 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A quasi-cyclic RNA nano-scale molecular object constructed using kink turns†
Lin
Huang
and
David M. J.
Lilley
*
Cancer Research UK Nucleic Acid Structure Research Group, MSI/WTB Complex, The University of Dundee, Dow Street, Dundee DD1 5EH, UK. E-mail: d.m.j.lilley@dundee.ac.uk; l.y.huang@dundee.ac.uk; Fax: +44-(0)1382-385893; Tel: +44-(0)1382-384243
First published on 10th August 2016
Abstract
k-Turns are widespread RNA architectural elements that mediate tertiary interactions. We describe a double-kink-turn motif comprising two inverted k-turns that forms a tight horse-shoe structure that can assemble into a variety of shapes by coaxial association of helical ends. Using X-ray crystallography we show that these assemble with two (dumbell), three (triangle) and four units (square), with or without bound protein, within the crystal lattice. In addition, exchange of a single basepair can almost double the pore radius or shape of a molecular assembly. On the basis of this analysis we synthesized a 114 nt self-complementary RNA containing six k-turns. The crystal structure of this species shows that it forms a quasi-cyclic triangular object. These are randomly disposed about the three-fold axis in the crystal lattice, generating a circular RNA of quasi D3 symmetry with a shape reminiscent of that of a cyclohexane molecule in its chair conformation. This work demonstrates that the k-turn is a powerful building block in the construction of nano-scale molecular objects, and illustrates why k-turns are widely used in natural RNA molecules to organize long-range architecture and mediate tertiary contacts.
Introduction
Interhelical junctions play a critical role in generating the folded architecture of functional RNA molecules, both large and medium scale. Junctions direct the trajectory of helical segments that typically make tertiary interactions and so build complex three-dimensional structures such as the ribosome. The kink-turn (k-turn) is a particularly common motif that behaves in this manner.1,2 This is observed in large RNA-protein assemblies, notably the ribosome,1,3–6 and in medium-scale RNA species such as many riboswitches7–14 and box C/D and H/ACA snoRNAs.15–18 The standard k-turn is formed in duplex RNA where a short bulge is followed by G·A and A·G basepairs, and folds to generate a kink with an included angle of 50° between the axes of the two helical arms19 (Fig. 1A and B). One of the arms is often relatively short, terminating in a loop that makes a long-range tertiary interaction elsewhere in the RNA molecule. The folded structure of the k-turn is stabilized by a two key cross-strand A-minor20 interactions, between the 2′-OH groups of L1 and -1n to ring nitrogen atoms of the conserved adenine nucleotides at the 1n and 2b positions respectively19,21–23 (the nomenclature of nucleotide positions is shown in Fig. 1A). The latter hydrogen bond may be accepted by either N3 or N1 of A2b, dividing the k-turns into two conformational classes.19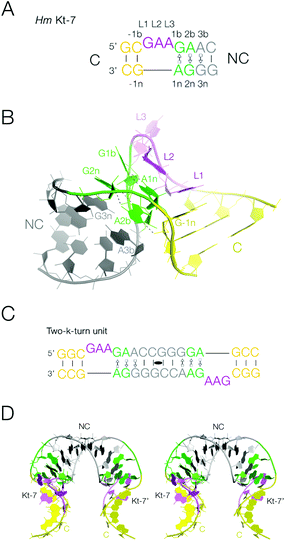 | ||
| Fig. 1 The k-turn, and the two-k-turn unit. A. The sequence of Kt-7, a standard simple k-turn. The standard nomenclature of the nucleotide positions is shown. The color scheme for the nucleotides is followed in all the structures show subsequently. B. The structure of a single Kt-7 k-turn, taken from structure PDB 4CS1. In the view shown the loop of the k-turn (colored purple) is at the back, and the NC helix with the G·A pairs is on the left. C. The sequence of the two-k-turn unit. A self-complementary RNA forms a duplex structure with two Kt-7 k-turns related by a central two-fold axis of rotational symmetry. The two k-turns are connected through their common NC helix (grey), and the two C helices (yellow) form the termini. D. A parallel-eye stereoscopic view of the structure of the two-k-turn unit taken from structure PDB 4CS1. | ||
Thus k-turns may be regarded as a natural nano-scale building block in RNA. RNA has previously been used as a material for engineering on the molecular scale, and helical bulges have been used to create regular structures of square or triangular shape.24–26 In principle such objects can enclose molecular pores of different sizes. k-Turns suggest themselves as a very natural choice of construction unit for this purpose, and Saito and colleagues27,28 have explored this previously.
In this work we have closely examined the packing of a double-stranded RNA containing two k-turns related by two-fold rotational symmetry in different crystal lattices (Fig. 1C and D). These two-k-turn elements adopt a horse-shoe shape, that pack together in a very precise way by coaxial end-to-end stacking of the RNA helices to form quasi-cyclic assemblies with different numbers of component units. This analysis suggested that three two-k-turn units (i.e. six k-turns in total, with three loops on each strand) within one double-stranded RNA molecule would close to form a triangular structure. We have synthesized this structure and demonstrated its quasi cyclic structure by X-ray crystallography. This illustrates the precision with which the k-turn organizes three-dimensional RNA structure, and introduces the two-k-turn unit as a valuable construction unit in nano-technological engineering of RNA structure.
Results and discussion
A double k-turn duplex RNA as a structural building block
We have previously described the construction of a duplex RNA comprising two head-to-head oriented k-turns of Kt-7 sequence2 where the loops lie on opposite strands (Fig. 1C).29 These are connected through their NC helices (i.e. the helices with the G·A pairs), with ten basepairs separating the two loops, including the G·A pairs (Fig. 1C). The structure has been determined by X-ray crystallography to 2.0 Å resolution (PDB 4CS1),30 showing it to adopt an overall horse-shoe shape with a two-fold rotational axis (Fig. 1D). The Kt-7 elements adopt the standard k-turn structure, of the N3 class19 expected when the 3b,3n sequence is A·G.29 Within the individual k-turns the helical axes are not coincident; the kink is not a simple bend, and the axes of the C and NC helices are displaced relative to each other. The double-k-turn unit encloses the deep major groove of the two NC helices, and splays out the shallow minor groove on the outer face of the horse-shoe. This structure forms a building block for the formation of molecular assemblies within different crystal lattices as we now discuss. Crystallographic statistics for new structures are presented in Table S1,† and all the structures discussed are summarized in Table S2.†Two forms of packing of the double k-turn duplex RNA within the crystal lattice
We have obtained crystals for the two-k-turn unit based on Kt-7 in two different forms with differing crystal symmetry. The Kt-7 structures adopt their standard N3 conformation within both forms, with 2b·2n basepairs connected by two hydrogen bonds. However, the units associate in two different ways in these alternative crystal forms (Fig. 2), both by end-to-end coaxial stacking through their C helices (the conventionally basepaired helical arms). The first had the tetragonal space group P4222, and the structure was determined to a resolution of 2.2 Å (PDB 5FJ0).29 Within the crystal lattice two-k-turn units interact through their termini end-to-end, giving an overall dumbell shape with two-fold rotational symmetry (Fig. 2A). Spacefilling representations of the structures of this and all the species discussed in this work are compared in Fig. S1.†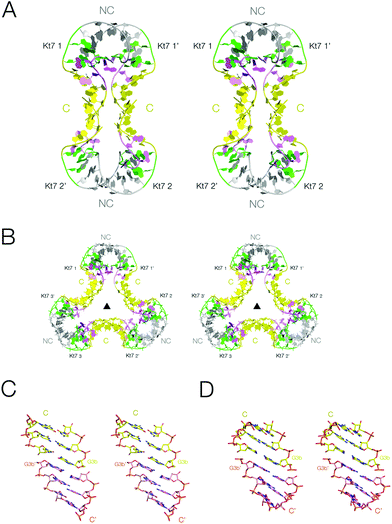 | ||
| Fig. 2 The association of two-k-turn units occurring in the lattice of two different crystal forms. Parallel-eye stereoscopic views of the assemblies found in different crystal lattices. A. Association of two two-k-turn units in a dumbell-shaped structure in PDB 5FJ0. B. Association of three two-k-turn units in a triangular-shaped structure in PDB 4CS1. A three-fold axis of rotational symmetry runs through the center of the triangle. C, D. End-to-end coaxial stacking between the C-helices in the association of two (C) and three (D) two-k-turn units. In these images the color of one of the C helices has been changed to orange in order to differentiate the two helices more easily. Note the perfect stacking between the associated helices. | ||
The second form (PDB 4CS1)30 contained an association of three two-k-turn units, forming a triangular association with three-fold rotational symmetry (Fig. 2B and S2†). As with the previous form, the individual two-k-turn units associate by coaxial stacking through their C helices. These crystals have hexagonal symmetry, with a P6322 space group, and data were collected to 2.0 Å resolution.
The end-to-end association of helices by which the two-k-turn units associate is closely similar in both structures (Fig. 2C and D). The terminal basepairs of the C helices are well stacked with a separation between base planes of 3.3 Å. The long axes (defined here by the C1′–C1′ vectors) of the terminal basepairs are close to being parallel, so departing from the normal rotation at basepair steps within an A-form helix. The conformations of the k-turns are closely similar in both structures. The k-turn of the triangular association (PDB 4CS1) superimposes with the three structures in the asymmetric unit of the dumbell crystal (PDB 5FJ0) with RMSD values of 0.488, 0.363 and 0.410 Å.29
Variation of crystal packing of the double k-turn duplex RNA as a function of 3b,3n sequence
We have shown that the 3b,3n sequence of k-turns plays a critical role in determining both folding characteristics (whether or not they can be folded by addition of metal ions alone)30 and conformation (whether they adopt the N3 or the N1 conformation).29 We have solved new crystal structures for two further forms of the two-k-turn unit in which the 3b,3n sequences of the Kt-7 k-turns were changed either to U·U or to GC. Both sequences confer the N3 conformation,29 but neither is well folded in the presence of metal ions alone.30 These were therefore co-crystallized in the presence of Archeoglobus fulgidus L7Ae protein (AfL7Ae) which drives the folding of k-turns31–33 irrespective of 3b,3n sequence.30The two-k-turn unit based on Kt-7 3bU,3nU crystallized with orthorhombic symmetry (space group P212121) with a resolution of 2.65 Å (PDB 5G4U). Three two-k-turn units bound to six L7Ae proteins formed a triangular assembly in the lattice with three-fold rotational symmetry (Fig. 3A). Once again the k-turns adopt the standard N3 conformation, and the conformation of the k-turns is similar in each of the structures presented here (Fig. S3†). Each is bound to one AfL7Ae molecule in the normal way (Fig. S4 and S5†), placing a basic helix into the major groove of the C helix, and covering the L2 nucleobase with the hydrophobic loop34 (Fig. S6†). For each two-k-turn unit the two AfL7Ae molecules are related by two fold rotational symmetry, so that three proteins lie on one side of the triangular assembly, three on the other. The overall conformation of the triangular association is similar to that based on Kt-7, with the same coaxial end-to-end interactions between C helices, but the structures are not identical. The Kt-7 3bU,3nU-based structure has a larger pore through the center of the triangle (seen clearly in the space-filling representations in Fig. S1†), the radius of which is increased from 7.5 to 14 Å (Table S3†). Clearly the formation of these trimolecular assemblies amplifies the conformational effect, revealing relatively subtle structural differences resulting from the change in 3b,3n sequence despite the k-turns remaining in the N3 conformation in both cases. This may be due to the influence of the altered 3b,3n sequence or the binding of the L7Ae proteins, or a combination of the two.
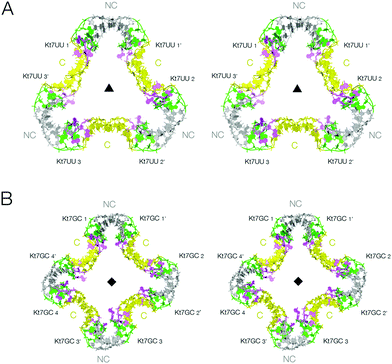 | ||
| Fig. 3 The lattice association of two-k-turn units based on Kt-7 variants in the 3b,3n position. Parallel-eye stereoscopic views of the assemblies are shown. A. Association of three two-k-turn units based on Kt-7 3bU,3nU, forming a triangular-shaped structure (PDB 5G4U). A three-fold axis of rotational symmetry runs through the center of the triangle. The structure differs from that of the triangular assembly formed by Kt-7 (Fig. 2B), having a more open central pore. This can be clearly seen in the spacefilling representations shown in Fig. S1.† B. Association of four two-k-turn units based on Kt-7 3bG,3nC, forming a square-shaped structure (PDB 5G4V). A four-fold axis of rotational symmetry runs through the center of the square. | ||
The two-k-turn unit based on Kt-7 3bG,3nC crystallized with monoclinic symmetry (space group C121) with a resolution of 2.87 Å (PDB 5G4V). In contrast to the 3b,3n = U·U structure, four two-k-turn units have associated in the lattice to form a square structure with four-fold rotational symmetry (Fig. 3B). Eight AfL7Ae molecules bind the square assembly each with standard interactions with the k-turns, four on each side. The four two-k-turn units interact sequentially around the square by coaxial end-to-end interactions between C helices as before.
Comparison of these two structures reveals the strong influence of the 3b,3n sequence. By alteration of just one basepair, the shape of the assembly changes significantly, and the pore radius almost doubles.
Construction and structure of a six-k-turn molecular object
Analysis of the packing of the two-k-turn units within crystal lattices demonstrates that the individual units readily associate by head-to-head coaxial stacking, to generate assemblies containing two, three or four units. We therefore considered the possibility that if three two-k-turn units were contained within the same double-stranded molecule this might fold to generate a closed quasi-continuous triangular species. We synthesized the 114 nt self-complementary RNA shown in Fig. 4A. This contains three two-k-turn units based on the unmodified Kt-7 k-turn sequence (i.e. with 3b,3n = A·G), covalently linked through their C helices. The 3′ cytosine nucleotides of the two-k-turn units were changed to uridine, so that the connected NC helices contain central U·G and G·U pairs. In our experience this adds torsional flexibility, improving the probability that a regular helical conformation could be maintained in the connection between the NC helices, and we found that our crystals diffracted to significantly higher resolution when these were included (Fig. S9†).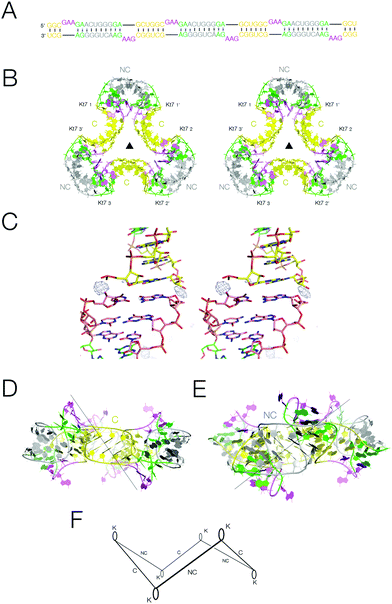 | ||
| Fig. 4 The structure of a quasi-cyclic six k-turn duplex RNA species. A. The sequence of the RNA. The self-complementary RNA hybridizes to create a duplex with six Kt-7 k-turns, alternating between strands and in polarity. B. Parallel-eye stereoscopic view of the crystal structure of the six k-turn duplex RNA (PDB 5G4T). Although one of the C-helices is not covalently continuous, these are randomly located within the crystal lattice, so there is a three-fold rotation axis through the center, and the asymmetric unit contains a single Kt-7 sequence. C. Parallel-eye stereoscopic view of an Fo − Fc omit map showing electron density for the phosphate groups at the center of the C-helices contoured at 3σ. D, E. Two side views of the triangular structure, rotated 60° relative to each other. The views are towards the C-helix (D) and the NC-helix (E). F. Schematic of the structure of the quasi-cyclic six k-turn duplex RNA species. The axes of the helices are depicted by single lines, and the loops of the k-turns by oval shapes. The alternation in direction between the axes of the C- and NC-helices generates a shape reminiscent of that of the cyclohexane molecule in its chair conformation. | ||
The RNA was crystallized in the absence of protein. We obtained crystals of hexagonal symmetry (space group P6322) and solved the structure at a resolution of 2.75 Å (Fig. 4B) (PDB 5G4T). As before, the k-turns adopt the standard N3 conformation with the normal hydrogen bonding (Fig. S7†). The complete six-k-turn molecule in the lattice has folded into a triangular shape closely similar to the packing of the three individual two-k-turn units based on Kt-7 discussed above. The ends of the RNA have associated by coaxial stacking. This is so perfect that these non-covalent ends are randomly located within the crystal lattice, i.e. there is a crystallographic three-fold rotational axis through the centre perpendicular to the plane of the triangle. This averages the covalent and non-covalent connections between two-k-turn units, so that each phosphate group connecting the two-k-turn units is populated with an occupancy of 2/3; an Fo − Fc omit map clearly shows electron density for the phosphate groups connecting the C helices (Fig. 4C). If the ends of the RNA were ligated the structure would become a covalently-closed 114 bp nanocircle with a linking number Lk = 3.
The overall shape of the six-k-turn object that has been effectively cyclized by crystal symmetry is a triangle with edges defined by the C helices and vertices formed by the NC helices (Fig. 4B). Three two-fold axes lie in the plane of the triangle, relating the two k-turns of each two-k-turn unit and passing through the centre of the connected C helices. These axes are perpendicular to the three-fold axis and all four axes intersect at the center, i.e. the symmetry point group is D3. The loops of the k-turn are directed sequentially above and below the triangular plane. The axes of the C and NC helices do not lie in the plane of the triangle, but rather alternate in direction. Thus if we view the structure with one k-turn loop directed upwards, the NC helix falls to the left and the C helix falls to the right (Fig. 4D). The overall structure is therefore reminiscent of the chair form of cyclohexane but with RNA helices in place of carbon–carbon bonds (Fig. 4F).
In conclusion
The crystal packing of the two-k-turn units demonstrates how the crystal lattice is an important resource pointing to the design of nanoparticles. It can form different shapes, and objects with different pore sizes which is normally a challenge for RNA or RNP. The self-assembly of the quasi-cyclic six-k-turn species demonstrates how precisely the k-turn organizes the longer-range architecture of the RNA, colocalizing the ends so exactly that they are accommodated into the crystal lattice indistinguishably from the covalently-connected sections. It indicates that the k-turn is a powerful building block in the construction of nano-scale molecular objects, and illustrates why k-turns are widely used in natural RNA molecules to organize long-range architecture and mediate tertiary contacts.Materials and methods
RNA synthesis
Ribooligonucleotides were synthesized using t-BDMS phosphoramidite chemistry,35 as described in Wilson et al.36 Oligoribonucleotides were deprotected in 25% ethanol/ammonia solution at 20 °C for 3 h, and evaporated to dryness. They were redissolved in 100 μl DMSO to which was added 125 μl 1 M triethylamine trihydrofluoride (TEA, 3HF) (Sigma-Aldrich) and incubated at 65 °C for 2.5 h to remove t-BDMS protecting groups. All oligonucleotides were purified by gel electrophoresis in polyacrylamide in the presence of 7 M urea. The full-length RNA product was visualized by UV shadowing. The band was excised and electroeluted using an Elutrap® Electroelution System (GE Healthcare) into 45 mM Tris-borate (pH 8.5), 5 mM EDTA buffer for 8 h. at 200 V at 4 °C. The RNA was precipitated with ethanol, washed once with 70% ethanol and suspended in water.Expression and purification of A. fulgidus L7Ae
The gene encoding full-length A. fulgidus L7Ae was cloned into a modified pET-Duet1 plasmid (Novagen).37 A hexahistidine-L7Ae fusion protein was expressed in E. coli BL21-Gold (DE3) pLysS cells (Stratagene). The protein was purified by chromatography on HisTrap (GE Healthcare). After removal of the N-terminal six-histidine tag the protein was further purified by chromatography on heparin, followed by gel filtration on Superdex 200.Crystallization, structure determination, and refinement
A mixture of 0.25 mM RNA and 0.25 mM L7Ae in 5 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM MgCl2 was incubated for 5 min at 37 °C. Crystals were grown by vapor diffusion using drops prepared by mixing 1.0 μL of the RNA–protein complex with 1 μL of a reservoir solution comprising 1.6 M tri-sodium citrate dihydrate (pH 6.5) at 18 °C. Crystals appeared after 5 days. The crystals were flash frozen by mounting in nylon loops and plunging into liquid nitrogen. The crystals were characterized in-house with a MicroMax HF007 copper rotating anode X-ray generator equipped with an ACTOR sample changer system and a Saturn 944HG+ CCD detector (Rigaku). Suitable crystals were stored and full datasets subsequently collected on beamline I02 at Diamond Light Source (Harwell, UK).
The crystals had space group P212121 and unit cell dimensions a = 261.3 Å, b = 69.2 Å, c = 85.4 Å. From the crystal density,38,39 three RNA–protein complexes were expected to be present in the asymmetric unit.
The structure was determined by molecular replacement, with PDB 4BW0 as the search model using the program PHASER.40 Ramachandran analysis shows that 98.7% of amino acid residues are in the most favored and additionally allowed regions. Model geometry and the fit to electron-density maps were monitored with MOLPROBITY41 and the validation tools in COOT.
A mixture of 0.3 mM RNA and 0.3 mM L7Ae in 5 mM Tris·HCl (pH 8.0), 100 mM NaCl, 10 mM MgCl2 was incubated for 5 min at 37 °C. Crystals were grown by vapor diffusion using drops prepared by mixing 1.0 μL of the RNA–protein complex with 1 μL of a reservoir solution comprising 0.2 M ammonium acetate, 10 mM calcium chloride, 50 mM sodium cacodylate (pH 6.5) and 10% w/v polyethylene glycol 4000 at 18 °C. Crystals appeared after 7 days. They were transferred to a solution containing an additional 30% w/v polyethylene glycol 200 for ∼2 s. The crystals were flash frozen by mounting in nylon loops and plunging into liquid nitrogen. Suitable crystals were stored and subsequently used to measure full datasets on beamline I02 of Diamond Light Source (Harwell, UK).
The crystals had space group C121 and unit cell dimensions a = 118.7 Å, b = 70.6 Å, c = 92.8 Å. From the crystal density,38,39 three RNA–protein complexes were expected to be present in the asymmetric unit.
The structure was determined by molecular replacement with PDB 4BW0 as the search model using the program PHASER.40 Ramachandran analysis shows that 99.8% of amino acid residues are in the most favored and additionally allowed regions. Model geometry and the fit to electron-density maps were monitored with MOLPROBITY41 and the validation tools in COOT.
This self-complementary sequence forms the structure shown in Fig. 4, containing six Kt-7 motifs. A solution of 1 mM RNA in 5 mM Tris·HCl (pH 8.0) and 100 mM NaCl was heated to 95 °C for 1 min. The solution was slowly cooled to 20 °C and MgCl2 was added to a final concentration of 10 mM. The hanging-drop vapor diffusion method was used for crystallization. A volume of 1.0 μL of RNA was mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with well solution comprising 3.5 M Na formate, 0.1 M Na acetate (pH 4.6) at 20 °C. Crystals (approximate dimensions 100 × 40 × 40 μm) with space group P6322 grew in about 10 days. Crystals were briefly washed in well solution supplemented with 30% glycerol. The crystals were flash frozen by mounting in nylon loops and plunging into liquid nitrogen. A 2.75 Å resolution dataset was collected on beamline ID23-1 of European Synchrotron Radiation Facility.
1 with well solution comprising 3.5 M Na formate, 0.1 M Na acetate (pH 4.6) at 20 °C. Crystals (approximate dimensions 100 × 40 × 40 μm) with space group P6322 grew in about 10 days. Crystals were briefly washed in well solution supplemented with 30% glycerol. The crystals were flash frozen by mounting in nylon loops and plunging into liquid nitrogen. A 2.75 Å resolution dataset was collected on beamline ID23-1 of European Synchrotron Radiation Facility.
The resolution cutoff for the data was determined by examining both CC1/2 and difference map of the magnesium ions, as described previously.42,43
The crystals had space group P6322 and unit cell dimensions a = 70.3 Å, b = 70.3 Å, c = 47.8 Å. From the crystal density,38,39 only one sixth of the duplex was expected to be present in the asymmetric unit.
The structure was determined by molecular replacement with H. marismortui Kt-7 (PDB 4CS1) as the search model using the program PHASER.40
Structural models were built in Coot44 and RCrane.45 The structure was refined with Refmac5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 46 from the CCP4 suite of programs47 and Phenix refine.48 Model geometry and the fit to electron-density maps were monitored with MOLPROBITY41 and the validation tools in COOT.
46 from the CCP4 suite of programs47 and Phenix refine.48 Model geometry and the fit to electron-density maps were monitored with MOLPROBITY41 and the validation tools in COOT.
Atomic coordinates and structure factor amplitudes have been deposited with the PDB with accession codes 5G4U, 5G4V and 5G4T.
Conflict of Interest
The authors declare no competing financial interests.Acknowledgements
We thank our colleagues for discussion, Diamond and ESRF for access to synchrotron beamlines, CRUK for program support and the Wellcome Trust for the in-house diffractometer.References
- D. J. Klein, T. M. Schmeing, P. B. Moore and T. A. Steitz, The kink-turn: a new RNA secondary structure motif, EMBO J., 2001, 20, 4214–4221 CrossRef CAS PubMed.
- T. A. Goody, S. E. Melcher, D. G. Norman and D. M. J. Lilley, The kink-turn motif in RNA is dimorphic, and metal ion dependent, RNA, 2004, 10, 254–264 CrossRef CAS PubMed.
- N. Ban, P. Nissen, J. Hansen, P. B. Moore and T. A. Steitz, The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution, Science, 2000, 289, 905–920 CrossRef CAS PubMed.
- B. S. Schuwirth, M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila-Sanjurjo, J. M. Holton and J. H. Cate, Structures of the bacterial ribosome at 3.5 A resolution, Science, 2005, 310, 827–834 CrossRef CAS PubMed.
- G. Blaha, G. Gurel, S. J. Schroeder, P. B. Moore and T. A. Steitz, Mutations outside the anisomycin-binding site can make ribosomes drug-resistant, J. Mol. Biol., 2008, 379, 505–519 CrossRef CAS PubMed.
- A. Ben-Shem, N. Garreau de Loubresse, S. Melnikov, L. Jenner, G. Yusupova and M. Yusupov, The structure of the eukaryotic ribosome at 3.0 A resolution, Science, 2011, 334, 1524–1529 CrossRef CAS PubMed.
- W. C. Winkler, F. J. Grundy, B. A. Murphy and T. M. Henkin, The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs, RNA, 2001, 7, 1165–1172 CrossRef CAS PubMed.
- R. K. Montange and R. T. Batey, Structure of the S-adenosylmethionine riboswitch regulatory mRNA element, Nature, 2006, 441, 1172–1175 CrossRef CAS PubMed.
- S. Thore, M. Leibundgut and N. Ban, Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand, Science, 2006, 312, 1208–1211 CrossRef CAS PubMed.
- S. Blouin and D. A. Lafontaine, A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control, RNA, 2007, 13, 1256–12567 CrossRef CAS PubMed.
- B. Heppell and D. A. Lafontaine, Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot, Biochemistry, 2008, 47, 1490–1499 CrossRef CAS PubMed.
- K. D. Smith, S. V. Lipchock, T. D. Ames, J. Wang, R. R. Breaker and S. A. Strobel, Structural basis of ligand binding by a c-di-GMP riboswitch, Nat. Struct. Mol. Biol., 2009, 16, 1218–1223 CAS.
- A. Peselis and A. Serganov, Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch, Nat. Struct. Mol. Biol., 2012, 19, 1182–1184 CAS.
- J. Zhang and A. R. Ferre-D'Amare, Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA, Nature, 2013, 500, 363–366 CrossRef CAS PubMed.
- T. Moore, Y. Zhang, M. O. Fenley and H. Li, Molecular basis of box C/D RNA-protein Interactions; Cocrystal structure of archaeal L7Ae and a box C/D RNA, Structure, 2004, 12, 807–818 CrossRef CAS PubMed.
- T. Hamma and A. R. Ferré-D'Amaré, Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution, Structure, 2004, 12, 893–903 CrossRef CAS PubMed.
- L. B. Szewczak, J. S. Gabrielsen, S. J. Degregorio, S. A. Strobel and J. A. Steitz, Molecular basis for RNA kink-turn recognition by the h15.5 K small RNP protein, RNA, 2005, 11, 1407–1419 CrossRef CAS PubMed.
- O. A. Youssef, R. M. Terns and M. P. Terns, Dynamic interactions within sub-complexes of the H/ACA pseudouridylation guide RNP, Nucleic Acids Res., 2007, 35, 6196–6206 CrossRef CAS PubMed.
- P. Daldrop and D. M. J. Lilley, The plasticity of a structural motif in RNA: structural polymorphism of a kink turn as a function of its environment, RNA, 2013, 19, 357–364 CrossRef CAS PubMed.
- P. Nissen, J. A. Ippolito, N. Ban, P. B. Moore and T. A. Steitz, RNA tertiary interactions in the large ribosomal subunit: The A-minor motif, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 4899–4903 CrossRef CAS PubMed.
- A. Lescoute, N. B. Leontis, C. Massire and E. Westhof, Recurrent structural RNA motifs, isostericity matrices and sequence alignments, Nucleic Acids Res., 2005, 33, 2395–2409 CrossRef CAS PubMed.
- J. Liu and D. M. Lilley, The role of specific 2′-hydroxyl groups in the stabilization of the folded conformation of kink-turn RNA, RNA, 2007, 13, 200–210 CrossRef CAS PubMed.
- K. Reblova, J. E. Sponer, N. Spackova, I. Besseova and J. Sponer, A-minor tertiary interactions in RNA kink-turns. Molecular dynamics and quantum chemical analysis, J. Phys. Chem. B, 2011, 115, 13897–13910 CrossRef CAS PubMed.
- S. M. Dibrov, J. McLean, J. Parsons and T. Hermann, Self-assembling RNA square, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6405–6408 CrossRef CAS PubMed.
- M. A. Boerneke, S. M. Dibrov and T. Hermann, Crystal-structure-guided design of self-assembling RNA nanotriangles, Angew. Chem., 2016, 55, 4097–4100 CrossRef CAS PubMed.
- H. Ohno and T. Inoue, Designed regular tetragon-shaped RNA-protein complexes with ribosomal protein L1 for bionanotechnology and synthetic biology, ACS Nano, 2015, 9, 4950–4956 CrossRef CAS PubMed.
- H. Ohno, T. Kobayashi, R. Kabata, K. Endo, T. Iwasa, S. H. Yoshimura, K. Takeyasu, T. Inoue and H. Saito, Synthetic RNA-protein complex shaped like an equilateral triangle, Nat. Nanotechnol., 2011, 6, 116–120 CrossRef CAS PubMed.
- H. Ohno, E. Osada and H. Saito, Design, assembly, and evaluation of RNA-protein nanostructures, Methods Molec. Biol., 2015, 1297, 197–211 Search PubMed.
- L. Huang, J. Wang and D. M. J. Lilley, A critical base pair in k-turns determines the conformational class adopted, and correlates with biological function, Nucleic Acids Res., 2016, 44, 5390–5398 CrossRef PubMed.
- S. A. McPhee, L. Huang and D. M. Lilley, A critical base pair in k-turns that confers folding characteristics and correlates with biological function, Nat. Commun., 2014, 5, 5127 CrossRef CAS PubMed.
- B. Turner, S. E. Melcher, T. J. Wilson, D. G. Norman and D. M. J. Lilley, Induced fit of RNA on binding the L7Ae protein to the kink-turn motif, RNA, 2005, 11, 1192–1200 CrossRef CAS PubMed.
- B. Turner and D. M. J. Lilley, The importance of G.A hydrogen bonding in the metal ion- and protein-induced folding of a kink turn RNA, J. Mol. Biol., 2008, 381, 431–442 CrossRef CAS PubMed.
- J. Wang, T. Fessl, K. T. Schroeder, J. Ouellet, Y. Liu, A. D. Freeman and D. M. J. Lilley, Single-molecule observation of the induction of k-turn RNA structure on binding L7Ae protein, Biophys. J., 2012, 103, 2541–2548 CrossRef CAS PubMed.
- L. Huang and D. M. Lilley, The molecular recognition of kink-turn structure by the L7Ae class of proteins, RNA, 2013, 19, 1703–1710 CrossRef CAS PubMed.
- S. L. Beaucage and M. H. Caruthers, Deoxynucleoside phosphoramidites - a new class of key intermediates for deoxypolynucleotide synthesis, Tetrahedron Lett., 1981, 22, 1859–1862 CrossRef CAS.
- T. J. Wilson, Z.-Y. Zhao, K. Maxwell, L. Kontogiannis and D. M. J. Lilley, Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme, Biochemistry, 2001, 40, 2291–2302 CrossRef CAS PubMed.
- L. Huang, P. Yin, X. Zhu, Y. Zhang and K. Ye, Crystal structure and centromere binding of the plasmid segregation protein ParB from pCXC100, Nucleic Acids Res., 2011, 39, 2954–2968 CrossRef CAS PubMed.
- B. W. Mathews, Solvent content of protein crystals, J. Mol. Biol., 1968, 33, 491–497 CrossRef.
- K. A. Kantardjieff and B. Rupp, Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals, Protein Sci., 2003, 12, 1865–1871 CrossRef CAS PubMed.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, Phaser crystallographic software, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS PubMed.
- V. B. Chen, W. B. Arendall, 3rd, J. J. Headd, D. A. Keedy, R. M. Immormino, G. J. Kapral, L. W. Murray, J. S. Richardson and D. C. Richardson, MolProbity: all-atom structure validation for macromolecular crystallography, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 12–21 CrossRef CAS PubMed.
- P. A. Karplus and K. Diederichs, Linking crystallographic model and data quality, Science, 2012, 336, 1030–1033 CrossRef CAS PubMed.
- Z. Luo, K. Rajashankar and Z. Dauter, Weak data do not make a free lunch, only a cheap meal, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2014, 70, 253–260 CAS.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Features and development of Coot, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501 CrossRef CAS PubMed.
- K. S. Keating and A. M. Pyle, RCrane: semi-automated RNA model building, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 985–995 CAS.
- G. N. Murshudov, P. Skubak, A. A. Lebedev, N. S. Pannu, R. A. Steiner, R. A. Nicholls, M. D. Winn, F. Long and A. A. Vagin, REFMAC5 for the refinement of macromolecular crystal structures, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 355–367 CrossRef CAS PubMed.
- M. D. Winn, C. C. Ballard, K. D. Cowtan, E. J. Dodson, P. Emsley, P. R. Evans, R. M. Keegan, E. B. Krissinel, A. G. Leslie and A. McCoy, et al., Overview of the CCP4 suite and current developments, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 235–242 CrossRef CAS PubMed.
- P. D. Adams, P. V. Afonine, G. Bunkoczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral and R. W. Grosse-Kunstleve, et al., PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 213–221 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: PDF file comprising eight figures and three tables of data. See DOI: 10.1039/c6nr05186c |
| This journal is © The Royal Society of Chemistry 2016 |
