Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An ion-controlled four-color fluorescent telomeric switch on DNA origami structures†
L.
Olejko
abc,
P. J.
Cywiński
de and
I.
Bald
*ac
aDepartment of Chemistry, Physical Chemistry, University of Potsdam, Karl-Liebknecht Str. 24-25, 14476 Potsdam, Germany. E-mail: olejko@uni-potsdam.de; bald@uni-potsdam.de
bSchool of Analytical Sciences Adlershof, Humboldt-Universität zu Berlin, Unter den Linden 6, 10099, Germany
cBAM Federal Institute for Materials Research and Testing, Richard-Willstätter Str. 11, 12489 Berlin, Germany
dFraunhofer IAP, Geiselbergstrasse 69, 14476 Potsdam, Germany. E-mail: piotrcyw@interia.pl
eInstitute of Physical Chemistry, Polish Academy of Sciences, Kasprzaka 44/53, 01-224 Warsaw, Poland
First published on 15th April 2016
Abstract
The folding of single-stranded telomeric DNA into guanine (G) quadruplexes is a conformational change that plays a major role in sensing and drug targeting. The telomeric DNA can be placed on DNA origami nanostructures to make the folding process extremely selective for K+ ions even in the presence of high Na+ concentrations. Here, we demonstrate that the K+-selective G-quadruplex formation is reversible when using a cryptand to remove K+ from the G-quadruplex. We present a full characterization of the reversible switching between single-stranded telomeric DNA and G-quadruplex structures using Förster resonance energy transfer (FRET) between the dyes fluorescein (FAM) and cyanine3 (Cy3). When attached to the DNA origami platform, the G-quadruplex switch can be incorporated into more complex photonic networks, which is demonstrated for a three-color and a four-color FRET cascade from FAM over Cy3 and Cy5 to IRDye700 with G-quadruplex-Cy3 acting as a switchable transmitter.
1. Introduction
The specific Watson–Crick base pairing and possibility for rather simple functionalization makes DNA an extremely versatile material, which can adopt virtually any nanoscale shape and simultaneously can be functionalized with different chemical entities such as metal nanoparticles or fluorophores.1–7 In such systems the optically-active functionalities act as signal reporters providing information about events occurring at the microscale and allow for detection, structure analysis and precise localization in sensing and imaging applications. Particularly interesting are nanophotonic programmable photonic networks or multifluorophoric cascades based on Förster resonance energy transfer (FRET) to control system functionality, readout or characterization. In such systems, excitons are transferred down in an array of fluorophores possessing successively lower excitation energies.8,9 DNA nanostructures enable the controlled arrangement of multiple fluorophores,10 which can be used for light harvesting, energy conversion, artificial photosynthesis, excitonic wires, switches, logic gates and optical communication.11,12By exploiting the dynamic function of DNA sequences, switchable systems can be designed, which further extend the potential applications of functional DNA nanostructures to more sophisticated excitonic systems such as DNA micromachines and bio-computers.13
Very prominent switchable DNA structures are guanine (G) quadruplexes. Linear G-rich telomeric DNA strands can fold into G-quadruplex structures in the presence of monovalent cations such as K+ or Na+. G-quadruplex structures consist of two or more stacked planar G-tetrads, which are formed from four G nucleobases assembled and stabilized by Hoogsteen-type hydrogen bonds.14 Basically, the G-quadruplex can unfold into single-stranded telomeric DNA when the central cation is removed from the G-quadruplex. Thus, G-quadruplex structures can be used to design DNA-nanoswitches sensitive to different chemical triggers.13,15 An electrochemical switch based on a K+ stabilized G-quadruplex structure has recently been developed.16 Fluorescent switches based on reversible folding and unfolding have also been reported for Pb2+-stabilized G-quadruplex structures17 as well as for Na+/K+ induced G-quadruplex structures.18–20 We have recently demonstrated ion-selective G-quadruplex folding on DNA origami structures using Förster resonance energy transfer (FRET), which can be used for selective K+ recognition even in the presence of 145 mM Na+.21
FRET is a non-radiative energy transfer, in which the energy is transferred from an excited donor molecule to an acceptor molecule through dipole–dipole interactions. Due to its prominent efficiencies of energy transfer at the corresponding distances, FRET is a process used in artificial photosynthesis and photonic wires or networks with different fluorophores attached e.g. to DNA.10,22,23 Furthermore, FRET plays an important role as a sensing tool for a variety of biologically relevant molecules such as antibodies, proteins, aptamers and even nanoparticles,24–27 and for structure elucidation of macromolecules such as double stranded DNA and DNA nanostructures.28–30 Especially for the study of macromolecular structures multi-color FRET plays a crucial role due to the possibility of modulating the optical signal over longer distances, i.e. beyond 2R0.31–34
Since FRET is a highly distance-dependent process, it is important to arrange the donor and acceptor molecules in a controlled way at well-defined distances. DNA origami nanostructures are excellent substrates to arrange and analyze different modifications with a high local control.35–37 DNA origami structures can adopt various 2D and 3D shapes by folding a circular single stranded virus DNA (scaffold strand, here: M13mp18) through hybridization with a suitable set of short single DNA strands (staple strands, about 200 different strands).1,35–37 Since every staple strand can be addressed separately and individually different modifications with predetermined distances can be introduced. Based on this property, DNA origami nanostructures have been used to investigate single-molecule chemical reactions,38,39 radiation induced DNA strand breaks in different DNA sequences40–43 and few-molecule detection using surface-enhanced Raman scattering (SERS).44,45 DNA origami frames have also been used to study the folding and unfolding events of G-quadruplex structures via atomic force microscopy (AFM).46–49
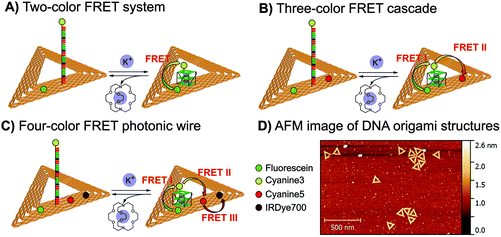
Here, we present the preparation and characterization of a reversibly switchable three and four-color FRET cascade realized on DNA origami nanostructures. The telomeric DNA 5′-(GGG ATT)4 is the middle element in the wire, and as already demonstrated, it assures reversible folding selective for K+.21 We also demonstrate that the ion-selectivity for K+ depends strongly on the DNA sequence. To this end we investigate the G-quadruplex formation in human telomeric DNA (5′-(GGG TTA)4, HumTel) and the reversed human telomeric DNA with the sequence 5′-(GGG ATT)4 (RevHumTel) both attached to DNA origami nanostructures.
2. Results and discussion
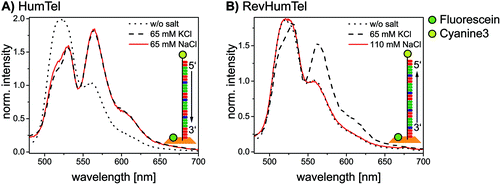
2.1. The importance of the telomere sequence for ion-selectivity
The telomeric DNA sequences (HumTel or RevHumTel) are functionalized with an acceptor molecule (here: cyanine3, Cy3) at their 5′-end and prolonged with a staple strand to place them on DNA origami triangles. The donor molecule fluorescein (FAM) is placed 3.3 nm away from the 5′-end of the telomeric DNA by modifying one staple strand internally. Fluorescence measurements have been carried out in solution using steady-state and time-resolved spectroscopy (fluorescence decays and the corresponding decay times are shown in ESI Fig. S1†). The steady-state fluorescence spectroscopy results for both sequences are shown in Fig. 1. The emission spectra of HumTel are shown in Fig. 1A without salt (dotted line) and after KCl (dashed line) and NaCl (red line) addition. The fluorescence emission spectrum before salt addition (dotted line) exhibits high intensity at 515 nm (FAM) and a weak shoulder at 565 nm (Cy3). After salt addition (KCl or NaCl) the emission at 515 nm drops and the emission at 565 nm rises as a consequence of FRET. The FRET efficiency increases because of a decrease in the distance between the fluorophores due to a conformational change of the telomeric DNA from a random coil to the compact G-quadruplex structure. When HumTel is used, the G-quadruplex formation is observed in the presence of both K+ and Na+; however, for the reversed sequence RevHumTel, the observed effect is remarkably different. As the fluorescence emission spectra in Fig. 1B indicate, the FRET process, and thus the G-quadruplex folding, is only turned on after KCl addition (dashed line, see also Fig. S2†), but not after NaCl addition (red line). That is, only for RevHumTel on DNA origami platforms, the G-quadruplex folding is K+ selective, whereas the HumTel sequence is sensitive for both Na+ and K+. The different selectivities can be ascribed to different 3D structures of the folded G-quadruplexes adopted by the two different sequences. Recently, it was demonstrated that subtle differences in the G-quadruplex forming sequence such as the sequence inversion from 5′–3′ to 3′–5′ can result in different G-quadruplex structures.50 Some of these structures might be suppressed on DNA origami substrates. Selective K+ sensing can be achieved even in solutions containing Na+ at a concentration as high as 145 mM, which represents the concentration usually occurring under physiological conditions.21 In the following experiments only RevHumTel is used. Other monovalent cations such as Na+, Li+, Cs+ and NH4+ do not influence the FRET efficiency and therefore do not induce G-quadruplex formation (Fig. S2†). Divalent cations such as Mg2+ and Ca2+ can basically influence the FRET efficiency (Fig. S2†) and stabilize G-quadruplex structures.51,52 However, since the MgCl2 concentration for the DNA origami preparation is as low as 10 mM in the present case and does not increase throughout the experiments, only a small constant number of Mg2+ induced G-quadruplexes should be present.2.2. Two-color FRET system
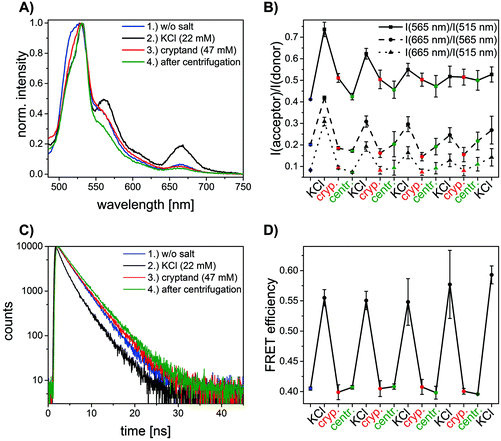
In order to explore the potential of RevHumTel to become a switch for a photonic cascade, the reversibility of G-quadruplex formation is extensively studied using fluorescence spectroscopy. To demonstrate that the G-quadruplex formation is reversible as shown in Fig. 2 we used a complexing agent, which has a higher affinity towards the central ion (here: K+) rather than towards the G-quadruplex. When the agent is added the G-quadruplex unfolds and returns to its initial conformation. Typical complexing agents are for example crown ethers or cryptands. 18-Crown-6 is a well-known substance which can encapsulate K+ (see ESI Fig. S3†). Since cryptands have a stronger binding to cations 4,7,13,16,21,24-hexaoxa-1,10-diazabicyclo[8.8.8]hexaco-sane, also known as [2.2.2]cryptand and in the following referred to as “cryptand”, is used in our study. Cryptands are bi- and oligocyclic multidentate ligands, which can bind specific ions. In general, they are three dimensional molecules similar to crown ethers, but containing additionally two nitrogen atoms connected via ethyleneoxy bridges. These compounds can encapsulate metal ions with high selectivity.53–55 The cryptand-induced switching phenomenon is analyzed in solution with steady-state (λex = 450 nm) and time-resolved fluorescence spectroscopy (λex = 490 nm, λem = 520 nm). The results for the two-color FRET system are shown in Fig. 3. In Fig. 3A the fluorescence emission spectra of the sample before and after KCl or cryptand addition are shown. The emission spectrum of the initial sample (blue spectrum, Fig. 3A) exhibits a strong peak at 515 nm from FAM. A very weak shoulder at 565 nm arising from Cy3 emission is also visible indicating that energy transfer occurs although the G-quadruplex is not folded, as the donor–acceptor distance is short enough for low-efficient FRET to occur. When 22 mM KCl is added to the sample the FAM emission is quenched, and at the same time the Cy3 emission is highly intensified (black spectrum, Fig. 3A) due to a shorter donor–acceptor distance resulting in a higher FRET efficiency. After adding 47 mM cryptand Cy3 emission decreases drastically and FAM emission increases slightly (red spectrum, Fig. 3A). The FRET process is weakened or rather switched off due to G-quadruplex unfolding. The FAM emission does not return to its initial intensity because the cryptand quenches the FAM fluorescence (see ESI Fig. S4†). To repeat the folding of the G-quadruplex on DNA origami structures potassium chloride and cryptand are removed using 10 kDa molecular weight cut-off centrifugal filters and washing three times with TAE (1×) buffer containing 10 mM MgCl2. The fluorescence spectrum of the sample after centrifugation is shown in Fig. 3A (green spectrum). The overall fluorescence intensity is lower because the total sample concentration decreases after centrifugation. It has to be noted that the peak at 530 nm is due to the Raman peak of water, which becomes visible due to the overall low fluorescence intensity of the sample. Decay associated spectra of the first switching cycle are shown in ESI Fig. S7 and S8.† The ratio of acceptor emission intensity (at 565 nm) to donor emission intensity (at 515 nm) changes due to the FRET process and can be used to monitor the changes occurring in the system (Fig. 3B). It increases due to G-quadruplex folding after the addition of KCl (black dots, Fig. 3B) and decreases due to G-quadruplex unfolding after cryptand addition (red dots, Fig. 3B). Because the cryptand quenches the FAM fluorescence, the acceptor–donor ratio does not return to its initial value after cryptand addition. Lower FAM emission intensity when compared to the initial point results in a higher ratio. After centrifugation KCl and cryptand are removed from the solution and the value virtually returns to its initial value. We demonstrate that the switching between the folded and unfolded state can be repeated 5 times on DNA origami structures in the two-color FRET system. The acceptor–donor ratio decreases step by step after each KCl/cryptand addition. This is attributed to a partial unfolding of the DNA origami structure induced by the cryptand, which might also capture Mg2+. The removal of Mg2+ ions from the DNA structure results in a destabilization of the DNA origami triangles and thus to more free FAM labeled DNA strands in solution, which do not participate in the FRET process (see ESI, Fig. S5†). Since the emission intensity is highly dependent on the total sample concentration, time-resolved measurements were carried out to overcome this drawback and to calculate the FRET efficiencies. Fluorescence decays are shown in Fig. 3C for a sample before (blue decay) and after KCl (black decay) and cryptand (red decay) addition. The FAM fluorescence decay time decreases drastically after KCl addition (black decay, Fig. 3C) due to higher FRET efficiencies and it increases again after cryptand addition (red decay, Fig. 3C). After centrifugation the fluorescence decay time is the same as the initial sample (green decay, Fig. 3C). The fluorescence decays have been fitted with a tri-exponential function with fixed τ-values (see ESI Table S1†). The decay time τ1 is set to 1.6 ns and is attributed to FAM fluorescence in the folded species (Rfold. = 6.0 nm), while τ2 is set to 2.7 ns assigned to FAM fluorescence in the unfolded species (Runfold. = 7.2 nm). The donor–acceptor distance in the unfolded state is still short enough for FRET to take place with a low efficiency. The decay time τ3 is equal to 4.4 ns and is due to the unquenched FAM, which does not take part in the FRET process (free strands or missing acceptor molecules on the DNA origami structure). The physical acceptor–donor distances R are determined using eqn (1) and (2).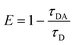 | (1) |
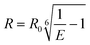 | (2) |
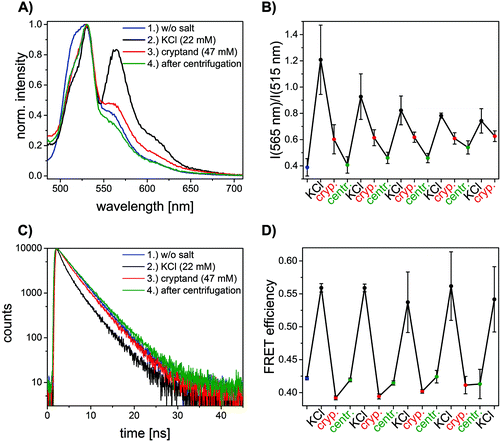 | ||
| Fig. 3 Results for the two-color FRET system with FAM and Cy3 as donor and acceptor molecules (color code: blue = initial sample, black = after KCl addition, red = after cryptand addition, green = after centrifugation). (A) Normalized emission spectra excited at 450 nm. The emission intensity of FAM at 515 nm decreases and at the same time Cy3 emission at 565 nm increases after KCl addition due to G-quadruplex formation. After cryptand addition the FAM emission increases and Cy3 emission decreases again due to unfolding of the G-quadruplex. After centrifugation the emission properties of the sample are similar when compared to the starting point but the overall intensity is lower (Raman peak of water becomes more dominant at 530 nm). The decay associated spectra of the first switching cycle are shown in ESI Fig. S7 and S8.† (B) Acceptor–donor intensity ratio for one sample after subsequent KCl and cryptand addition. The ratio rises after KCl addition and drops after cryptand addition. The FRET process is turned on and off due to folding and unfolding of the G-quadruplex. (C) FAM fluorescence decay time (λex = 490 nm, λem = 520 nm) decreases after KCl addition due to G-quadruplex formation. After cryptand addition the decay time increases again because the FRET process is turned off. The system is restored after centrifugation. (D) FRET efficiencies calculated with average FAM fluorescence decay times for one sample with subsequent KCl and cryptand addition. The switchable FRET system is based on G-quadruplex folding and unfolding. The G-quadruplex formation can be repeated 5 times on DNA origami structures. | ||
2.3. Switchable three-color FRET cascade
In the next step we introduced a third dye to realize a three-color FRET cascade, which can be switched on and off using the reversible G-quadruplex folding and unfolding. In this three-color FRET cascade FAM, Cy3 and cyanine5 (Cy5) are used as a donor, a transmitter and an acceptor, respectively (Fig. 2B). Cy3 is attached to the 5′-end of the telomeric DNA and FAM and Cy5 are placed 3.3 nm away from it on opposite sides (Fig. 2B). Since the spectral overlap between FAM and Cy5 is very small (see ESI Fig. S9†) FRET is unlikely to take place between these two dyes. Additionally, the donor–acceptor distance is around 7 nm, which makes FRET highly inefficient (see ESI Fig. S10A†). For FRET to take place, a third molecule (transmitter molecule) has to be introduced (here: Cy3). Because FAM/Cy3 and Cy3/Cy5 FRET pairs have larger spectral overlaps, FRET can take place from FAM to Cy3 followed by an energy transfer from Cy3 to Cy5 (see ESI Fig. S9†). This results in a FRET cascade, which can be excited at 450 nm (excitation wavelength for FAM) and can emit at 665 nm (Cy5). With such a FRET cascade longer inter-molecular distances (here: FAM-Cy5 ≈ 7 nm) can be achieved and a larger spectral range can be screened.The results for the three-color FRET cascade on DNA origami structures are depicted in Fig. 4. In Fig. 4A the emission spectra of one sample before and after KCl and cryptand addition are shown. For the initial sample (without KCl), the FAM emission at 515 nm is rather strong and the Cy3 at 565 nm and Cy5 at 665 nm emissions are very weak (blue spectrum, Fig. 4A). After KCl addition (black spectrum, Fig. 4A) the FAM emission is quenched and the Cy3 and Cy5 emissions strongly increase due to FAM-to-Cy3 and Cy3-to-Cy5 FRET. Since the G-quadruplex is formed, the distance between each FRET pair decreases resulting in higher energy transfer efficiency. After cryptand addition the emission of Cy3 and Cy5 is clearly turned off (red spectrum, Fig. 4A) and the FAM emission becomes stronger. As observed for the two-color system, the FAM emission does not return to the initial value because of the quenching effect induced by the cryptand. After a centrifugation step (green spectrum, Fig. 4A) KCl and cryptand are removed and the system is restored, but the overall fluorescence intensity decreases. As was described before for the two-color FRET system the water Raman peak becomes more pronounced at 530 nm because the overall sample concentration is very low and the fluorescence emission intensity is weaker. In Fig. 4B the acceptor–donor intensity ratios determined for each FRET pair are shown (Cy3/FAM (squares) = I(565 nm)/I(515 nm); Cy5/Cy3 (circles) = I(665 nm)/I(565 nm); Cy5/FAM (triangles) = I(665 nm)/I(515 nm)). The different FRET pair curves exhibit a similar tendency. After the addition of 22 mM KCl (black points, Fig. 4B) the ratio increases drastically due to an increase in FRET efficiency. After the addition of 47 mM cryptand (red points, Fig. 4B) the ratio decreases because of the G-quadruplex unfolding. Similar to what was described before for the two-color FRET system, the intensity ratio for all FRET pairs does not return to its initial value after cryptand addition. The FAM fluorescence decays for one folding–unfolding cycle are depicted in Fig. 4C. The fluorescence decay time decreases after the addition of KCl (black decay, Fig. 4C) compared to the fluorescence decay time of the initial sample (blue decay, Fig. 4C). After cryptand addition the fluorescence decay time increases again (red decay, Fig. 4C) because of G-quadruplex unfolding and increasing donor–transmitter–acceptor distances. The FAM fluorescence decay curves were fitted just as the two-color FRET system and the amplitudes for each decay time and the average decay times are shown in Table S1 in the ESI.† The Cy3 fluorescence decay times are shown in Fig. S11 of the ESI.† The FRET efficiencies were calculated using eqn (1) and are shown in Fig. 4D. The FRET efficiency rises after the addition of KCl (black dots, Fig. 4D) and the FRET cascade is turned off after the addition of cryptand (red dots, Fig. 4D) since the transmitter dye is moved away from the acceptor. The three color FRET cascade is again turned on 5 times.
2.4. Four-color FRET photonic wire
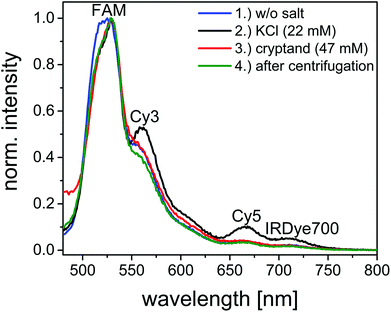
Finally, we introduce a fourth dye to the FRET cascade to exploit the full potential of the telomeric FRET switch on DNA origami structures. The design of the four-color FRET photonic wire is basically the same as the three-color FRET cascade but with IRDye700 as an additional acceptor molecule placed 3.3 nm away from Cy5 (see Fig. 2). In this photonic wire the energy is transferred from FAM to Cy3 followed by an energy transfer to Cy5 and finally to IRDye700 (see also ESI Fig. S12 and S13†). With such a photonic wire an inter-molecular distance of ca. 10 nm is achieved. This system is excited at 450 nm (FAM excitation) and it emits at 710 nm (IRDye700 emission) when the G-quadruplex is folded.The results of one switching cycle based on the four-color FRET photonic wire are shown in Fig. 5 and S12† to show the basic mechanism. For the initial sample, FAM emission at 515 nm is dominant and the emission intensities of Cy3 at 565 nm, Cy5 at 665 nm and IRDye700 at 710 nm are rather weak (blue spectrum, Fig. 5). The FRET cascade is turned on after KCl addition leading to a decrease of FAM emission and at the same time an increase in Cy3, Cy5 and IRDye700 emissions (black spectrum, Fig. 5). The distances between each FRET pair are reduced resulting in higher FRET efficiencies. The energy transfer cascade is turned off again after cryptand addition due to unfolding of the G-quadruplex. Thus, the Cy3, Cy5 and IRDye700 emissions are turned off. The FAM emission does not return to its initial value (as already observed for the two- and three-color FRET systems) because the cryptand quenches the FAM emission (red spectrum, Fig. 5). The system is restored after a centrifugation step (green spectrum, Fig. 5). Again, the water Raman peak becomes visible at 530 nm because the overall sample concentration decreases after centrifugation and therefore the overall fluorescence intensity decreases.
3. Conclusions
In this paper, we present a comprehensive comparison of two different telomeric DNA strands (HumTel and RevHumTel) with respect to their ion-selective G-quadruplex formation using FRET. We have shown that HumTel, when immobilized on DNA origami structures, folds into G-quadruplex structures both in the presence of Na+ and K+. RevHumTel on the other hand is selective for K+ only when attached to DNA origami triangles. Furthermore, we have analyzed the switching between folded and unfolded G-quadruplex structures for RevHumTel. We have realized a highly ion-selective and restorable, reversible nanophotonic FRET switch on DNA origami structures based on a two-color FRET system (FAM–Cy3–FRET). Additionally, we developed a switchable photonic wire on DNA origami structures using three and four different fluorophores (FAM-to-Cy3-to-Cy5-to-IRDye700 FRET cascade). We have shown that the G-quadruplex formation can be repeated up to 5 times. The unique properties of telomeric DNA (conformational switching and ion-selectivity) combined with the excellent possibilities of DNA origami structures to assemble different moieties at defined distances give the opportunity to design and further extend the applications of functional DNA nanostructures towards new nanophotonic systems such as light harvesting antennae, photonic wires and networks and chemically triggered logic gates or nanosensors with an optical readout.4. Experimental section
4.1. Chemicals
Unmodified oligonucleotides (used as staple strands) were purchased from Integrated DNA Technologies (Leuven, Belgium). The viral genome M13mp18 (scaffold strand) was purchased from tilbit nanosystems GmbH (Garching, Germany). The oligonucleotides modified with the organic dyes (fluorescein, cyanine3, cyanine5) were acquired from Metabion International AG (Planegg/Steinkirchen, Germany). The modified oligonucleotides have been purified by the manufacturer using HPLC and have been used as delivered. Magnesium chloride, potassium chloride, sodium chloride, 4,7,13,16,21,24-hexaoxa-1,10-diazabicyclo[8.8.8]hexacosane and Tris acetate-EDTA buffer (TAE buffer) were acquired from Sigma Aldrich (Taufkirchen, Germany). Diluted TAE buffer (1×) (pH = 8.2) contained 40 mM Tris-acetate and 1 mM EDTA.4.2. DNA origami preparation
The DNA origami structures were prepared by mixing the staple strands (150 nM) with the single stranded M13mp18 viral genome (5 nM) in TAE buffer (10×) containing 100 mM MgCl2 and ultrapure water (Millipore). This mixture was heated up to 80 °C and then slowly cooled down to 8 °C in 2 h (80 °C–66 °C: 1 °C every 30 s; 66 °C–25 °C: 1 °C every 2 min, 25 °C–8 °C: 1 °C every 1 min) using a thermal cycler (PEQLAB, Germany). After the annealing process, the solutions were purified via a 100 kDa molecular weight cut-off centrifugal filter (Millipore) with TAE buffer (1×) containing 10 mM MgCl2 (4 × 3214g for 10 min).4.3. G-quadruplex folding/unfolding
After the preparation of DNA origami triangles a KCl, NaCl or cryptand solution (c = 200 mM, 2 M in ultrapure water (Millipore)) was added and the samples were shaken for 15 min at 40 °C to fold (KCl, NaCl) and unfold (cryptand) the G-quadruplexes on DNA origami structures. After KCl/cryptand addition the sample was purified via a 10 kDa molecular weight cut-off centrifugal filter (Millipore) with TAE buffer (1×) containing 10 mM MgCl2 (3 × 3800g for 7 min) to remove KCl and cryptand. Steady-state and time-resolved fluorescence spectroscopy was performed after each KCl and cryptand addition and centrifugation step at DNA origami concentration of ca. 5 nM.4.4. Atomic force microscopy (AFM)
To investigate the correctly formed DNA origami structures, AFM was performed for each sample after the preparation. AFM imaging was done with a Flex AFM (Nanosurf, Germany). A Tap150 Al-G cantilever (Budget Sensors, Sofia, Bulgaria) with a resonance frequency of (125–160) kHz and a spring constant of 5 N m−1 was used to visualize the DNA origami structures. The samples were prepared on freshly cleaved mica (Plano GmbH, Germany). For this, 2 μl of the sample (ca. 20 nM) and 33 μl of TAE (1×) containing 10 mM MgCl2 were incubated for 30 s and subsequently washed twice with 1 ml of ultrapure water (Millipore). Afterwards, the fluid was removed with compressed air. The measurements were performed in air using the phase contrast mode. The measured triangles have a length of 100–150 nm and a height of 1–2 nm (see Fig. 2). The yield of correctly formed DNA origami structures has been determined from AFM images to be about 95%.4.5. Steady-state fluorescence spectroscopy
Steady-state fluorescence spectroscopy measurements were done using a FluoromaxP fluorescence spectrophotometer (HORIBA Jobin Yvon GmbH, Germany) with 3 mm quartz cuvettes. The measurements were performed in a 90° angle acquisition using the system-internal quantum correction. The excitation wavelength for all emission spectra was set to 450 nm. For the measurements an increment of 1 nm and integration time of 0.2 s were chosen and the bandpass was set to 5 nm for both emission and excitation.4.6. Time-resolved fluorescence spectroscopy
Time-correlated single photon counting (TCSPC) measurements were performed on a FLS920 Fluorescence spectrophotometer (Edinburgh Instruments, UK) using 3 mm quartz cuvettes. The samples were measured in a 90° setup using a white light source (SC-400-PP supercontinuum-source, Fianium: 0.5–20 MHz, 400 nm < l < 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm, pulse width: ca. 30 ps) as the excitation source and a Multi-Channel-Plate (ELDY EM1-132/300, Europhoton GmbH, Berlin) as the detector. The excitation wavelength was set to 490 nm and the emission wavelength was set to 520 nm. The fluorescence decay curves were fitted with a tri-exponential function (eqn (3)) using FAST software (Edinburgh Instruments, UK).
000 nm, pulse width: ca. 30 ps) as the excitation source and a Multi-Channel-Plate (ELDY EM1-132/300, Europhoton GmbH, Berlin) as the detector. The excitation wavelength was set to 490 nm and the emission wavelength was set to 520 nm. The fluorescence decay curves were fitted with a tri-exponential function (eqn (3)) using FAST software (Edinburgh Instruments, UK).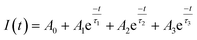 | (3) |
![[small tau, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0d4.gif) DA was calculated with eqn (4).
DA was calculated with eqn (4).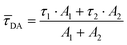 | (4) |
Acknowledgements
This research was supported by the Deutsche Forschungs-gemeinschaft (DFG), a Marie Curie FP7 Integration Grant within the 7th European Union Framework Programme, by the University of Potsdam, the Federal Institute for Materials Research (BAM), and the DFG project GSC 1013 (SALSA).References
- W. K. P. Rothemund, Nature, 2006, 440, 297–302 CrossRef PubMed.
- Z.-G. Wang and B. Ding, Acc. Chem. Res., 2014, 47, 1654–1662 CrossRef CAS PubMed.
- T. Tørring, N. V. Voigt, J. Nangreave, H. Yan and K. V. Gothelf, Chem. Soc. Rev., 2011, 40, 5636–5646 RSC.
- Y. R. Yang, Y. Liu and H. Yan, Bioconjugate Chem., 2015, 26, 1381–1395 CrossRef CAS PubMed.
- R. Schreiber, J. Do, E.-M. Roller, T. Zhang, V. J. Schüller, P. C. Nickels, J. Feldmann and T. Liedl, Nat. Nanotechnol., 2014, 9, 74–78 CrossRef CAS PubMed.
- Z. Deng, A. Samanta, J. Nangreave, H. Yan and Y. Liu, J. Am. Chem. Soc., 2012, 134, 17424–17427 CrossRef CAS PubMed.
- T. L. Doane, R. Alam and M. M. Maye, Nanoscale, 2015, 7, 2883–2888 RSC.
- C. M. Spillmann, M. G. Ancona, S. Buckhout-White, W. R. Algar, M. H. Stewart, K. Susumu, A. L. Huston, E. R. Goldman and I. L. Medintz, ACS Nano, 2013, 7, 7101–7118 CrossRef CAS PubMed.
- C. M. Spillmann, S. Buckhout-White, E. Oh, E. R. Goldman, M. G. Ancona and I. L. Medintz, Chem. Commun., 2014, 50, 7246 RSC.
- S. Buckhout-White, C. M. Spillmann, W. R. Algar, A. Khachatrian, J. S. Melinger, E. R. Goldman, M. G. Ancona and I. L. Medintz, Nat. Commun., 2014, 5, 5615 CrossRef CAS PubMed.
- W. Su, V. Bonnard and G. A. Burley, Chem. – Eur. J., 2011, 17, 7982–7991 CrossRef CAS PubMed.
- B. Albinsson, J. K. Hannestad and K. Börjesson, Coord. Chem. Rev., 2012, 256, 2399–2413 CrossRef CAS.
- F. Wang, X. Liu and I. Willner, Angew. Chem., Int. Ed., 2015, 54, 1098–1129 CrossRef CAS PubMed.
- S. Neidle and G. N. Parkinson, Curr. Opin. Struct. Biol., 2003, 13, 275–283 CrossRef CAS PubMed.
- Y. Krishnan and F. C. Simmel, Angew. Chem., Int. Ed., 2011, 50, 3124–3156 CrossRef CAS PubMed.
- B. Ge, Y. C. Huang, D. Sen and H.-Z. Yu, Angew. Chem., Int. Ed., 2010, 49, 9965–9967 CrossRef CAS PubMed.
- T. Li, S. Dong and E. Wang, J. Am. Chem. Soc., 2010, 132, 13156–13157 CrossRef CAS PubMed.
- B. Saccà, B. Siebers, R. Meyer, M. Bayer and C. M. Niemeyer, Small, 2012, 8, 3000–3008 CrossRef PubMed.
- Y. Hu, F. Wang, C.-H. Lu, J. Girsh, E. Golub and I. Willner, Chem. – Eur. J., 2014, 20, 16203–16209 CrossRef CAS PubMed.
- L. Hu, X. Liu, A. Cecconello and I. Willner, Nano Lett., 2014, 14, 6030–6035 CrossRef CAS PubMed.
- L. Olejko, P. J. Cywinski and I. Bald, Angew. Chem. Int. Ed., 2015, 54, 673–677 CAS.
- P. D. Cunningham, A. Khachatrian, S. Buckhout-White, J. R. Deschamps, E. R. Goldman, I. L. Medintz and J. S. Melinger, J. Phys. Chem. B, 2014, 118, 14555–14565 CrossRef CAS PubMed.
- P. K. Dutta, R. Varghese, J. Nangreave, S. Lin, H. Yan and Y. Liu, J. Am. Chem. Soc., 2011, 133, 11985–11993 CrossRef CAS PubMed.
- P. J. Cywiński, L. Olejko and H.-G. Löhmannsröben, Anal. Chim. Acta, 2015, 887, 209–215 CrossRef PubMed.
- P. J. Cywinski, T. Hammann, D. Hühn, W. J. Parak, N. Hildebrandt and H.-G. Löhmannsröben, J. Biomed. Opt., 2014, 19, 101506 CrossRef PubMed.
- P. J. Cywiński, K. Nchimi Nono, L. J. Charbonnière, T. Hammann and H.-G. Löhmannsröben, Phys. Chem. Chem. Phys., 2014, 16, 6060–6067 RSC.
- H. Härmä, S. Pihlasalo, P. J. Cywinski, P. Mikkonen, T. Hammann, H.-G. Löhmannsröben and P. Hänninen, Anal. Chem., 2013, 85, 2921–2926 CrossRef PubMed.
- S. Kalinin, T. Peulen, S. Sindbert, P. J. Rothwell, S. Berger, T. Restle, R. S. Goody, H. Gohlke and C. A. M. Seidel, Nat. Methods, 2012, 9, 1218–1225 CrossRef CAS PubMed.
- I. H. Stein, V. Schüller, P. Böhm, P. Tinnefeld and T. Liedl, ChemPhysChem, 2011, 12, 689–695 CrossRef CAS PubMed.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, M. M. Golas, B. Sander, H. Stark, L. P. C Oliveira, J. S. Pedersen, V. Birkedal, F. Besenbacher, K. V. Gothelf and J. Kjems, Nature, 2009, 459, 73–76 CrossRef CAS PubMed.
- S. Hohng, C. Joo and T. Ha, Biophys. J., 2004, 87, 1328–1337 CrossRef CAS PubMed.
- A. Kienzler, R. Flehr, R. A. Kramer, S. Gehne, M. U. Kumke and W. Bannwarth, Bioconjugate Chem., 2011, 22, 1852–1863 CrossRef CAS PubMed.
- S. Gehne, R. Flehr, A. Altevogt, M. Berg, W. Bannwarth and M. U. Kumke, J. Phys. Chem. B, 2012, 116, 10798–10806 CrossRef CAS PubMed.
- I. H. Stein, C. Steinhauer and P. Tinnefeld, J. Am. Chem. Soc., 2011, 133, 4193–4195 CrossRef CAS PubMed.
- M. Endo, Y. Yang and H. Sugiyama, Biomater. Sci., 2013, 1, 347 RSC.
- A. Rajendran, M. Endo and H. Sugiyama, Angew. Chem. Int. Ed., 2012, 51, 874–890 CrossRef CAS PubMed.
- I. Bald and A. Keller, Molecules, 2014, 19, 13803–13823 CrossRef PubMed.
- S. Helmig, A. Rotaru, D. Arian, L. Kovbasyuk, J. Arnbjerg, P. R. Ogilby, J. Kjems, A. Mokhir, F. Besenbacher and K. V. Gothelf, ACS Nano, 2010, 4, 7475–7480 CrossRef CAS PubMed.
- N. V. Voigt, T. Tørring, A. Rotaru, M. F. Jacobsen, J. B. Ravnsbaek, R. Subramani, W. Mamdouh, J. Kjems, A. Mokhir, F. Besenbacher and K. V. Gothelf, Nat. Nanotechnol., 2010, 5, 200–203 CrossRef CAS PubMed.
- S. Vogel, J. Rackwitz, R. Schürmann, J. Prinz, A. R. Milosavljevic, M. Refregiers, A. Giuliani and I. Bald, J. Phys. Chem. Lett., 2015, 6, 4589–4593 CrossRef CAS PubMed.
- A. Keller, J. Rackwitz, E. Cauët, J. Liévin, T. Körzdörfer, A. Rotaru, K. V. Gothelf, F. Besenbacher and I. Bald, Sci. Rep., 2014, 4, 7391 CrossRef CAS PubMed.
- A. Keller, J. Kopyra, K. V. Gothelf and I. Bald, New J. Phys., 2013, 15, 83045 CrossRef.
- A. Keller, I. Bald, A. Rotaru, E. Cauët, K. V. Gothelf and F. Besenbacher, ACS Nano, 2012, 6, 4392–4399 CrossRef CAS PubMed.
- J. Prinz, C. Heck, L. Ellerik, V. Merk and I. Bald, Nanoscale, 2016, 8, 5612–5620 RSC.
- J. Prinz, B. Schreiber, L. Olejko, J. Oertel, J. Rackwitz, A. Keller and I. Bald, J. Phys. Chem. Lett., 2013, 4, 4140–4145 CrossRef CAS.
- Y. Sannohe, M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 16311–16313 CrossRef CAS PubMed.
- A. Rajendran, M. Endo, K. Hidaka, P. L. T. Tran, J.-L. Mergny, R. J. Gorelick and H. Sugiyama, J. Am. Chem. Soc., 2013, 135, 18575–18585 CrossRef CAS PubMed.
- A. Rajendran, M. Endo, K. Hidaka and H. Sugiyama, Angew. Chem., 2014, 126, 4191–4196 CrossRef.
- A. Rajendran, M. Endo, K. Hidaka, P. L. T. Tran, M.-P. Teulade-Fichou, J.-L. Mergny and H. Sugiyama, RSC Adv., 2014, 4, 6346 RSC.
- M. Marušič and J. Plavec, Angew. Chem., Int. Ed., 2015, 54, 11716 CrossRef PubMed.
- Y.-Y. Yan, J. Lin, T.-M. Ou, J.-H. Tan, D. Li, L.-Q. Gu and Z.-S. Huang, Biochem. Biophys. Res. Commun., 2010, 402, 614–618 CrossRef CAS PubMed.
- B. Kankia, Sci. Rep., 2015, 5, 12996 CrossRef CAS PubMed.
- K. E. Krakowiak, J. S. Bradshaw, H.-Y. An and R. M. Izatt, Pure Appl. Chem., 1993, 65, 511–516 CrossRef CAS.
- P. Müller, Pure Appl. Chem., 1994, 66, 1077–1184 CrossRef.
- IUPAC Compendium of Chemical Terminology, ed. M. Ničet al., IUPAC, Research Triangle Park, NC, 2009 Search PubMed.
- T. Förster, Ann. Phys., 1948, 437, 55–75 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6nr00119j |
| This journal is © The Royal Society of Chemistry 2016 |