C. Ducrocq, J.-M. Lhoste and E. Bisagni, J. Org. Chem., 1980, 45, 2176 CrossRef CAS.
- C. Rivalle, C. Ducrocq, J.-M. Lhoste, F. Wendling, E. Bisagni and J.-C. Cherman, Tetrahedron, 1981, 37, 2097 CrossRef CAS.
- K. M. Keeling, D. Wang, Y. Dai, S. Murugesan, B. Chenna, J. Clark, V. Belakhov, J. Kandasamy, S. E. Velu, T. Baasov and D. M. Bedwell, PLoS One, 2013, 8, 60478 Search PubMed.
- A. Vilsmeier and A. Haack, Chem. Ber., 1927, 60, 119 CrossRef.
- G. Lai, X. R. Bu, J. Santos and E. A. Mintz, Synlett, 1997, 1275 CrossRef CAS.
- A. Rieche, H. Gross and E. Höft, Chem. Ber., 1960, 93, 88 CrossRef CAS.
- C. Schneider, D. Gueyrard, F. Popowycz, B. Joseph and P. G. Goekjian, Synlett, 2007, 2237 CAS.
- B. O. Lindgren and T. Nilsson, Acta Chem. Scand., 1973, 27, 888 CrossRef CAS.
- B. S. Bal, W. E. Childers and H. W. Pinnick, Tetrahedron, 1981, 37, 2091 CrossRef CAS.
- L. Colombo, C. Gennari, M. Santandrea, E. Narisano and C. Scolastico, J. Chem. Soc., Perkin Trans. 1, 1980, 136 RSC.
- A. El-Faham, R. S. Funosas, R. Prohens and F. Albericio, Chem. – Eur. J., 2009, 15, 9404 CrossRef CAS PubMed.
- G. Ricci, R. Ruzziconi and E. Giorgio, J. Org. Chem., 2005, 70, 1011 CrossRef CAS PubMed.
- J. K. Laha, P. Petrou and G. D. Cuny, J. Org. Chem., 2009, 74, 3152 CrossRef CAS PubMed.
- A. Alder and D. Bellus, J. Am. Chem. Soc., 1983, 105, 6712 CrossRef CAS.
- R. G. Micetich, R. Singh and C. C. Shaw, J. Org. Chem., 1986, 51, 1811 CrossRef CAS.
- R. G. Micetich, R. Singh, M. P. Singh and C. C. Shaw, Synth. Commun., 1986, 16, 453 CrossRef CAS.
- J. Zhang, X. Sun, Y. Qian and L. E. Maquat, RNA, 1998, 4, 801 CrossRef CAS PubMed.
- L. E. Maquat and A. J. Kinniburgh, Nucleic Acids Res., 1985, 13, 2855 CrossRef CAS PubMed.
- J. Cheng, M. Fogel-Petrovic and L. E. Maquat, Mol. Cell. Biol., 1990, 10, 5215 CrossRef CAS PubMed.
- K. R. Prowse and H. Baumann, Mol. Cell. Biol., 1988, 8, 42 CrossRef CAS PubMed.
- X. Dong, P. Stothard, I. J. Forsythe and D. S. Wishart, Nucleic Acids Res., 2004, 32, W660 CrossRef CAS PubMed.
- T. F. Brazão, J. Demmers, W. van IJcken, J. Strouboulis, M. Fornerod, L. Romão and F. G. Grosveld, FEBS Lett., 2012, 586, 1101 CrossRef PubMed.
- L. Linde, S. Boelz, G. Neu-Yilik, A. E. Kulozik and B. Kerem, Eur. J. Hum. Genet., 2007, 15, 1156 CAS.
- V. Ramu, M. R. Gill, P. J. Jarman, D. Turton, J. A. Thomas, A. Das and C. Smythe, Chem. – Eur. J., 2015, 21, 9185 CrossRef CAS PubMed.
- F. Usuki, A. Yamashita, I. Higuchi, T. Ohnishi, T. Shirigashi, M. Osame and S. Ohno, Ann. Neurol., 2004, 55, 740 CrossRef CAS PubMed.
- H. Jungwirth, H. Bergler and G. Högenauer, J. Biol. Chem., 2001, 276, 36419 CrossRef CAS PubMed.
- Y. Dang, W.-K. Low, J. Xu, N. H. Gehring, H. C. Dietz, D. Romo and J. O. Liu, J. Biol. Chem., 2009, 284, 23613 CrossRef CAS PubMed.
- S. Gonzalez-Hilarion, T. Beghyn, J. Jia, N. Debreuck, G. Berte, K. Mamchaoui, V. Mouly, D. C. Gruenert, B. Déprez and F. Lejeune, Orphanet J. Rare Dis., 2012, 7, 58 CrossRef PubMed.
- J. Wengrod, L. Martin, D. Wang, P. Frischmeyer-Guerrerio, H. C. Dietz and L. B. Gardner, Mol. Cell. Biol., 2013, 33, 2128 CrossRef CAS PubMed.
- L. Martin, A. Grigoryan, D. Wang, J. Wang, L. Breda, S. Rivella, T. Cardozo and L. B. Gardner, Cancer Res., 2014, 74, 3104 CrossRef CAS PubMed.
- M. Bhuvanagiri, J. Lewis, K. Putzker, J. P. Becker, S. Leicht, J. Krijgsveld, R. Batra, B. Turnwald, B. Jovanovic, C. Hauer, J. Sieber, M. W. Hentze and A. E. Kulozik, EMBO Mol. Med., 2014, 6, 1593 CrossRef CAS PubMed.
- K. M. Keeling, D. Wang, S. E. Conard and D. M. Bedwell, Crit. Rev. Biochem. Mol. Biol., 2012, 47, 444 CrossRef CAS PubMed.
- H. L. Lee and J. P. Dougherty, Pharmacol. Ther., 2012, 136, 227 CrossRef CAS PubMed.
- http://cbligand.org/PAINS/login.php .
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and spectroscopic data. See DOI: 10.1039/c5ob02482j |
| This journal is © The Royal Society of Chemistry 2016 |