Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Folate-based single cell screening using surface enhanced Raman microimaging†
C.
Fasolato
ab,
S.
Giantulli
c,
I.
Silvestri
c,
F.
Mazzarda
a,
Y.
Toumia
d,
F.
Ripanti
a,
F.
Mura
e,
F.
Luongo
a,
F.
Costantini
e,
F.
Bordi
af,
P.
Postorino
a and
F.
Domenici
*ad
aDipartimento di Fisica, Università Sapienza, P.le Aldo Moro 5, Rome, Italy. E-mail: fabiodomenici@gmail.com; Fax: +39064463158/+390672594328; Tel: +390649913403/+390672594454
bCenter for Life Nanoscience, Istituto Italiano di Tecnologia, V.le Regina Elena 291, Rome, Italy
cDipartimento di Medicina Molecolare, Università Sapienza, P.le Aldo Moro 5, Rome, Italy
dDipartimento di Scienze e Tecnologie Chimiche, Università di Roma Tor Vergata, Via della Ricerca Scientifica, Rome, Italy
eDipartimento di Chimica, Università Sapienza, P.le Aldo Moro 5, Rome, Italy
fCNR-ISC UOS Roma, Sapienza Università di Roma, P.le A. Moro 5, 00185 Roma, Italy
First published on 5th September 2016
Abstract
Recent progress in nanotechnology and its application to biomedical settings have generated great advantages in dealing with early cancer diagnosis. The identification of the specific properties of cancer cells, such as the expression of particular plasma membrane molecular receptors, has become crucial in revealing the presence and in assessing the stage of development of the disease. Here we report a single cell screening approach based on Surface Enhanced Raman Scattering (SERS) microimaging. We fabricated a SERS-labelled nanovector based on the biofunctionalization of gold nanoparticles with folic acid. After treating the cells with the nanovector, we were able to distinguish three different cell populations from different cell lines (cancer HeLa and PC-3, and normal HaCaT lines), suitably chosen for their different expressions of folate binding proteins. The nanovector, indeed, binds much more efficiently on cancer cell lines than on normal ones, resulting in a higher SERS signal measured on cancer cells. These results pave the way for applications in single cell diagnostics and, potentially, in theranostics.
Introduction
Over the last few years, much interest in the practice of oncology has been paid to early cancer detection, i.e. the capability to reveal the presence of cancer at the first stage of its progression, which could open the way to more effective treatments.1 For this reason, a growing need has been expressed for the development of highly sensitive and specific detection methodologies. The current challenge is indeed to reveal the presence of extremely few cancer cells in small, stable biological samples, granting low invasiveness, sensing reproducibility and high versatility with respect to the different tumor types and metastatic spread.2,3 However, because of tumor heterogeneity, cancer cells can often elude even specific analyses: for this reason, the implementation of novel detection methodologies4,5 can be crucial to address effective diagnosis.6In cellular diagnostics, screening consists of discriminating cancer and normal cells either by recognizing specific cancer cell characteristics, e.g. morphological7 or mechanical8 properties, or by selectively targeting cancer cells with traceable markers, such as fluorescent or spectroscopically active molecules.4,9 This traceability can be exploited for cell sorting, for example in flow-cytometry assays.10,11 These methods rely on the differential bioaffinity of the cell for specific molecules, which can be related to a higher density of certain receptors on cancer cell membranes.9 They have already been exploited for diagnostics, by targeting the receptors with objects functionalized with specific proteins or antibodies.12,13
Nevertheless, such macromolecules often present stability issues, being highly sensitive to temperature, pH and other environmental conditions, and often tend to form assemblies with other proteins in human serum,14 inhibiting both sensitivity and specificity of the biorecognition.15 Small and more stable molecules, such as folic acid (FA), represent a promising alternative to proteins.16 FA binding proteins are, indeed, differently expressed depending on the cell type.17 Cancer cells tend to express a large number of FA-receptors owing to the pivotal role of FA in the cell metabolism and replication process17 whereas there is a relatively low expression level in healthy cells.16,18,19
The folate interaction with cells is mediated by two widely expressed facilitative transporters, solute carrier (SLC) proteins, that are the reduced folate carrier (RFC) and the proton-coupled folate transporter (PCFT), and via a family of high affinity folate receptors (FRs), that transport folate via endocytosis.20 There are three isoforms of folate receptors: among them, FRα is highly expressed in specific malignant tumors of epithelial origin (for example, it is strongly expressed in cervical uterine and ovarian carcinomas, while it is less frequent in prostate carcinoma) when compared to normal cells.21 The basic idea is therefore to identify a suitable technique to highlight the different densities or types of FA receptors that can be found on different cells.
In this regard, Surface Enhanced Raman Scattering (SERS) methodology22,23 offers a great opportunity towards a simplified, ultrasensitive detection of biomolecules involved in a specific cellular interaction.24,25 The SERS effect is based on the localized plasmon resonance in metallic nanostructures: the collective excitation of electrons in a nanoparticle (Np) illuminated by light in the visible/infrared spectral range increases the intensity of the electromagnetic field at the Np surface, causing a remarkable enhancement of the Raman signal of the molecules close to the Np.26 Gold nanocolloids have been widely used to design biosensors for tumor markers,25 based on their peculiar properties, such as a high surface-to-volume ratio, which allows the possibility of suitable biomolecular conjugation,27,28 desirable chemical stability29 and relatively low cytotoxicity.30 Exploiting their signal enhancement capabilities, suitably functionalized Nps31,32 can thus be used to highlight the differences among the cells in terms of density and types of specific receptors,33 providing the basis for cancer detection at the single cell level.34–36
Here we report a novel approach that combines the high sensitivity of SERS with the selective recognition of FA operated by several types of cells through the different expression of their molecular receptors.37,38 By treating the cells with a folate-based SERS-active nanovector, our idea is to distinguish the different cell populations based on the measurement of the overall SERS signal intensity from a single cell. We thus realized a SERS active nanovector, employing gold Nps, covered with the Raman active bifunctional linker 4-aminothiophenol (4ATP)39–43 and further functionalized with FA.44 We performed a thorough analysis of the overall SERS response on different treated cell types. The treatment with the folate nanovector, under physiological conditions, was carried out on three different cell lines known for showing different folate receptor expressions, in terms of both density and types.45–47 Two of them are well studied and stable tumor models: PC-3 (prostatic cancer cells) and HeLa (cervical cancer cells), the latter exhibiting higher expression of FRα.20,47–49 For a comparative study we chose HaCaT immortalized keratinocytes, previously used as a control cell line in fluorescence-based experiments on bioaffinity with folate.33 It is noteworthy that these experiments were unable to detect folate nanocarriers bound to HaCaT biorecognized receptors, possibly because of the low density of folate binding proteins, as recently demonstrated from the PCFT gene expression.50 On the contrary, in the present experiment, we were able to detect nanovectors on the HaCaT membrane, demonstrating the higher sensitivity of SERS imaging.
A direct comparison of the SERS intensity from single cells clearly discriminates between normal (HaCaT) and cancer (PC-3 and HeLa) cell lines. Moreover, a statistical analysis of the SERS signal finally proves that the present approach, without affecting cell viability, permits one to distinguish the PC-3 from the HeLa cancer line, owing to a more efficient interaction of the nanovector with the latter.
Experimental
Nanovector preparation
All the preparation steps were conducted in water, ensuring a good biocompatibility of the nanocolloidal dispersion. The fabrication was successfully monitored at each step with different techniques: UV-visible (UV-Vis) absorption, infrared (IR) spectroscopy, dynamic light scattering (DLS) and Z potential and atomic force microscopy (AFM). Citrate stabilized, gold Nps of 60 nm diameter in aqueous solution were purchased from Ted Pella (Redding, CA). All the other chemicals were purchased from Sigma-Aldrich and used without further purification. The functionalization of Nps with 4ATP via the S–Au bond took place in the Np aqueous solution after the addition of a small amount of ethanol solution of 4ATP (1% in volume at 5 mg mL−1 concentration).51 The kinetics of the chemical reaction were followed via UV-visible absorption measurements, which confirmed that the reaction was complete after 3 hours of incubation. Residual 4ATP molecules not bound to the Nps were removed by dialysis for 24 hours as described by Fasolato et al.51 Afterwards, folic acid carboxylic groups were activated with the subsequent action of 1-ethyl-3-[dimethylaminopropyl]carbodiimidehydrochloride (EDC) and N-hydroxy-succinimide (NHS)44,52 (see also section S1 of the ESI†) in order to allow the covalent functionalization of the Nps with 4ATP molecules. It is important to stress that the 4ATP-folate binding reaction does not occur with a stoichiometric ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, due to the stereochemical hindrance of folic acid. Nevertheless, it is an oriented binding which ensures the conservation of folate biofunctionality, thanks to the molecular structure: the covalent bond involves indeed one of the two carboxylic groups, and leaves the pteroyl active part exposed and available for the interaction with cellular receptors53 (see Fig. S1.4 in the ESI†).
1, due to the stereochemical hindrance of folic acid. Nevertheless, it is an oriented binding which ensures the conservation of folate biofunctionality, thanks to the molecular structure: the covalent bond involves indeed one of the two carboxylic groups, and leaves the pteroyl active part exposed and available for the interaction with cellular receptors53 (see Fig. S1.4 in the ESI†).
UV-visible absorption measurements
The stepwise molecular assembly of FA–(4ATP–Np) has been checked by monitoring the resonant absorption bands due to the collective oscillations of the conductive electrons of gold Nps, called Localized Surface Plasmons (LSPs). Absorbance measurements were performed at room temperature (RT) by using a double beam Jasco V-570 spectrophotometer, set with a 0.5 nm bandwidth.Dynamic light scattering
The size of the Nps was characterized by means of dynamic light scattering (DLS) measurements using a Malvern NanoZetaSizer apparatus equipped with a 5 mW HeNe laser (Malvern Instruments Ltd, UK). This system uses quasi-backscatter detection, i.e. the scattered light is collected at an angle of 173°, the backscattering being less sensitive to multiple scattering effects. In order to obtain the size distributions, the measured autocorrelation functions were analyzed by using the CONTIN algorithm. Decay times are used to determine the distribution of the diffusion coefficients D of the particles, which in turn are converted to a distribution of apparent hydrodynamic radii RH using the Stokes Einstein relationship RH = KBT/6pηD, where KBT is the thermal energy and η the solvent viscosity. The reported radius values are the average of several measurements and are obtained from intensity-weighted distributions.Z potential measurements
The Z potential of the suspended Nps was measured in water and in the cell physiological medium at 25.0 °C temperature (0.1 °C, T accuracy), adopting the Phase Analysis Light Scattering (PALS) technique of a Malvern NanoZetaSizer apparatus (Malvern Instruments Ltd, UK). This enables the measurement of the electrophoretic mobility, and from this the Z potential, which we provided herein in the Helmholtz/Smoluchowski approximation. The measurements were performed employing a palladium electrode dip cell ZEN 1002 (Malvern, UK). The runs were set up in triplicate, each consisting of at least 30 sub-runs.Infrared absorption
The infrared characterization was performed at RT on 150 × 150 μm2 microsized regions using a JASCO Irtron IRT-30 Fourier Transform infrared (FTIR) microscope, equipped with a nitrogen cooled MCT detector, a Cassegrain objective 16×. The acquisition was set up in transmission mode, 512 scans, and at a spectral resolution of 4 cm−1. The microscope is coupled with a FTIR/410 Jasco spectrometer equipped with a conductive ceramic coil mounted in a water-cooled copper jacket source and a KBr beamsplitter. The optical path was purged continuously with gaseous nitrogen. For the microFTIR measurements, the Np solution was dried at 37 °C on CaF2 substrates.Raman/SERS
Raman measurements were performed employing a Horiba HR-Evolution microspectrometer, equipped with a 25 mW He–Ne laser (632.8 nm wavelength) and a set of neutral power attenuating filters. The spectrometer is coupled with a confocal microscope equipped with a set of objectives at different magnifications (50× – 0.50 NA and 100× – 0.8 NA were used for this experiment). A 600 lines per mm diffraction grating ensured a spectral resolution better than 3 cm−1. The sample can be moved with a software-controlled mapping stage with a sub-micrometric precision for collecting automatized spectroscopic mapping. Exploiting the high confocality, micrometric sampling along the optical axes can be achieved. The data reported in Fig. 6 were acquired by varying the focus along the vertical axis using a 50 μm confocal hole aperture. All the spectra are here presented after a polynomial (3rd degree) baseline subtraction performed using LabSpec software.Cell proliferation assay
All cell lines used in our experiments were obtained from Interlab Cell Line Collection (ICLC) (Istituto Nazionale per la Ricerca sul Cancro, Genoa, Italy). HaCat, a human normal keratinocyte cell line, HeLa, a human cervical cancer cell line, and PC-3, an androgen-independent human prostate cancer cell line were grown in DMEM (Euroclone, Life Science Division, GB, Pero, Italy). All media were supplemented with 100 U mL−1 penicillin, 100 μg mL−1 250 streptomycin and 10% fetal bovine serum (Euroclone). Cells were maintained in a tissue culture incubator at 37 °C, 5% CO2. The experiments were performed by plating the cells at a density of 5 × 103 mL−1 in 96-well flat plates in the culture medium and then incubated at 37 °C under 5% CO2. Following 24 hours, different concentrations of FA–4ATP–Nps, ranging from 1 pM to 2 pM, were added in triplicate. We performed different time courses: a 2 hours pulse and 24 hours of treatment. The cells were then incubated with MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium) (Sigma-Aldrich, Saint Louis, Missouri, USA) for 4 hours. The formed crystals were dissolved in 100 μL of DMSO and further incubated for 15 minutes. The absorbance was measured in a plate reader spectrophotometer (Labsystem Multiskan MS), using a test wavelength of 540 nm and a reference wavelength of 690 nm. The cells incubated with culture medium alone represented the controls, and wells containing the medium alone served as blanks. All the results were analyzed by ANOVA. The significance was evaluated by the Tukey honestly significant difference (HSD) post hoc test and data were expressed as means ± standard deviation (SD) of independent samplings from different experiments. The level of significance was established at p value <0.05.Results and discussion
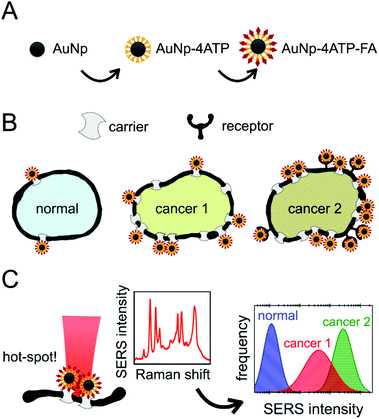
The steps of this experiment are summarized in Fig. 1. Herein, we will firstly describe the stepwise preparation of the nanovector (A): a careful characterization was performed after each step and at the end of the biofunctionalization. We will then illustrate the results obtained with the treatment of the three cell populations (B), and the monitoring of the SERS intensity that allowed the cell recognition (C).Nanovector preparation & characterization
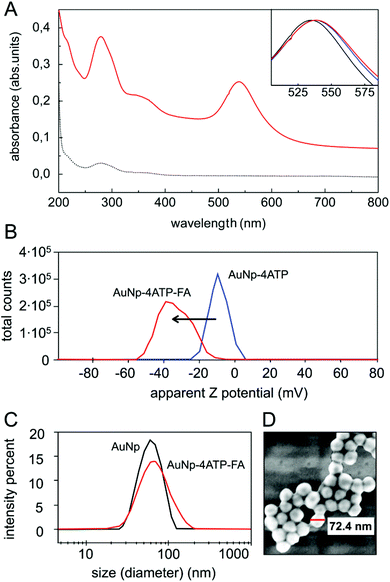
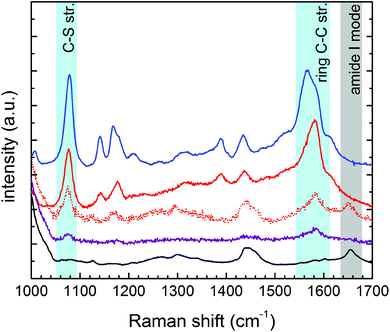
The core of our nanovector consists of gold Nps of 60 nm diameter, which were functionalized with a two-step process, as described in the previous section. The results of UV-visible spectroscopy are shown in Fig. 2(A). Note the presence of folate absorption bands and the shift of the Np plasmonic peak, which is related to the change in the dielectric environment at the Np surface. The colloidal features of the obtained folate nanovector were tested via both Z potential and dynamic light scattering (DLS) measurements. As shown in Fig. 2(B), upon conjugation with folic acid, the Np surface charge reached a high negative value (−38 mV) which prevented the Np aggregation through electrostatic repulsion. This was also confirmed by DLS measurements, which provided a size distribution centred around 70 nm diameter (Fig. 2(C)), in agreement with the estimate obtained by SEM imaging (see Fig. 2(D)). Similar results imply the stability of the nanovector also in cell culture medium (see section S2.1 of the ESI†).The spectroscopic characterization was completed with Fourier transform infrared absorption measurements. The extra peak in the spectrum of the complete nanovector (Fig. 3(C)), when compared to the spectrum of 4ATP functionalized Nps (Fig. 3(B)), can be related to the presence of the folate shell bound to the Nps (Fig. 3(A)). Indeed, the IR absorption band centered around 1700 cm−1 is assigned to the carbonyl group (–CONH– in the case of FA–4ATP–Np complex).54 Slight intensity modification and frequency shifts in the nanovector spectrum with respect to the bulk are ascribed to the gold Np surface enhanced infrared absorption effect.55 Titration experiments using the Coomassie Brilliant Blue (CBB) dye allowed for a quantitative estimation of the number of folate molecules bound to the single Np.56 The result shows an average number of 1900 ± 200 folate molecules (see section S3 of the ESI†). This is in good agreement with the approximated, theoretical estimate (∼2000 molecules) formulated by considering folate stereochemical hindrance (around 2000 molecules).
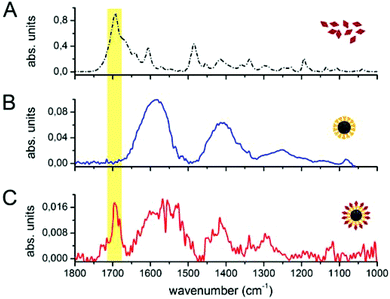 | ||
| Fig. 3 FTIR measurements on bulk folic acid (A), 4ATP–Nps (B) and FA–4ATP–Nps (C). The yellow region highlights the carbonyl group band of folic acid. | ||
SERS features of the nanovector
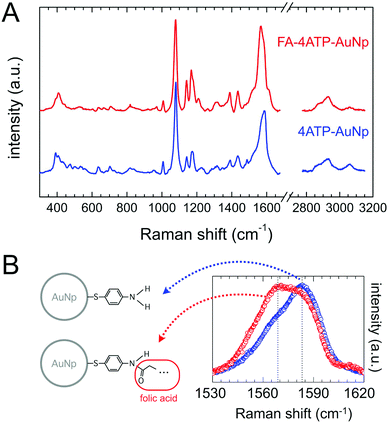
The typical SERS spectrum of the nanovector is shown in Fig. 4(A): the spectroscopic fingerprint is highly reproducible and can be mainly ascribed to the Raman reporter 4ATP.57,58 This can be due to several reasons, among which the higher distance of folate molecules from the Np surface, the low Raman cross section of folic acid (see Fig. S2 in the ESI†), and the fact that, according to our estimate, the covalent attachment between FA and 4ATP onto the Np surface occurred with a stoichiometric ratio of around 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 (section S3 of the ESI†). Nevertheless, weak but clear and reproducible modifications of the 4ATP SERS spectrum upon functionalization witness the occurrence of folic acid binding. Indeed, according to the literature, a few cm−1 frequency shift and/or a slight change in the line-shape of some peaks can be reliable in witnessing chemical conjugation in the case of 4ATP.59,60 In particular, the phenyl ring C–C stretching Raman band results in a strong intense SERS signal observed around 1580 cm−1 and susceptible to be influenced by the type of chemical substituent and its electronic properties.41,42,61–63 Considering this, we performed a comparative high-resolution spectral analysis on this band with respect to folate biofunctionalization. The average spectra corresponding to 4ATP–Np and FA–4ATP–Np are summarized in Fig. 4(B). There is a strong change in the relative weight of the two components of the band, whose central frequencies are highlighted in the spectrum of Fig. 4(B), in favor of the low frequency component for the biofunctionalized sample (see section S4.2 of the ESI† for details), the latter suggesting that some of the NH2 substituents of 4ATP were actually modified according to section S1 of the ESI.†
6 (section S3 of the ESI†). Nevertheless, weak but clear and reproducible modifications of the 4ATP SERS spectrum upon functionalization witness the occurrence of folic acid binding. Indeed, according to the literature, a few cm−1 frequency shift and/or a slight change in the line-shape of some peaks can be reliable in witnessing chemical conjugation in the case of 4ATP.59,60 In particular, the phenyl ring C–C stretching Raman band results in a strong intense SERS signal observed around 1580 cm−1 and susceptible to be influenced by the type of chemical substituent and its electronic properties.41,42,61–63 Considering this, we performed a comparative high-resolution spectral analysis on this band with respect to folate biofunctionalization. The average spectra corresponding to 4ATP–Np and FA–4ATP–Np are summarized in Fig. 4(B). There is a strong change in the relative weight of the two components of the band, whose central frequencies are highlighted in the spectrum of Fig. 4(B), in favor of the low frequency component for the biofunctionalized sample (see section S4.2 of the ESI† for details), the latter suggesting that some of the NH2 substituents of 4ATP were actually modified according to section S1 of the ESI.†
Overall, the characterization outcome (by UV-Vis, Z potential, IR, CBB titration assay) coherently points out the presence of folate in the functionalized Np sample, even after a long dialysis, suggesting a tight binding between folate and 4ATP. This is well evidenced also by SERS spectra: a change in the band shape of the phenyl ring C–C vibration can be ascribed to a significant change in the charge delocalization of the aromatic ring of 4ATP due to the occurrence of a typical mesomeric effect63,64 (see section S4.2 and Fig. S4.4 of the ESI† for a more detailed discussion). Note that different biofunctionalization procedures (e.g. diazotization reaction28) were here avoided, with the folate derivatization involved in its pteroyl active part.53
Cell treatment and SERS investigation
The cell treatment consisted of incubating the nanovector aqueous solution in the live cell culture for 2 hours. Nps unbound to the cell receptors were washed away with PBS after incubation. MTT viability tests confirmed the negligible cytotoxicity induced by the nanovector, which was detected for 24 hours of incubation (see section S7 in the ESI†).According to the Experimental section, the cellular samples were stabilized for Raman/SERS measurements by drying them under a laminar flow at a controlled temperature of 37 °C, without any further chemical fixing treatment. All cell lines, treated and not treated, were measured with Raman spectroscopy. Some representative spectra obtained on the PC-3 cancer cells and on the HaCaT normal cells in the region between 1000 and 1700 cm−1 are shown in Fig. 5 (see section S3 in the ESI† for assignment). The spectrum of an untreated cell is then compared with the spectra collected on the PC-3 treated cells (low and high SERS intensity) and on the HaCaT normal cells. The reference spectrum of the nanovector is also presented. It is straightforward to ascribe the strong bands revealed on treated cells, and in particular the ones evidenced in light blue, to the presence of the nanovector. In the case of low SERS signals, it is still possible to distinguish the cell signal, not masked by SERS peaks (see the grey region).
These spectral features thus allow one to clearly identify the signal arising from the nanovector in the spectrum collected from each cell. As a first qualitative comparison, we noticed that the nanovector signal arising from acquisitions performed on the PC-3 and HeLa cells is much bigger than that measured on HaCaT, the latter cells expressing PCFT only.50 This finding confirms the original idea about the possibility of developing a spectroscopic procedure to discriminate cancer and normal cells, based on their different FA receptor expression levels. Before discussing in detail the SERS-based protocol we developed for single cell screening (see the next subsection), we want to stress that these spectroscopic measurements allow for a detailed investigation of the nanovector–cell interaction. In particular, Raman imaging allowed us to understand where and how the nanoparticles bind to the cells. Indeed, by exploiting the high confocality of our microspectrometer we were able to optimize the spatial resolution along the optical axes to a few microns, collecting the signal from a very thin focus region. Combining SERS with the micrometric resolution either along the vertical axes or over the sample surface allows investigating the distribution of the Nps all over the cell surface. The representative results for a PC-3 cell are shown in Fig. 6. The very good correspondence between the optical image and the cell Raman map (integrated intensity of the lipid CH2–CH3 stretching peaks vs. horizontal position) in Fig. 6(D) makes us confident to fully exploit spectroscopic imaging. SERS intensity maps (integrated intensity of a nanovector specific peak vs. horizontal position) shown in Fig. 6(A) are compared with optical images (Fig. 6(B)). Maps and relative images are collected moving the focus plane of a few microns along the optical axes as shown by the schematic in panel (C), where a detailed stratigraphic analysis of the cell is also shown. The integrated intensities (normalized) of the nanovector-, cell- and substrate-specific peaks vs. vertical position z are shown in the graphic in Fig. 6(C). The maximum of these three signals is found at different vertical positions, being that of the nanovector located ∼1 μm above that of the cell, which in turn is located ∼2 μm above that of the substrate. This vertical arrangement suggests that the nanovector is located out of the cell but close to the membrane. This picture is in agreement with FESEM imaging (see Fig. S5.4 and experimental details in the ESI†) showing that the great majority of the Nps are clearly visible outside the cell membrane, while only a few of them are apparently located inside the cell.
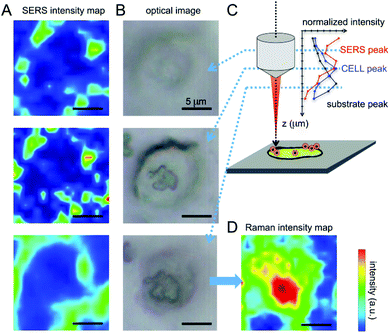 | ||
| Fig. 6 (A) SERS imaging obtained by integrating the C–S stretching peak of 4ATP and plotted in the thermal scale is correlated with the optical image of the cell (B). The measurements were performed varying the focus of the objective at different values of z. Nps seem to be located at the margins of the cell, as pointed out by Boca-Farcau et al.37 (C) Intensity of SERS and Raman peaks relative to the different components of the system acquired varying the objective focus (versus z). (D) The imaging of the cell performed integrating the lipid membrane CH2–CH3 stretching peaks shows a good correspondence with the optical image, giving information on the location of the cell nucleus (double membrane and higher signal). | ||
It is also worth noting that illuminating the Nps with the red beam (633 nm) leads to the sensitivity to the Np density, as the SERS signal coming from “hot-spots” resulting from Np aggregation is considerably more intense than the one coming from a single, isolated Np, because of the changes in the shape of the surface plasmon peak arising from Np aggregation.29 The fact that aggregation takes place in the presence of a dense receptor distribution was confirmed by scanning electron microscopy imaging, which allowed us to monitor the successful binding of Np to the cell membrane (see section S5 in the ESI†).
SERS screening & diagnostics
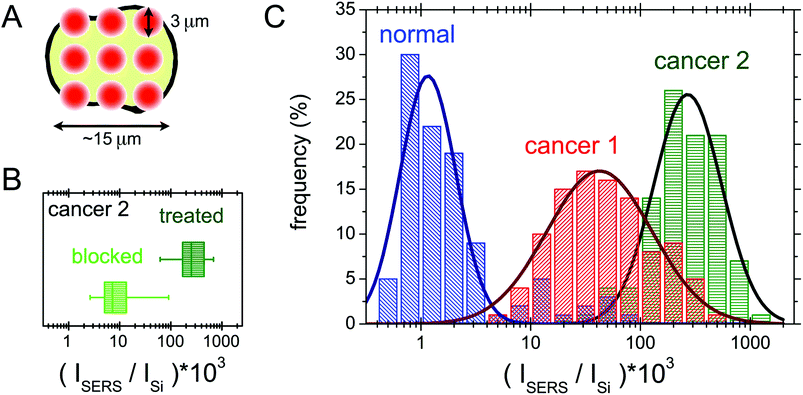
In this section the protocol developed for the SERS biorecognition of single cells is described. Once focused on a cell treated with the SERS active folate nanovector, we carried out a fast, automatized spectroscopic sampling of the cell membrane by collecting 9 spectra all over the cell area (see Fig. 7(A)). In order to obtain a single, simple control parameter from each cell, the integrated intensity of the nanovector-specific SERS band (centred at 1580 cm−1), ISERS, was calculated for all the spectra and its value was normalized to the substrate integrated intensity, Isubstrate, to account for slightly different experimental conditions from cell to cell. Further details of this procedure are given in section S6 of the ESI.† Values obtained for the control parameter, actually defined by C = 1000 × ISERS/Isubstrate, confirmed the qualitative finding mentioned above, that is the selective targeting of the different cell populations induced by folate specific biorecognition. The average control parameter value indeed systematically increases going from HaCaT (non-cancer, 〈C〉 = 5) to PC-3 (cancer, 〈C〉 = 75) to HeLa (cancer, with the highest overexpression of FA receptors, 〈C〉 = 196).A blocking experiment was also performed, to be sure that the above results are really induced by folate specific biorecognition. To this aim, HeLa cells were incubated with the nanovector, in the presence of a high concentration of unbound folate molecules in solution. After performing the usual protocol (see Experimental for details), a population of 100 cells from the ordinarily treated sample was screened and compared with a 100-cell population of the blocking experiment sample. The obtained results are compared in the box plot shown in Fig. 7(B). As it is shown, there is a neat decrease of the signal on folate-blocked cells, indicating a competition between the free folate molecules and folate functionalized nanoparticles for the molecular binding sites on the cell membrane, resulting in a lower density of Nps on blocked cells, and therefore in a lower intensity of the overall SERS signal.
The results of SERS screening obtained for three 100-cell populations of HeLa, PC-3 and HaCaT cells are reported in a histogram in Fig. 7(C). The occurrence frequencies of different intervals of the parameter C are evident. The logarithmic binning was chosen because of the wide range of C values measured. The data for the three cell populations follow the same statistical behaviour, which we chose to model as a lognormal distribution,65 but with largely different parameters. The optimal fitting lognormal curves are plotted in a solid line over the histograms. Differences can be noticed among the three populations: in the case of the HaCaT line, a distribution peaked around small values of C reflects the very low density of FA binding proteins on the surface of normal cells. Moreover, the two populations of cancer cells can be distinguished, with the expected value of the PC-3 control parameter being quite lower than the one in the case of HeLa cells.
A semi-quantitative interpretation of the data in terms of the density and the type of folate binding proteins on the cell membrane is possible.65 Indeed, biological investigation on the HeLa cell line showed an undiscussed overexpression of FRα, in addition to a higher density on the membrane of the folate solute carriers, as RFC and PCFT.47,48 The case of PC-3 is more controversial, as there are different hypotheses on the expression of folate binding proteins on these cells.18,46,47,66 Most probably, the overexpression of the folate solute carriers is the one responsible for the enhanced targeting of the PC-3 cells with respect to the HaCaT population.50 The clearly higher signal on the HeLa cells is in good agreement with the idea of the overexpression of the folate receptor FRα, in addition to the solute carrier overexpression on the PC-3 cells. The statistical interpretation of our data, although it may be improved, paves the way for the implementation of hypothesis testing for new data.
Conclusions
In recent years, the need to develop methodologies for early cancer detection has significantly grown. In this context, the sensitivity and the timing of diagnosis need to be improved in order to lead towards a more effective treatment. In this framework, several folic acid mediated cell targeting strategies have been recently proposed.67 Indeed, the degree of over- and hypo-expression of folate binding proteins on the cell membrane enables the discrimination between normal and cancer cells and, among the latter, it could provide insight into the different stages of the disease (as in the case of metastatic tumors), when those transformations and progress imply a change in the expression level of folate receptors and carriers.68 Nevertheless, these methods, which rely on cellular selective recognition, suffer the absence of analysis methodologies with suitable sensitivity and reproducibility.Surface enhanced Raman microspectroscopy has proved its potentiality in biosensing applications, although the high sensitivity and selectivity of this technique come together with non-negligible reproducibility issues, which have been widely reported.57,69 Despite the advantages in using SERS for biophysical investigations, to the best of our knowledge, SERS based screening of multiple and different cell lines, based on the receptor expression, has not yet been reported.38 For this reason, we focused on the differential expression of membrane folate binding proteins of different cell lines, in order to develop a SERS-biorecognition strategy leading to single cell screening.
To this aim, we assembled a simple SERS active bio-nanoparticle platform where folic acid was interfaced to the plasmonic gold Np coated with the Raman reporter 4ATP. As shown in Fig. 2 and 3, the functionalization of the nanovector was successfully performed in water, ensuring a good biocompatibility of the system, and proved with different experimental techniques. The stability of the nanovector in cell culture medium, resulting from the negative surface charge given by the carboxylic groups (Fig. S1.4 in the ESI†) and witnessed by the size distribution measurements (see Fig. S2.2 in the ESI†), prevents the formation of an adhesion layer of proteins around the nanovector, which could impact on the selectivity, as happens in some cases with protein-based functionalizations.15 The titration experiments allowed one to ascertain that the 4ATP–Nps can be biofunctionalized efficiently with an average number of ∼1900 molecules of FA (see section S3 of the ESI†). The SERS activity of the system was proved with a thorough investigation of the features of the SERS spectrum of the Np–4ATP–folate adduct, as compared to the well-known Np–4TP system,51 and further confirmed the functionalization that occurred.
The experiment of cell biorecognition was performed with the incubation of three different cell lines, chosen for their different levels of expression of folate receptors.20,21 The selective binding of the nanoparticles to the molecular receptors is favoured by the orientation of folic acid molecules on the Np surface, which exposes the heterocyclic part of the molecule for the interaction with the cell.53 This selectivity was also proved with a blocking experiment (see the subsection SERS screening & diagnostics). The design of a screening procedure, based on the SERS spectroscopic imaging of the cells, allowed for monitoring the overall SERS response, at the single cell level, on 100-cell populations. With proper statistical analysis, we were able to distinguish the typical SERS signals belonging to human cell models of the non-tumorigenic keratinocyte HaCat line, metastatic prostate cancer PC-3 line, and uterine cervical cancer HeLa line. The detectable, low signal measured on the HaCaT cells proves the sensitivity of our method, and the capability to detect the presence, on HaCaT membranes, of the few folate solute PCFT carriers expressed, which are necessary for the cellular folate supply.50 The difference between the response of the HeLa and PC-3 cells is ascribed mainly to the presence of the FRα receptor, which is known to be present in a comparatively higher density on the HeLa membrane.20,21,47
Among the advantages of our method, we must recall the rather rapid time of incubation with the nanovector, when compared to similar studies:49 for this reason, we can state that there is no change in the cell population during the treatment. Treatment can be realized under physiological conditions (cell culture medium, 37 °C temperature), without preventing the selective binding of the nanoparticle to the folate receptors. Cytotoxicity measurement ensured that, during the treatment, the viability of the cell culture remains unvaried by the presence of the nanovector, granting the reliability of the procedure (see section S7 of the ESI†). We stabilized the cellular sample only by drying it, without further addition of fixing agents, which could affect the spectroscopic response of the system.
It has to be emphasized that the characteristics of this nanovector make it suitable as a membrane receptor label for studies of near field microscopy, scanning probe and electronic microscopy techniques (see for example section S5 in the ESI†). According to our estimate on the HeLa cell line, the treatment gave rise to approximately 100 Np-receptor binding events per cell. This is in good agreement with the Np density that can be inferred from SEM measurements (see section S5 of the ESI†), and with the expected high (approximately nM) folate-receptor affinity. The use of a red laser for SERS excitation, while avoiding the damage of the sample and therefore ensuring a high reproducibility of the analysis, with sensitivity to the presence of nanoparticle aggregates rather than single nanostructures.29 This means in turn that the SERS screening shown in Fig. 7 is related to the spatial distribution of the folate receptor density rather than to the presence of single receptors. On one hand, this reduces the risk of single, non-specific binding events affecting the measurement; on the other hand, this prevents to detect the presence of many single, isolated Nps (see Fig. S5.4 and S5.5 in the ESI†). In order to further enhance the diagnostic sensitivity, it could be useful to induce the nanovector ad hoc aggregation (e.g. in dimers) before treating the cells.
The overall results prove the possibility of reaching statistically relevant responses starting from rather small cell populations, if compared with the statistics required for example by fluorescence-based methods. Although we have not yet realized in vivo studies, the small size, the stability and the negligible cytotoxicity of the nanovector make our method a good candidate for such investigations, also in view of possible theranostic applications.
Given the positive correlation between the FRα expression and tumor stage and grade, the role of FA in tumor progression was indeed evidenced.21 In this respect, folate binding competitors (also known as antifolate drugs, e.g., aminopterin and methotrexate), have been developed over several decades for the treatment of cancer and inflammatory diseases.18,19,53,70 To date, antifolates approved for clinical use enter cells via RFC (primarily) and PCFT solute carriers, and by folate receptors.19,48 As an overexpression of FRα seems a typical characteristic of cancer cells, a predominant internalization of antifolate loaded nanovectors via FRα would be desirable in order to improve the treatment specificity, while limiting collateral damage on normal cells. For this reason we are currently testing the same Np functionalization protocol substituting/including folate and antifolate drug aminopterin (AMT). From our tests is emerging an intriguing significant increase in HeLa cell mortality after incubation with the AMT-nanovector, suggesting a valuable role of SERS folate-Nps to design valuable traceable multi-therapeutics carriers.
Acknowledgements
Prof. Gaio Paradossi group, Department of Chemical Science and Technology at the University of Rome Tor Vergata, is acknowledged with thanks for providing lab facilities for the chemical titration experiments and for useful discussions and suggestions.Notes and references
- A. R. Smith, et al. , Ca-Cancer J. Clin., 2015, 65(1), 30–54 CrossRef PubMed.
- V. Backman, et al. , Nature, 2000, 406(6791), 35–36 CrossRef CAS PubMed.
- M. G. Harisinghani, et al. , N. Engl. J. Med., 2003, 348(25), 2491–2499 CrossRef PubMed.
- A. B. Chinen, et al. , Chem. Rev., 2015, 115(19), 10530–10574 CrossRef CAS PubMed.
- Nanotechnology-Based Precision Tools for the Detection and Treatment of Cancer, ed. C. A. Mirkin, et al., Springer International Publishing, 2015 Search PubMed.
- D. L. Cao, et al. , Prostate, 2011, 71(7), 700–710 CrossRef CAS PubMed.
- A. Bocking, J. Stockhausen and D. Meyer-Ebrecht, Cell. Oncol., 2004, 26(1–2), 73–80 CAS.
- S. E. Cross, et al. , Nat. Nanotechnol., 2007, 2(1), 780–783 CrossRef CAS PubMed.
- M. Mitsunaga, et al. , Nat. Med., 2011, 17(12), 1685–1691 CrossRef CAS PubMed.
- B. Barlogie, et al. , Cancer Res., 1983, 43(9), 3982–3997 CAS.
- J. Krüger, et al. , J. Micromech. Microeng., 2002, 12(4), 486 CrossRef.
- K. Fan, et al. , Nat. Nanotechnol., 2012, 7(7), 459–464 CrossRef CAS PubMed.
- R. Bazak, et al. , J. Cancer Res. Clin. Oncol., 2012, 141(5), 769–784 CrossRef PubMed.
- M. S. Ehrenberg, et al. , Biomaterials, 2009, 30(4), 603–610 CrossRef CAS PubMed.
- A. Salvati, et al. , Nat. Nanotechnol., 2013, 8(2), 137–143 CrossRef CAS PubMed.
- M. Lucock, Mol. Genet. Metab., 2000, 71(1), 121–138 CrossRef CAS PubMed.
- S. D. Weitman, et al. , Cancer: Res., 1992, 52(12), 3396–3401 CAS.
- N. Parker, et al. , Anal. Biochem., 2005, 338(2), 284–293 CrossRef CAS PubMed.
- A. S. Wibowo, et al. , Proc. Natl. Acad. Sci. U. S. A., 2013, 110(38), 15180–15188 CrossRef CAS PubMed.
- D. Feng, et al. , Anal. Chem., 2013, 85(13), 6530–6535 CrossRef CAS PubMed.
- L. E. Kelemen, Int. J. Cancer, 2006, 119(2), 243–250 CrossRef CAS PubMed.
- S. Nie and S. R. Emory, Science, 1997, 275(5303), 1102–1106 CrossRef CAS PubMed.
- C. L. Haynes, A. D. McFarland and R. P. Van Duyne, Anal. Chem., 2005, 77(17), 338-A CrossRef CAS.
- T. Vo-Dinh, H. N. Wang and J. Scaffidi, J. Biophotonics, 2010, 3(1-2), 89–102 CrossRef CAS PubMed.
- J. C. Y. Kah, et al. , Int. J. Nanomed., 2007, 2(4), 785 CAS.
- X. M. Qian and S. M. Nie, Chem. Soc. Rev., 2008, 37(5), 912–920 RSC.
- F. Domenici, A. R. Bizzarri and S. Cannistraro, Int. J. Nanomed., 2011, 6, 2033–2042 CAS.
- F. Domenici, A. R. Bizzarri and S. Cannistraro, Anal. Biochem., 2012, 421(1), 9–15 CrossRef CAS PubMed.
- S. T. Ghosh and P. Tarasankar, Chem. Rev., 2007, 107(11), 4797–4862 CrossRef CAS PubMed.
- I. Fratoddi, et al. , Nano Res., 2015, 8(6), 1771–1799 CrossRef CAS.
- J. Kneipp, et al. , J. Phys. Chem. C, 2010, 114(16), 7421–7426 CAS.
- Y. Li, et al. , Biomaterials, 2015, 53, 25–31 CrossRef CAS PubMed.
- M. J. Kang, et al. , Int. J. Nanomed., 2013, 8, 1155 CrossRef PubMed.
- J. Ando, et al. , Nano Lett., 2011, 11(12), 5344–5348 CrossRef CAS PubMed.
- A. Pallaoro, et al. , ACS Nano, 2015, 9(4), 4328–4336 CrossRef CAS PubMed.
- M. E. Werner, et al. , Biomaterials, 2011, 32(33), 8548–8554 CrossRef CAS PubMed.
- S. Boca-Farcau, et al. , Mol. Pharm., 2013, 11(2), 391–399 CrossRef PubMed.
- A. Pallaoro, G. B. Braun and M. Moskovits, Proc. Natl. Acad. Sci. U. S. A., 2011, 108.40, 16559–16564 CrossRef PubMed.
- M. Osawa, et al. , J. Phys. Chem., 1994, 98.(48), 12702–12707 CrossRef.
- N. Curreli, et al. , J. Appl. Polym. Sci., 1997, 66.(8), 1433–1438 CrossRef.
- L. S. Jiao, et al. , Electrochem. Commun., 2005, 7.(2), 219–222 CrossRef.
- M. Baia, et al. , Chem. Phys. Lett., 2006, 422(1), 127–132 CrossRef CAS.
- A. R. Bizzarri and S. Cannistraro, Nanomedicine: Nanotechnology, Biology and Medicine, 2007, 3(4), 306–310 CrossRef CAS PubMed.
- Q. Li, et al. , Small, 2013, 9(6), 927–932 CrossRef CAS PubMed.
- M. Wu, W. Gunning and M. Ratnam, Cancer Epidemiol., Biomarkers Prev., 1999, 8(9), 775–782 CAS.
- Y. Hattori and Y. Maitani, Cancer Gene Ther., 2005, 12(10), 796–809 CrossRef CAS PubMed.
- H. C. Joshi, S. N. Vangapandu and R. Aneja, U.S. PatentNo. 8426398, 2013 Search PubMed.
- S. K. Desmoulin, et al. , Cancer Biol. Ther., 2012, 13(14), 1355–1373 CrossRef CAS PubMed.
- G. A. Mansoori, K. S. Brandenburg and A. Shakeri-Zadeh, Cancers, 2010, 2(4), 1911–1928 CrossRef CAS PubMed.
- R. Fujiwara, et al. , Sci. Rep., 2014, 4, 5251 CAS.
- C. Fasolato, et al. , Appl. Phys. Lett., 2014, 105(7), 073105 CrossRef.
- F. Costantini, et al. , Lab Chip, 2010, 10(24), 3407–3412 RSC.
- C. Chen, et al. , Nature, 2013, 500(7463), 486–489 CrossRef CAS PubMed.
- A. Shakeri-Zadeh, et al. , Dyn. Biochem. Process Biotechnol. Mol. Biol., 2010, 4(1), 06–12 Search PubMed.
- M. Baia, et al. , ChemPhysChem, 2009, 10(7), 1106–1111 CrossRef CAS PubMed.
- Y. Toumia, et al. , ACS Appl. Mater. Interfaces, 2016, 8(25), 16465–16475 CAS.
- F. Domenici, et al. , Colloids Surf., A, 2016, 498, 168–175 CrossRef CAS.
- A. Kokaislová, et al. , J. Raman Spectrosc., 2014, 45(9), 750–757 CrossRef.
- K. W. Kho, et al. , ACS Nano, 2012, 6(6), 4892–4902 CrossRef CAS PubMed.
- M. Gühlke, Z. Heiner and J. Kneipp, Phys. Chem. Chem. Phys., 2015, 17(39), 26093–26100 RSC.
- R. A. Davies, N. S. Chong and B. G. Ooi, Opt. Photonics J., 2013, 3(5A), 36994 Search PubMed.
- E. D. Schmid, P. Schlenker and R. R. M. Brand, J. Raman Spectrosc., 1977, 6(6), 314–318 CrossRef CAS.
- G. N. R. Tripathi and R. H. Schuler, J. Phys. Chem., 1988, 92(18), 5129–5133 CrossRef CAS.
- T. W. G. Solomons and C. B. Fryhle, Organic Chemistry, John Wiley & Sons Inc., 6th edn, 2000 Search PubMed.
- J. C. Weddell and P. I. Imoukhuede, PLoS One, 2014, 9(5), e97271 Search PubMed.
- R. Singh, et al. , Drug Dev. Ind. Pharm., 2015, 41(11), 1888–1901 CrossRef CAS PubMed.
- P. S. Low and S. A. Kularatne, Curr. Opin. Chem. Biol., 2009, 13(3), 256–262 CrossRef CAS PubMed.
- M. M. Doucette and V. L. Stevens, J. Nutr., 2001, 131(11), 2819–2825 CAS.
- R. Tantra, R. J. C. Brown and M. J. T. Milton, J. Raman Spectrosc., 2007, 38(11), 1469–1479 CrossRef CAS.
- S. Farber, et al. , N. Engl. J. Med., 1948, 238(23), 787–793 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6nr05057c |
| This journal is © The Royal Society of Chemistry 2016 |