Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Graphene-stabilized lipid monolayer heterostructures: a novel biomembrane superstructure
Lia M. C.
Lima
,
Wangyang
Fu
,
Lin
Jiang
,
Alexander
Kros
 and
Grégory F.
Schneider
*
and
Grégory F.
Schneider
*
Leiden University, Faculty of Science, Leiden Institute of Chemistry, Einsteinweg 55, 2333CC Leiden, The Netherlands. E-mail: g.f.schneider@chem.leidenuniv.nl
First published on 31st August 2016
Abstract
Chemically defined and electronically benign interfaces are attractive substrates for graphene and other two-dimensional materials. Here, we introduce lipid monolayers as an alternative, structurally ordered, and chemically versatile support for graphene. Deposition of graphene on the lipids resulted in a more ordered monolayer than regions without graphene. The lipids also offered graphene a more uniform and smoother support, reducing graphene hysteresis loop and the average value of the charge neutrality point under applied voltages. Our approach promises to be effective towards measuring experimentally biochemical phenomena within lipid monolayers and bilayers.
1. Introduction
Graphene1 is typically supported – sometimes sandwiched – with other two-dimensional materials to promote higher mobility,2 to ensure the reproducibility in electrical performances,3 and to prevent environmental contamination.4 Frequently composed of inorganic, hard and crystalline materials, the so called van der Waals heterostructures have emerged as a route to design new and remarkably complex layer-by-layer films of 2D materials, including graphene.5 One challenge associated with 2D materials as supporting and sandwiching layers is their limited chemical diversity, functions, and inherent inorganic nature. The possibility of combining graphene with soft, dynamic and molecular self-assembled monolayers is therefore of high interest as an organic alternative to inorganic 2D materials and could provide a versatile platform for applications, such as biosensors, drug delivery systems or cellular devices.6Lipids – main constituents of cell membranes – are amphiphilic molecules that can self-assemble and form stable quasi two dimensional fluidic membrane structures.7 Lipids can spread on graphene,8 however little is known on the formation, stability and molecular structure of lipid molecules surrounding graphene.8–15 Mainly, studies focused on graphene oxide (GO), as both lipid vesicles and GO form stable suspensions in aqueous environments.16,17 GO is an easily accessible form of graphene, suitable to study the influence of oxidation states on the chemical characteristics of GO-lipid assemblies, at the cost of lower electron mobility, higher chemical reactivity, oxygen doping, and surface/edge inhomogeneities. Being negatively charged, GO has a particular affinity with positively charged lipid head groups,18 highlighting the importance of electrostatic interactions in the assembly process.19 Pristine graphene, however, does not contain charges on the basal plane therefore minimizing electrostatic interactions and favoring hydrophobic interactions between lipid tails and graphene at the interface.20 To understand and quantify the interactions between lipid tails and graphene, an approach is that graphene crowd surfs directly on the lipid chains, and measure the molecular structure of the lipids and the corresponding electrical properties of graphene.
In this paper we investigate the stability and structure of a lipid monolayer placed underneath graphene and report that graphene affects the conformation of a lipid monolayer, yielding a rearrangement of the lipids into a more ordered and compact supramolecular conformation. Remarkably, attenuated total reflectance infrared spectroscopy (ATR-IR) and ellipsometry demonstrate an increase of the absorbance intensity and of the thickness of the lipid monolayer in presence of graphene, respectively. Our finding suggests a high affinity between the lipid tails and the graphene basal plane promoting a favorable heterostructure for biosensing applications, and represents the first step towards embedding graphene into a lipid bilayer as proposed by recent molecular dynamics simulations.21,22
2. Results and discussion
2.1. Langmuir–Blodgett technique: sample preparation
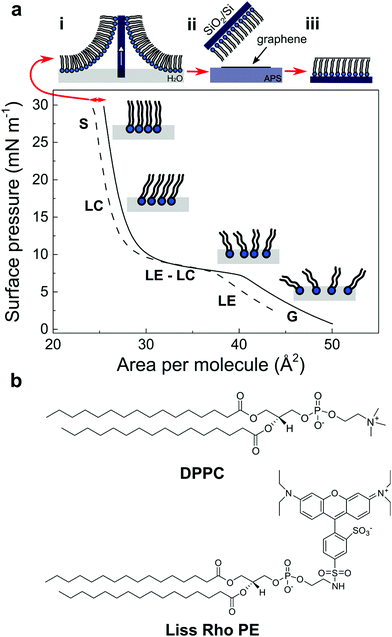
One straightforward approach to form and study a graphene–lipid monolayer interface is to pre-form a well-packed monolayer of lipids on a known substrate, such as SiO2/Si and transfer a graphene layer on top. A well-established route to build an ordered lipid monolayer is by applying the Langmuir–Blodgett technique23 using 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1% of 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Liss Rhod PE) (Fig. 1b). First, the mixture of lipids is dissolved in an organic solution of chloroform/methanol (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and is subsequently deposited dropwise at the air/water interface of the Langmuir trough. By further compressing this unordered lipid phase (i.e. in a so called gas phase (G), Fig. 1a) to specific surface pressures, a very compact lipid monolayer can be formed and transferred to any arbitrary substrate.24 In this study, the lipids were compressed until a surface pressure (π) of 30 mN m−1 – to form a compact and stable monolayer – and thereafter transferred to a SiO2/Si substrate by retracting the substrate out of the trough at maximum compression. A CVD graphene layer was then transferred above the lipid film by bringing into contact the lipid film with graphene floating on ammonium persulfate solution (APS). The sample was immediately rinsed with ultra-pure water to remove traces of APS (Fig. 1a, top; see experimental section).
1) and is subsequently deposited dropwise at the air/water interface of the Langmuir trough. By further compressing this unordered lipid phase (i.e. in a so called gas phase (G), Fig. 1a) to specific surface pressures, a very compact lipid monolayer can be formed and transferred to any arbitrary substrate.24 In this study, the lipids were compressed until a surface pressure (π) of 30 mN m−1 – to form a compact and stable monolayer – and thereafter transferred to a SiO2/Si substrate by retracting the substrate out of the trough at maximum compression. A CVD graphene layer was then transferred above the lipid film by bringing into contact the lipid film with graphene floating on ammonium persulfate solution (APS). The sample was immediately rinsed with ultra-pure water to remove traces of APS (Fig. 1a, top; see experimental section).
Fig. 1a shows the surface pressure (π) – area compression isotherm of DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Liss Rhod PE (99
Liss Rhod PE (99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (solid line) on a pure water sub-phase and the subsequent decompression of the Langmuir film (dashed line) after the transfer of the lipid monolayer onto the SiO2/Si substrate. The lipid monolayer was compressed until a π of 30 mN m−1 passing through distinct separate phases characteristic of phospholipid molecules. In the first step, the lipids spread at the air/water interface yielding the gaseous state (G) due to the small forces exerted by the large distance between molecules. As the mobile barriers of the Langmuir trough start to compress, the available area per molecule and the intermolecular distance between the lipids decreases, resulting in the transition from the gaseous to a liquid expanded state (LE). After further compression, the molecules undergo a phase transition from a fluidic to a condensed phase. This liquid expanded (LE)–liquid condensed (LC) phase transition is characterized by a different aggregation state where the lipids present a strong lateral cohesion and a well-defined orientation. Finally, when the available area of the monolayer is further reduced, the molecules self-organize in a perfectly ordered and stable monolayer, called the solid state (S).25 At this stage, the well-packed lipid monolayer is transferred to the SiO2/Si substrate, resulting in a shift in the compression isotherms from which the transfer ratio of the lipids on the substrate is determined (Fig. 1a, dashed line, red arrow; see experimental section).
1) (solid line) on a pure water sub-phase and the subsequent decompression of the Langmuir film (dashed line) after the transfer of the lipid monolayer onto the SiO2/Si substrate. The lipid monolayer was compressed until a π of 30 mN m−1 passing through distinct separate phases characteristic of phospholipid molecules. In the first step, the lipids spread at the air/water interface yielding the gaseous state (G) due to the small forces exerted by the large distance between molecules. As the mobile barriers of the Langmuir trough start to compress, the available area per molecule and the intermolecular distance between the lipids decreases, resulting in the transition from the gaseous to a liquid expanded state (LE). After further compression, the molecules undergo a phase transition from a fluidic to a condensed phase. This liquid expanded (LE)–liquid condensed (LC) phase transition is characterized by a different aggregation state where the lipids present a strong lateral cohesion and a well-defined orientation. Finally, when the available area of the monolayer is further reduced, the molecules self-organize in a perfectly ordered and stable monolayer, called the solid state (S).25 At this stage, the well-packed lipid monolayer is transferred to the SiO2/Si substrate, resulting in a shift in the compression isotherms from which the transfer ratio of the lipids on the substrate is determined (Fig. 1a, dashed line, red arrow; see experimental section).
2.2. Effect of graphene on the structure of the lipid monolayer
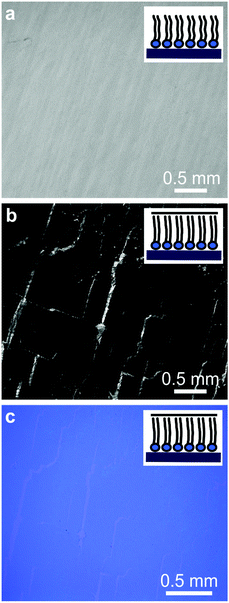
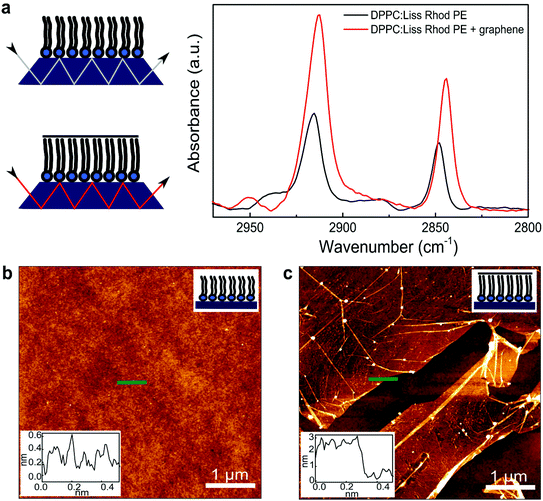
Optical and fluorescence microscopy measurements of the lipid monolayer and the subsequent lipid–graphene assembly are shown in Fig. 2. A homogenous and continuous fluorescence lipid layer is observed on SiO2/Si substrate (Fig. 2a). Next, graphene was transferred onto the lipid monolayer resulting in a strong fluorescence quenching of the rhodamine B dye. Note that millimeter sized graphene domains are observed, even without the need of a polymer such as PMMA for the transfer, as shown by Fig. 2b and c.26 The cracks on the basal plane of graphene are advantageous to image graphene using fluorescence quenching microscopy.27 Remarkably, after rinsing with ultra-pure water (to remove APS traces), the lipids underneath the graphene area remained intact, as confirmed by infrared spectroscopy and ellipsometry (Fig. 3a), suggesting that graphene acts as a shield that prevent the lipids from getting rinsed off and protecting them from the environment (Fig. 2b).In order to characterize the molecular structure and organization of the lipids, we performed attenuated total reflectance infrared (ATR-IR) spectroscopy measurements. Fig. 3a shows the absorption bands characteristic for the stretching vibrations of the lipid acyl chains for the lipid monolayer (black line) and for the lipid–graphene assembly (red line).28 The presence of these peaks confirms that the lipids remain underneath the graphene. Depending on whether the lipids are in contact or not with graphene, a shift in the peaks maxima is observed, characteristic for changes in the lipid conformation.29 Additionally, a shift was observed in the asymmetric methylene vibration (CH2) from ∼2915 to 2912 cm−1 and in the symmetric methylene vibration (CH2) from ∼2848 to 2844 cm−1, respectively. Furthermore the intensity of the asymmetric and symmetric CH2 bands of the lipid–graphene assembly increased. The observed shift is attributed to a change of the physical properties of the lipids film, where the frequencies of CH2 stretching vibrations are known to decrease if lipids are well packed. An elongation of the lipid acyl chains will also yield an increase of intensity of the band. As the CH2 frequency decreases, the lipid hydrocarbon chain order increases, suggesting a change from gauche to trans conformation of the lipid chains.30 The molecules are closer to each other, have less freedom to vibrate and therefore leading to a decrease of the wavenumber. Thus, the lipids underneath graphene present a crystalline structure with presumably very restricted diffusional mobility.29–31 At this stage of the experiment, we therefore consider that graphene enters into – what we call – a ‘crowd surfing’ mode, either static or dynamic.
To obtain further information on the change of conformation of the lipid molecules upon their interaction with graphene, we determined the thickness of the different layers composing the thin film by ellipsometry (see experimental section). The thickness of the SiO2 layer was determined 282.9 ± 1.4 nm.32 Then, the deposited lipid monolayer increased the thickness by 2.5 ± 0.4 nm. As expected, a further increase of the lipid thickness after transferring graphene was observed. The lipid monolayer increased to 3.9 ± 0.9 nm. This expansion of the lipid acyl chains, as confirmed previously by ATR-IR, corresponds to the formation of a more ordered structure. The interactions of the hydrophobic lipid tails with the hydrophobic graphene lying on top are most probably remarkably favorable.8,21 This increase of 1.4 nm, could also be expected if a bilayer would form, but this hypothesis is excluded as no more lipids were in contact with the sample at that time of the sample processing. Although an absolute increase in thickness of 1.4 nm is surprising giving the expected head-to-tail length of DPPC, it is evident that the lipids rearrange in a more organized layer after interacting with graphene, as confirmed previously by ATR-IR. The graphene thickness measured was 0.4 ± 0.2 nm, supporting the transfer of a single monolayer graphene.
2.3. Topography study of the lipid–graphene heterostructure
The atomic force microscopy (AFM) images and the corresponding height profiles of the lipid monolayer in air at room temperature before and after transferring graphene showed the presence of an homogenous monolayer of lipids (Fig. 3b), and for this reason, no significant step height differences could be measured on the lipid monolayer. These results confirm the formation of a stable and compacted lipid layer on SiO2/Si substrate (Fig. 1a). The transferred graphene sheet on top of the lipid monolayer showed very flat and continuous domains (Fig. 3c) with only a few wrinkles and cracks observed, as shown by fluorescence microscopy (Fig. 2b).332.4. Raman spectroscopy of graphene on the lipids structure
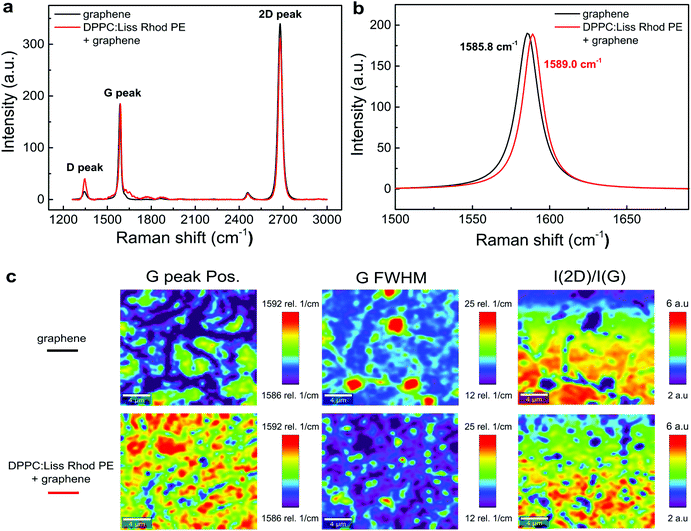
Fig. 4a shows the averaged Raman spectra for graphene on a SiO2/Si substrate (black line) and for graphene transferred above the lipid monolayer in air at room temperature (red line). The sharp, symmetric and intensive 2D peak (∼2680 cm−1) and G peak (∼1580 cm−1) indicates the presence of single layer graphene.34 The weak D peak at 1350 cm−1 is commonly present in CVD graphene, revealing a reasonable graphene quality. Specifically, the G band has been generally considered as an important indicator of doping effect in graphene.35,36 In Fig. 4b, the G peak of graphene (1585.8 cm−1) blue shifts if lipids are present underneath (1589.0 cm−1). Such blue shift can be attributed to the known p-doping of lipids,34 or due to the presence of an adsorbed water layer37 at the interface between graphene and the substrate.The imaging of the G peak position, its full width half maximum (FWHM) and the intensity ratio of I(2D)/I(G) are summarized in the mapping data shown in Fig. 4c. The G peak shows a larger blue shift and a much narrower width in the presence of the lipids as seen by the overall more reddish plots. Furthermore, the ratio of I(2D)/I(G) is less for the lipid–graphene assembly compare to graphene on SiO2/Si substrate. All of these evidences confirm the p-doping effect from the lipids underneath graphene.38,39 It is also worth to notice that the lipid-graphene assembly presents a more even color distribution than those of graphene on SiO2/Si, which suggests that the lipid monolayer underneath represents a more uniform and smooth support for graphene.
2.5. Field-effect characterization of graphene on the lipids structure
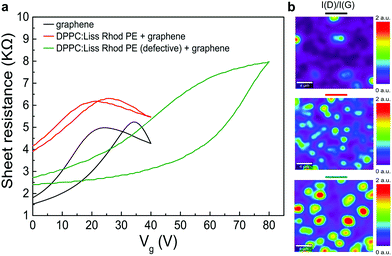
For electrical characterization of the lipid–graphene assembly, metal electrodes (chromium, 30 nm) were deposited on the CVD graphene above the lipid monolayer using a physical mask. As a control, we fabricated another graphene device directly on the SiO2/Si substrate using a PMMA assisted transfer method.26The graphene device on bare SiO2/Si substrate exhibits a hysteresis of 10 V with an average charge neutrality point (CNP), VCNP of about +30 V (Fig. 5a, black line). The relatively large hysteresis and VCNP can be ascribed to the well-known charge trap and p-doping effect of the SiO2/Si substrate.40 Remarkably, the lipid monolayer favored the screening of the silicon substrate (Fig. 5a, red line). As a result, the electrical performances of the lipid–graphene device were improved with a reduced hysteresis loop (4 V) and a smaller average VCNP (23 V). We note here that the decrease of the estimated carrier mobility of graphene on the lipid monolayer (430 cm2 V−1 s−1 compared to 640 cm2 V−1 s−1 on bare substrate) most likely originates from the incomplete surface coverage of graphene on the lipid monolayer. Another possible origin of this lower mobility is maybe the presence of some wrinkles as depicted in Fig. 3c. The graphene coverage with the lipid-assisted transfer method was below 80%, whereas the graphene coverage on bare substrate was larger than 95% (observed by optical images). In addition, occasionally we also observed in our experiments that defective lipids (Langmuir–Blodgett transfer ratio <1) degraded the performance of graphene device by introducing even more p-doping effect (with VCNP > 80 V) and larger hysteresis (>25 V) (Fig. 5a, green line). We have repeated the electrical measurements for another lipid–graphene sample. This sample exhibited a carrier mobility of 570 cm2 V−1 s−1, a hysteresis of 1.5 V, and a VCNP of 8 V, closely resembling what we showed in Fig. 5a. We noted here that we tested also a few defective samples. The carrier mobility, hysteresis, and VCNP of these defective samples demonstrated a random and wide distribution of ∼100–700 cm2 V−1 s−1, ∼6–11 V, and ∼11–45 V, respectively.
In Fig. 5b, the intensity ratio between the D peak and G peak I(D)/I(G) revealed the disorder and defects on graphene, which can be ascribed to the substrate effects. These substrate effects are primarily due to the roughness of SiO2/Si substrate, the strain induced by the lipid monolayer underneath, and the possible adsorbed water layer at the interface. Compared to graphene on the lipid monolayer (Fig. 5b, middle), the I(D)/I(G) ratio of graphene on defective lipids (Fig. 5b, bottom) is more intense. A higher I(D)/I(G) ratio is in line with the larger field-effect hysteresis (25 V in case of defective lipids supporting graphene compared to 4 V in case of graphene on lipid monolayer) and the higher p-doping (i.e., larger average VCNP of 80 V compared to 23 V for graphene on lipid monolayer). We note here that the I(D)/I(G) ratio of graphene on bare substrate (Fig. 5b, top) was indeed less intensive than the one for graphene on a lipid monolayer (Fig. 5b, middle). Nevertheless, we observed a less pronounced field-effect hysteresis (4 V in case of lipid monolayer supporting graphene compared to 10 V in case of graphene on bare substrate) and a smaller average VCNP (23 V compared to 30 V for graphene on bare substrate), which might be ascribed to the PMMA assisted transfer method we used for transferring graphene on bare substrate.
3. Conclusion
The observation of a unique re-ordering of lipid molecules in the presence of graphene reveals an increase in the packing of the lipid monolayer as graphene crowd surfs on the lipid monolayer. Remarkably, lipids ameliorated the electrical performances of the graphene, which is of high interest for using chemically versatile soft materials as alternative substrates for graphene. Additionally, the direct contact between graphene and lipids is particularly attracting for measuring biochemical phenomena, for example in situ a lipidic layer. Future experiments investigating different lipids with different charges, different phase transition temperatures, and different lipid packing will be essential to elucidate the potential sensitivity of graphene to even more subtle changes in (bio)molecular conformations. A practical benefit of the lipid monolayer is also that large millimeter sized and continuous graphene domains are supported, avoiding polymers or any contaminants usually used during typical graphene transfers.We believe that interfacing graphene with lipid molecules offers a new sensing platform to chemically modulate the electrical properties of graphene by varying the lipids structure and is the first step towards sandwiching graphene within the hydrophobic core of a lipid bilayer.
4. Experimental section
4.1. Langmuir–Blodgett technique
DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Liss Rhod PE (99
Liss Rhod PE (99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) lipids (Avanti Polar Lipids Inc.) with a concentration of 1 mg mL−1 were prepared in CHCl3/CH3OH 3
1) lipids (Avanti Polar Lipids Inc.) with a concentration of 1 mg mL−1 were prepared in CHCl3/CH3OH 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 vol%. The lipid solution was dropwise deposited on ultra-pure water at 25 °C by means of a microliter syringe, and the solvent was allowed to evaporate for 15 min. The measurements were performed in a Minitrough 2, KSV Instruments using KSV Research Lab v2.01 software. A Teflon trough was equipped with hydrophilic barriers made of Delrin. The surface pressure (π) was measured by a microbalance platinum plate, with the lipids passing through different phases while compressing. The SiO2/Si substrate was primarily inserted in the ultra-pure water and after the compression of the lipids until a π of 30 mN m−1, it was slowly retracted from the trough while simultaneously keeping the surface pressure constant. Consequently, the lipid monolayer is transferred onto the SiO2/Si substrate and the lipids transfer ratio can be determined. The transfer ratio is defined by the ratio between the drop in the monolayer area during deposition and the area of the substrate. In all the samples prepared, the transfer ratio was 1.0 ± 0.1, signifying a homogeneous lipid film deposited. Multiple samples, at least 10, were tested to confirm reproducibility of the experiments.
1 vol%. The lipid solution was dropwise deposited on ultra-pure water at 25 °C by means of a microliter syringe, and the solvent was allowed to evaporate for 15 min. The measurements were performed in a Minitrough 2, KSV Instruments using KSV Research Lab v2.01 software. A Teflon trough was equipped with hydrophilic barriers made of Delrin. The surface pressure (π) was measured by a microbalance platinum plate, with the lipids passing through different phases while compressing. The SiO2/Si substrate was primarily inserted in the ultra-pure water and after the compression of the lipids until a π of 30 mN m−1, it was slowly retracted from the trough while simultaneously keeping the surface pressure constant. Consequently, the lipid monolayer is transferred onto the SiO2/Si substrate and the lipids transfer ratio can be determined. The transfer ratio is defined by the ratio between the drop in the monolayer area during deposition and the area of the substrate. In all the samples prepared, the transfer ratio was 1.0 ± 0.1, signifying a homogeneous lipid film deposited. Multiple samples, at least 10, were tested to confirm reproducibility of the experiments.
4.2. Lipid-assisted transfer method
The monolayer CVD graphene grown on a copper foil (Graphenea) is placed in a solution of ammonium persulfate (0.5 M) (APS) – copper etchant – and after etching, the graphene is transferred (‘fished’) above the lipid monolayer on SiO2/Si substrate prepared before. Immediately after, the sample is rinsed with ultra-pure water to remove the eventual residues of the etchant, obtaining a SiO2/Si–lipid–graphene heterostructure. Raman spectroscopy confirms the monolayer form of graphene after transfer above the lipid monolayer. This method is polymer free and therefore yields minimal contamination on graphene surface and also avoids water trapping between the substrate and graphene.4.3. Imaging set-up
Optical images were performed on a Leica DM 2700 M microscope and bright field fluorescence images were obtained on a Zeiss Axiovert 200 with a 5× objective. 1 mol% Liss Rhod PE was mixed with DPPC for fluorescence measurements. The lipids were labeled to characterize the lipid monolayer assembly and the quenching of graphene after the transfer.4.4. ATR-IR
Spectra were collected with a BIO-RAD Excalibur Series spectroscopy using a silicon ATR crystal 25 reflection with an angle of incidence of 45° (Specac) and a spectra range of 8.000–0 cm−1. Each spectrum was collected for 128 scans with a resolution of 4 cm−1 and the chamber was continuously purged with dry air. The data were processed using Origin-Pro. The bare ATR crystal was collected as the background. The sample preparation on the ATR crystal was the same as on the SiO2/Si substrate, using a bigger CVD monolayer graphene on Cu (60 × 40 mm from Graphenea). All the lipids spectra were performed before and after transferring the graphene layer.4.5. Ellipsometry
The layers thickness were obtained using a WVASE ellipsometer M-2000F EC-400 from J. A. Woollam Co. Inc. The data were collected with the WVase32 software. Each sample was measured at least in 3 different positions with 5 angles of incidence (65°, 70°, 75°, 80° and 85°), at wavelengths ranging between 250 and 1000 nm. The thicknesses were measured step-by-step, as ellipsometry models were built layer-by-layer: (i) first we measured the bare SiO2/Si substrate, (ii) then the Langmuir lipid monolayer deposited and, (iii) finally the graphene layer. The collected data were fitted with a distinct model specific for each material. Each material has a set of optical constants containing the complex refractive index n and the extinction coefficient k. The bare SiO2 substrate was fitted with SiO2. JAW model (n = 1.4554–1.5157; k = 0) and the lipid monolayer with a CAUCHY model. The CAUCHY optical constants were n = 1.46–1.615 and k were set at 0. An, Bn and Cn were 1.45, 0.01 and 0, respectively. An, Bn and Cn were kept constant. The monolayer graphene model was taken from the literature.414.6. AFM microscopy
AFM images were performed on a JPK NanoWizard Ultra Speed machine and the images were processed using a JPK SPM Data Processing software. The experiments were performed using a silicon probe (AC240TS, Asylum Research) with 70 kHz nominal resonance frequency. The images were scanned in an intermittent contact mode in air at room temperature with 512 × 512 pixels.4.7. Raman spectroscopy
Raman spectra and imaging were measured on a WITec confocal spectrometer with a 532 nm laser and a 100× objective with the lateral resolution of 200–300 nm. The laser power was are finely tuned to be below 1 mW to avoid damages to graphene and to the lipids underneath.4.8. Electrical measurements
The transistor characteristics of the graphene field-effect transistor devices on different substrates were tested using a home-made setup. A SR830 DSP lock-in amplifier with narrow filters was used to recover weak signal from a noisy background. The back gate voltage Vg (up to 200 V) was applied using a Keithley 2400 multimeter. The defective lipid monolayers had a transfer ratio of ∼0.5 using the Langmuir–Blodgett technique, meaning that the surface of SiO2/Si substrate was not fully covered with the lipid monolayer.Acknowledgements
This research was gratefully funded by the European Research Council under the European Union's Seventh Framework Programme (FP/2007–2013)/ERC Grant Agreement no. 335879 project acronym ‘Biographene’ and the Netherlands Organization for Scientific Research (NWO-VIDI 723.013.007). W. F. gratefully acknowledges financial support by the Netherlands Organization for Scientific Research (NWO-VENI 722.014.004) and the Swiss National Science Foundation (SNSF P300P2_154557). We thank M. Rabe for all the initial experimental help and expertise, and H. Arjmandi-Tash and Y. Cesa for all the helpful discussions. We thank F. Galli for her support. AFM measurements were performed in the AFM Lab of the Leiden Institute of Physics.References
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Science, 2004, 306, 666–669 CrossRef CAS PubMed
.
- C. R. Dean, A. F. Young, I. Meric, C. Lee, L. Wang, S. Sorgenfrei, K. Watanabe, T. Taniguchi, P. Kim, K. L. Shepard and J. Hone, Nat. Nanotechnol., 2010, 5, 722–726 CrossRef CAS PubMed
.
- J. Xue, J. Sanchez-Yamagishi, D. Bulmash, P. Jacquod, A. Deshpande, K. Watanabe, T. Taniguchi, P. Jarillo-Herrero and B. J. Leroy, Nat. Mater., 2011, 10, 282–285 CrossRef CAS PubMed
.
- L. Wang, I. Meric, P. Y. Huang, Q. Gao, Y. Gao, H. Tran, T. Taniguchi, K. Watanabe, L. M. Campos, D. A. Muller, J. Guo, P. Kim, J. Hone, K. L. Shepard and C. R. Dean, Science, 2013, 342, 614–617 CrossRef CAS PubMed
.
- A. K. Geim and I. V. Grigorieva, Nature, 2013, 499, 419–425 CrossRef CAS PubMed
.
- A. C. Ferrari, F. Bonaccorso, V. Fal'ko, K. S. Novoselov, S. Roche, P. Boggild, S. Borini, F. H. L. Koppens, V. Palermo, N. Pugno, J. A. Garrido, R. Sordan, A. Bianco, L. Ballerini, M. Prato, E. Lidorikis, J. Kivioja, C. Marinelli, T. Ryhanen, A. Morpurgo, J. N. Coleman, V. Nicolosi, L. Colombo, A. Fert, M. Garcia-Hernandez, A. Bachtold, G. F. Schneider, F. Guinea, C. Dekker, M. Barbone, Z. Sun, C. Galiotis, A. N. Grigorenko, G. Konstantatos, A. Kis, M. Katsnelson, L. Vandersypen, A. Loiseau, V. Morandi, D. Neumaier, E. Treossi, V. Pellegrini, M. Polini, A. Tredicucci, G. M. Williams, B. Hee Hong, J.-H. Ahn, J. Min Kim, H. Zirath, B. J. van Wees, H. van der Zant, L. Occhipinti, A. Di Matteo, I. A. Kinloch, T. Seyller, E. Quesnel, X. Feng, K. Teo, N. Rupesinghe, P. Hakonen, S. R. T. Neil, Q. Tannock, T. Lofwander and J. Kinaret, Nanoscale, 2015, 7, 4598–4810 RSC
.
- G. van Meer, D. R. Voelker and G. W. Feigenson, Nat. Rev. Mol. Cell Biol., 2008, 9, 112–124 CrossRef CAS PubMed
.
- M. Hirtz, A. Oikonomou, T. Georgiou, H. Fuchs and A. Vijayaraghavan, Nat. Commun., 2013, 4, 2591 Search PubMed
.
- Y. Y. Wang, T. D. Pham, K. Zand, J. Li and P. J. Burke, ACS Nano, 2014, 8, 4228–4238 CrossRef CAS PubMed
.
- P. K. Ang, M. Jaiswal, C. H. Y. X. Lim, Y. Wang, J. Sankaran, A. Li, C. T. Lim, T. Wohland, Ö. Barbaros and K. P. Loh, ACS Nano, 2010, 4, 7387–7394 CrossRef CAS PubMed
.
- L. S. Connelly, B. Meckes, J. Larkin, A. L. Gillman, M. Wanunu and R. Lal, ACS Appl. Mater. Interfaces, 2014, 6, 5290–5296 CAS
.
- K. Yamazaki, S. Kunii and T. Ogino, J. Phys. Chem. C, 2013, 117, 18913–18918 CAS
.
- S. R. Tabaei, W. B. Ng, S.-J. Cho and N.-J. Cho, ACS Appl. Mater. Interfaces, 2016, 8, 11875–11880 Search PubMed
.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan, H. Fang and R. Zhou, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef CAS PubMed
.
- B. Luan, T. Huynh and R. Zhou, Nanoscale, 2016, 8, 5750–5754 RSC
.
- H. P. Boehm, A. Clauss, G. O. Fischer and U. Hofmann, Z. Anorg. Allg. Chem., 1962, 316, 119–127 CrossRef CAS
.
- W. S. Hummers and R. E. Offeman, J. Am. Chem. Soc., 1958, 80, 1339–1339 CrossRef CAS
.
- S. H. Li, A. J. Stein, A. Kruger and R. M. Leblanc, J. Phys. Chem. B, 2013, 117, 16150–16158 CAS
.
- R. Frost, G. E. Jonsson, D. Chakarov, S. Svedhem and B. Kasemo, Nano Lett., 2012, 12, 3356–3362 CrossRef CAS PubMed
.
- L. Rodriguez-Perez, M. A. Herranz and N. Martin, Chem. Commun., 2013, 49, 3721–3735 RSC
.
- A. V. Titov, P. Kral and R. Pearson, ACS Nano, 2010, 4, 229–234 CrossRef CAS PubMed
.
- J. Mao, R. Guo and L.-T. Yan, Biomaterials, 2014, 35, 6069–6077 CrossRef CAS PubMed
.
- Z.-W. Yu, J. Jin and Y. Cao, Langmuir, 2002, 18, 4530–4531 CrossRef CAS
.
- M. Hirtz, H. Fuchs and L. Chi, J. Phys. Chem. B, 2008, 112, 824–827 CrossRef CAS PubMed
.
- G. Oncins, L. Picas, J. Hernández-Borrell, S. Garcia-Manyes and F. Sanz, Biophys. J., 2007, 93, 2713–2725 CrossRef CAS PubMed
.
- X. Li, Y. Zhu, W. Cai, M. Borysiak, B. Han, D. Chen, R. D. Piner, L. Colombo and R. S. Ruoff, Nano Lett., 2009, 9, 4359–4363 CrossRef CAS PubMed
.
- J. Kim, L. J. Cote, F. Kim and J. Huang, J. Am. Chem. Soc., 2010, 132, 260–267 CrossRef CAS PubMed
.
- A. Blume and A. Kerth, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 2294–2305 CrossRef CAS PubMed
.
- S. A. Tatulian, Biochemistry, 2003, 42, 11898–11907 CrossRef CAS PubMed
.
- R. N. A. H. Lewis and R. N. McElhaney, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 2347–2358 CrossRef CAS PubMed
.
- D. M. Small, J. Lipid Res., 1984, 25, 1490–1500 CAS
.
- W. A. J. C. M. Herzinger and J. A. Woollam, J. Appl. Phys., 1998, 86, 3323–3336 CrossRef
.
- P. Nemes-Incze, Z. Osváth, K. Kamarás and L. P. Biró, Carbon, 2008, 46, 1435–1442 CrossRef CAS
.
- A. Das, S. Pisana, B. Chakraborty, S. Piscanec, S. K. Saha, U. V. Waghmare, K. S. Novoselov, H. R. Krishnamurthy, A. K. Geim, A. C. Ferrari and A. K. Sood, Nat. Nanotechnol., 2008, 3, 210–215 CrossRef CAS PubMed
.
- S. Pisana, M. Lazzeri, C. Casiraghi, K. S. Novoselov, A. K. Geim, A. C. Ferrari and F. Mauri, Nat. Mater., 2007, 6, 198–201 CrossRef CAS PubMed
.
- J. Yan, Y. Zhang, P. Kim and A. Pinczuk, Phys. Rev. Lett., 2007, 98, 166802 CrossRef PubMed
.
- T. Tsukamoto, K. Yamazaki, H. Komurasaki and T. Ogino, J. Phys. Chem. C, 2012, 116, 4732–4737 CAS
.
- M. Bruna, A. K. Ott, M. Ijäs, D. Yoon, U. Sassi and A. C. Ferrari, ACS Nano, 2014, 8, 7432–7441 CrossRef CAS PubMed
.
- D. M. Basko, S. Piscanec and A. C. Ferrari, Phys. Rev. B: Condens. Matter, 2009, 80, 165413 CrossRef
.
- M. Lafkioti, B. Krauss, T. Lohmann, U. Zschieschang, H. Klauk, K. V. Klitzing and J. H. Smet, Nano Lett., 2010, 10, 1149–1153 CrossRef CAS PubMed
.
- F. J. Nelson, V. K. Kamineni, T. Zhang, E. S. Comfort, J. U. Lee and A. C. Diebold, Appl. Phys. Lett., 2010, 97, 253110 CrossRef
.
| This journal is © The Royal Society of Chemistry 2016 |