Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Elemental bioimaging and speciation analysis for the investigation of Wilson's disease using μXRF and XANES
Oliver
Hachmöller
a,
Ana Guilherme
Buzanich
b,
Michaela
Aichler
c,
Martin
Radtke
b,
Dörthe
Dietrich
a,
Kristina
Schwamborn
d,
Lisa
Lutz
e,
Martin
Werner
e,
Michael
Sperling
af,
Axel
Walch
c and
Uwe
Karst
*a
aWestfälische Wilhelms-Universität Münster, Institute of Inorganic and Analytical Chemistry, Corrensstraße 30, 48149 Münster, Germany. E-mail: uk@uni-muenster.de; Fax: +49-251/83-36013; Tel: +49-251/83-33141
bFederal Institute for Materials Research and Testing (BAM), Richard-Willstätter-Straße 11, 12489 Berlin, Germany
cHelmholtz Zentrum München, Research Unit Analytical Pathology, Ingolstädter Landstraße 1, 85764 Neuherberg, Germany
dTechnische Universität München, Institut für Allgemeine Pathologie und Pathologische Anatomie, Trogerstraße 18, 81675 München, Germany
eUniversitätsklinikum Freiburg, Institut für Klinische Pathologie, Breisacher Straße 115a, 79106 Freiburg, Germany
fEuropean Virtual Institute for Speciation Analysis (EVISA), Mendelstraße. 11, 48149 Münster, Germany
First published on 10th March 2016
Abstract
A liver biopsy specimen from a Wilson's disease (WD) patient was analyzed by means of micro-X-ray fluorescence (μXRF) spectroscopy to determine the elemental distribution. First, bench-top μXRF was utilized for a coarse scan of the sample under laboratory conditions. The resulting distribution maps of copper and iron enabled the determination of a region of interest (ROI) for further analysis. In order to obtain more detailed elemental information, this ROI was analyzed by synchrotron radiation (SR)-based μXRF with a beam size of 4 μm offering a resolution at the cellular level. Distribution maps of additional elements to copper and iron like zinc and manganese were obtained due to a higher sensitivity of SR-μXRF. In addition to this, X-ray absorption near edge structure spectroscopy (XANES) was performed to identify the oxidation states of copper in WD. This speciation analysis indicated a mixture of copper(I) and copper(II) within the WD liver tissue.
Introduction
Wilson's disease (WD), also referred to as “progressive hepatolenticular degeneration”, is a rare genetic disorder of copper metabolism, showing a prevalence between one in 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and one in 100
000 and one in 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individuals.1,2 This disease is based on the defective gene ATP7B, which encodes for a metal-transporting ATPase, responsible for the excretion of excess copper via the bile.3–6 In the case of WD, excess copper is not excreted, but accumulated within the liver and the central nervous system, causing various hepatic, neurological, and psychiatric symptoms.7,8 Because of these unspecific symptoms, there is no universal diagnosis for WD and although genetic testing is available, the determination of hepatic copper by a liver biopsy is still required in many cases due to various mutations of ATP7B.8 By means of histological staining of these liver biopsy specimens, it is well known that the hepatic copper distribution presents, especially in later stages of WD, large fluctuations within the liver tissue.8,9 However, there is a poor level of knowledge concerning the metallome in WD. For that reason, spatially resolved methods to determine the elemental distribution in WD liver samples as well as tools for the elucidation of the present copper species are required.
000 individuals.1,2 This disease is based on the defective gene ATP7B, which encodes for a metal-transporting ATPase, responsible for the excretion of excess copper via the bile.3–6 In the case of WD, excess copper is not excreted, but accumulated within the liver and the central nervous system, causing various hepatic, neurological, and psychiatric symptoms.7,8 Because of these unspecific symptoms, there is no universal diagnosis for WD and although genetic testing is available, the determination of hepatic copper by a liver biopsy is still required in many cases due to various mutations of ATP7B.8 By means of histological staining of these liver biopsy specimens, it is well known that the hepatic copper distribution presents, especially in later stages of WD, large fluctuations within the liver tissue.8,9 However, there is a poor level of knowledge concerning the metallome in WD. For that reason, spatially resolved methods to determine the elemental distribution in WD liver samples as well as tools for the elucidation of the present copper species are required.
Non-destructive methods based on X-ray fluorescence like energy dispersive X-ray spectroscopy (EDX) and micro-X-ray fluorescence spectroscopy (μXRF) are applied for the determination of the elemental distribution within different liver tissues. Hayashi et al. showed several copper and iron overload patterns in the liver from WD patients using an electron-microscope with EDX.10 Although this approach provides an excellent spatial resolution, only a moderate sensitivity in the upper μg g−1 is given.11,12 In comparison to this, synchrotron radiation (SR)-based μXRF offers trace element sensitivity and submicron spatial resolution.13,14 Kinoshita et al. determined the iron distribution in the lobule of the human liver showing elevated iron intensity in diseased liver samples from patients with hepatitis or cirrhosis.15 The non-destructive visualization of the copper distribution in hepatocytes in an animal model for WD by means of SR-μXRF was also applied by Ralle et al., revealing similar copper accumulation within a liver lobule.16 Due to the fact that analysis time at synchrotron facilities is strongly limited, μXRF as a bench-top instrument represents an alternative for elemental mapping under laboratory conditions.17 However, this set-up is showing a lower resolution and sensitivity in comparison to SR-μXRF. Therefore, bench-top μXRF can be utilized for a coarse scan to identify regions of interest (ROI) for the further analysis by a more elaborate technique like SR-μXRF. Applications for elemental mapping in biological samples by means of bench-top μXRF are for example the investigation of elemental distribution in teeth or bone tissue, where high analyte concentrations are present.18,19 Parallel to the elemental distribution in WD, the elucidation of the present copper species is crucial for a better understanding of WD. For this purpose, X-ray absorption near edge structure spectroscopy (XANES) is a suitable tool for the determination of the present oxidation states. The advantage of this method is the fact that no complex sample preparation is required in comparison to hyphenated techniques. In the literature, several applications of XANES for copper speciation in proteins, cells as well as in animal and human samples are described.16,20–23 However, the copper species present in the liver from WD patients have not been unambiguously identified to date.
In this paper, elemental bioimaging by means of μXRF is applied for a paraffin-embedded liver biopsy specimen of a WD patient. In order to acquire a coarse scan of the elemental distribution and to define a ROI, a thin section of the liver sample was analyzed by means of bench-top μXRF. Afterwards, elemental mapping of the ROI was carried out by SR-μXRF with a higher resolution and sensitivity. Additionally, XANES was applied to identify the present oxidation states of copper in the human WD liver sample.
Experimental
WD liver sample
The WD liver sample was collected by a needle biopsy of the liver within a medical investigation and embedded in paraffin. For the analysis by means of laboratory bench-top μXRF, a section with a thickness of 3 μm was prepared using a microtome and mounted on a glass microscopic slide. A parallel section with a thickness of 10 μm for the analysis by means of SR-μXRF was placed on Kapton® foil (8 μm 3511 Kapton® film, SPEX SamplePrep, Metuchen, NJ, USA), which was clamped in a frame made of polyethylene. A further parallel section was stained with hematoxylin and eosin (H&E) to visualize the structure of the liver tissue. Microscopy images were recorded using a BZ-9000 inverted fluorescence/bright field microscope (Keyence, Osaka, Japan).Analysis by means of bench-top μXRF
For the non-destructive analysis of a paraffin-embedded section of the WD liver sample, a laboratory bench-top μXRF spectrometer M4 Tornado (Bruker Nano GmbH, Berlin, Germany) was applied. The Rh-anode micro-focus X-ray tube was set at a voltage of 50 V and an anode current of 600 μA. The device was equipped with polycapillary optics to focus the incident X-ray beam on a size of approximately 25 μm for the Mo–K line. For the detection of the emitted X-ray fluorescence, a silicon drift detector (SDD, XFlash® 5030, Bruker Nano GmbH) was utilized. In addition to this, the analyses were carried out in an evacuated sample chamber at 20 mbar. An area of 4.88 mm × 2.67 mm of the WD sample was analyzed. The step size was set at 25 μm and each spot was analyzed for 100 ms. Data processing was performed using the software ESPRIT HyperMap (Bruker Nano GmbH) and images were generated using the software ImageJ (National Institute of Health, Bethesda, MD, USA).Analysis by means of SR-μXRF
In order to obtain higher spatial resolution and sensitivity, analyses by means of SR-μXRF were performed at the BAMline at BESSY II (Helmholtz-Zentrum Berlin, Germany).24 A double multilayer monochromator set was applied for the emission of a parallel beam, which was focused by a compound refractive lens (CRL, Institute of Microstructure Technology, Karlsruhe, Germany) down to a spot size of 4 μm onto the sample. According to the nominal energy of the CRL, an excitation energy of 9.7 keV was acquired. The emitted X-ray fluorescence was detected using a SDD (XFlash® 5030, Bruker Nano GmbH). Between the incident beam and the detector, a 90° geometry was applied and measurements were carried out at atmospheric pressure. For this purpose, a short distance between the sample and the SDD of less than 10 mm was utilized. An area of 484 μm × 536 μm of the sample was analyzed. The step size was set at 4 μm and an analysis time of 12 s per spot was applied. A full XRF spectrum was obtained in each analyzed spot and fitted in the AXIL software package.25 Afterwards, the fitted peak areas were used to generate elemental distribution maps using the software ImageJ (National Institute of Health). The following X-ray fluorescence transitions were used for 2D distribution maps: Mn-Kα, Fe-Kα, Cu-Kα, and Zn-Kα.XANES analysis
To distinguish between copper(I) and copper(II) species, XANES measurements were carried out at the BAMline at BESSY II (Helmholtz-Zentrum Berlin, Germany). Monochromatic radiation was generated using a Si(111) double-crystal monochromator. XANES experiments were performed in fluorescence mode and a 90° geometry between the incident and the SDD (XFlash® 5030, Bruker Nano GmbH) was utilized. Again, measurements were carried out at atmospheric pressure and a distance between the sample and the detector of less than 10 mm was applied. The Cu-K edge (8979 eV) was scanned, using the following energy step sizes for the different ranges: 10 eV between 8954 eV and 8964 eV, 1 eV between 8965 eV and 9029 eV, 5 eV between 9030 eV and 9064 eV, and 10 eV between 9065 eV and 9079 eV. For each step, the analysis time was 100 s. Copper foil, copper(I) oxide, and copper(II) oxide (all Sigma Aldrich, Steinheim, Germany) were analyzed as standard materials for the respective oxidation states. Copper(I) oxide and copper(II) oxide were mounted in Kapton® foil. Two different areas of the WD sample showing elevated intensities for copper were investigated with a beam size of approximately 4 mm × 3 mm.In-house software (BAM, Berlin, Germany) was used for data processing. Afterwards, the produced XANES spectra were treated and analyzed using the software ATHENA from the IFEFFIT package.26
Results and discussion
Element bioimaging by bench-top μXRF
The results by means of laboratory bench-top μXRF for the WD sample are presented in Fig. 1. The microscopy image shows two adjacent liver tissue samples from needle biopsies. The H&E stained parallel section of the WD sample, shown in Fig. 1b, presents an inhomogeneous liver tissue. The progressing disease leads to the formation of nodules consisting of hepatocytes, which are marked with black arrows in Fig. 1b. In accordance with the H&E stained parallel section, copper and iron are distributed inhomogeneously within the liver tissue (Fig. 1c and 1d). The copper distribution shows ring-like structures, which are marked with white arrows in Fig. 1c. These structures correspond to the microscopy image and higher copper intensities can be detected at the outer areas of the hepatocytes. In contrast to this, iron shows a different distribution pattern and high iron intensities can be detected within areas of hepatocytes in the liver tissue (white arrows in Fig. 1d). The copper and iron distribution show an inverse correlation: areas with higher iron intensities are surrounded by areas with high copper intensities (Fig. 1e). Further elements besides copper and iron could not be detected within the liver tissue by means of bench-top μXRF.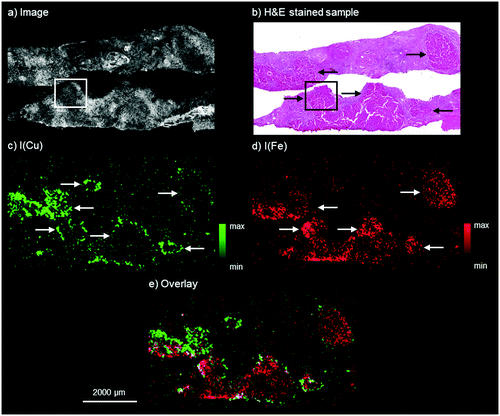 | ||
| Fig. 1 Autofluorescence microscopic image of the WD liver sample investigated by bench-top μXRF (a), bright field microscopy image of a H&E stained parallel section of the WD liver sample (b), distribution maps of copper (c) and iron (d), and overlay of the copper and iron distribution (e). The area marked with the white box in a and the black box in b corresponds to the area of the parallel section analyzed later by means of SR-μXRF (Fig. 2). | ||
Similar results of a combined copper and iron overload are described in previous studies.10,27 Iron overload associated with WD was for example described by Hayashi et al., who analyzed liver biopsy specimens of patients with WD by means of electron microscopy with EDX and detected several distribution patterns from isolated to evenly distributed combined copper and iron overload.10 The authors suggested an accumulation of iron for patients with hypoceruloplasminemia, which occurs in WD.10 In addition to this, an inhomogeneous distribution of copper with nodules showing no histochemically detectable copper next to nodules with abundant copper in later stages of WD is well known.8
The results indicate the suitability of bench-top μXRF for the analysis of thin sections of biological materials like liver tissue. Although copper concentrations are elevated in WD, a sample with a thickness of 3 μm means a very small sample volume and therefore a small amount of analyte for the analysis by means of bench-top μXRF. Normally, hepatic copper concentrations between 70 and 250 μg g−1 of dry weight liver are detected in WD, while healthy patients show concentrations below 50 μg g−1.8,28,29 The results obtained by non-destructive bench-top μXRF can be used for a coarse scan for further analysis by techniques offering a higher sensitivity and spatial resolution like laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) or SR-μXRF.
Elemental bioimaging by SR-μXRF
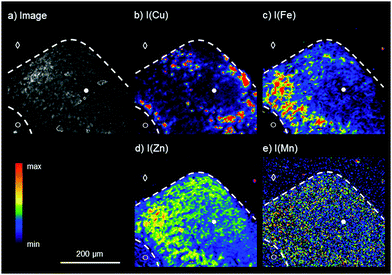
For the analysis of the liver tissue at a cellular level, a higher sensitivity and spatial resolution are needed, which can be provided by SR-μXRF. Based on the results obtained by laboratory bench-top μXRF, a ROI within the liver tissue was determined for the elemental bioimaging by means of SR-μXRF (white box in Fig. 1a). For the analysis, a parallel section of 10 μm was prepared and mounted on Kapton® foil.Fig. 2 shows the results for the elemental bioimaging by means of SR-μXRF. The analyzed area shown in the microscopy image (Fig. 2a) consists of hepatocytes (●), surrounding liver tissue (○), and paraffin (◇). In comparison to the laboratory μXRF, further elements to copper and iron like zinc and manganese are detected (Fig. 2(b–e)). Again, an inverse correlation of copper and iron is detected. In addition to this, zinc is showing a similar distribution pattern as iron and correlates inversely with copper. The three elements copper, iron, and zinc reveal an elevated intensity within the hepatocytes in comparison to surrounding liver tissue. This is in agreement with a previous study on elemental bioimaging by means of LA-ICP-MS: Boaru et al. showed a correlation between the accumulation of copper, iron, and zinc in liver tissue originating from an animal model for WD as well as in human liver tissue from WD patients.30 In contrast to these elements, manganese is evenly distributed within the hepatocytes and the surrounding liver tissue. In addition to the presented human WD liver sample, two control samples were analyzed by means of SR-μXRF during the beam time: a human liver of a patient with a liver metastasis and a rat liver. The elemental imaging by means of SR-μXRF revealed a homogeneous elemental distribution with lower intensities in both control samples in comparison to the analyzed WD liver sample; the results are not shown here.
In sum, SR-μXRF offers the possibility of a higher spatial resolution, owed to the use of a CRL lens, and a higher sensitivity for the detection of various elements in thin sections of biological materials, due to higher signal-to-background ratios. In this paper, liver tissue from a liver biopsy of a WD patient was analyzed and the distribution of the four elements copper, iron, zinc, and manganese was shown with a spatial resolution of 4 μm. This enables the presentation of the elemental distributions in the liver tissue at a cellular level.
Speciation analysis by XANES
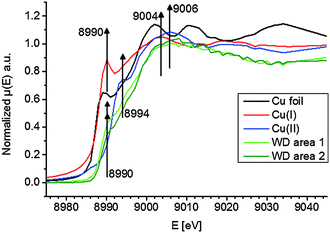
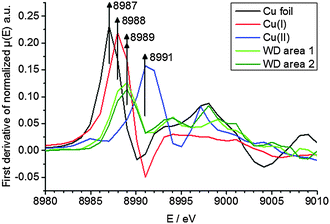
To elucidate the present copper species in WD, copper standards with different oxidation states and two parts of the WD liver sample showing elevated copper intensities were analyzed by means of XANES. The respective copper K-edge XANES spectra of the three standards copper foil (black), copper(I) oxide (red), and copper(II) oxide (blue) and the two analyzed areas of the WD samples (both green) are shown in Fig. 3. The copper(I) spectrum features a peak at 8990 eV due to 1s → 4p electronic transitions. In contrast to this, the copper(II) spectra show a peak at 8994 eV due to 1s → 4p electronic transitions. Furthermore, the white lines are featured at 9004 eV and 9006 eV for copper(I) and copper(II), respectively. The XANES spectra of the two analyzed areas of the WD liver samples show the same characteristics and a shoulder at 8990 eV. The maxima of the first derivative, which are shown in Fig. 4, can be applied as a marker for the energy position of the edge. For copper(I) oxide, this is located at 8988 eV, while the maximum of copper(II) oxide is located at 8991 eV. For the two areas of the WD sample, the maxima of the first derivative XANES spectra are located at 8989 eV. This indicates a mixture of copper(I) and copper(II) within the liver tissue. In addition to this, two control samples were analyzed by means of XANES: a human liver of a patient with liver metastasis and a rat liver. Due to low copper concentrations within these samples in comparison to the WD liver sample, no definite copper absorption edge was detected by the applied instrumental set-up. Because of this reason, the present copper oxidation states within the samples could not be specified.To date, there has been only little knowledge about the protein system in the liver, which is responsible for the copper disposition, and it is not described in the literature, which copper oxidation states are present in WD. However, it is known that copper(I) is mostly located intracellular, while copper(II) is mostly located extracellular.31 In the liver, there are specific enzymes, which bind for example copper(I), like the protein ATP7B, which shows mutations in WD, or which bind copper(II), like the ubiquitous copper protein COMMD1, which is associated with chronic copper overload in Bedlington terriers.31 While copper(II) is probably the preferred oxidation state of copper for elimination from the cell, it is unclear whether there is a specific role of copper(II) in intracellular processes in WD.31
However, for universally valid statements about the copper oxidation state, further WD samples will have to be analyzed by means of XANES. At this point of investigation, only an indication for both oxidation states is provided. Nevertheless, this work shows the applicability of XANES for such studies.
Parallel to the determined oxidation state of the present copper species, information about the ligands and binding atoms of copper would be of interest. Binding atoms like nitrogen or sulfur could be determined by means of extended X-ray absorption fine structure spectroscopy (EXAFS) to clarify whether copper is bound to proteins. In addition to this, low concentrations within the samples as well as the availability of suitable sample materials are challenging. Concerning the last point, further investigations could be extended on an animal model for WD.
Conclusions
In this work, the suitability of μXRF and XANES for the analysis of WD liver samples was demonstrated. A coarse scan of a thin section was performed using bench-top μXRF under laboratory conditions to define a ROI showing high copper and iron intensities for further investigations. In a next step, SR-μXRF provided the elemental distribution of copper, iron, zinc, and manganese with a spatial resolution of 4 μm, enabling the analysis at the cellular level. Interestingly, areas with high copper intensities and areas with high iron and zinc intensities showed an inverse correlation. Additionally, XANES revealed the presence of copper(I) and copper(II) species within the liver sample. However, only an indication of the present oxidation states can be provided at this stage. An expansion on an animal model for WD would be useful in order to obtain a larger number of samples and statistically significant data.Abbreviations
| EDX | Energy dispersive X-ray spectroscopy |
| EXAFS | Extended X-ray absorption fine structure spectroscopy |
| LA-ICP-MS | Laser ablation-inductively coupled plasma-mass spectrometry |
| μXRF | Micro-X-ray fluorescence spectroscopy |
| ROI | Region of interest |
| SDD | Silicon drift detector |
| SR | Synchrotron radiation |
| WD | Wilson's disease |
| XANES | X-ray absorption near edge structure spectroscopy |
Acknowledgements
The authors thank the Helmholtz-Zentrum Berlin for the allocation of synchrotron radiation beam time (proposals 15101957-ST and 15202487-ST/R). Parts of this study were supported by the Ministry of Education and Research of the Federal Republic of Germany (BMBF) (Grant No. 01ZX1310B) to Axel Walch and by the Cells in Motion Cluster of Excellence (CiM – EXC 1003), Münster, Germany (project FF-2013-17), to Uwe Karst.References
- S. A. K. Wilson, Brain, 1912, 34, 295–509 CrossRef.
- A. Ala, A. P. Walker, K. Ashkan, J. S. Dooley and M. L. Schilsky, Lancet, 2007, 369, 397–408 CrossRef CAS.
- P. C. Bull, G. R. Thomas, J. M. Rommens, J. R. Forbes and D. W. Cox, Nat. Genet., 1993, 5, 327–337 CrossRef CAS PubMed.
- R. E. Tanzi, K. Petrukhin, I. Chernov, J. L. Pellequer, W. Wasco, B. Ross, D. M. Romano, E. Parano, L. Pavone, L. M. Brzustowicz, M. Devoto, J. Peppercorn, A. I. Bush, I. Sternlieb, M. Pirastu, J. F. Gusella, O. Evgrafov, G. K. Penchaszadeh, B. Honig, I. S. Edelman, M. B. Soares, I. H. Scheinberg and T. C. Gilliam, Nat. Genet., 1993, 5, 344–350 CrossRef CAS PubMed.
- Y. Yamaguchi, M. E. Heiny and J. D. Gitlin, Biochem. Biophys. Res. Commun., 1993, 197, 271–277 CrossRef CAS PubMed.
- J. D. Gitlin, Gastroenterology, 2003, 125, 1868–1877 CrossRef.
- D. Huster, Best Pract. Res., Clin. Gastroenterol., 2010, 24, 531–539 CrossRef CAS PubMed.
- E. A. Roberts and M. L. Schilsky, Hepatology, 2008, 47, 2089–2111 CrossRef CAS PubMed.
- G. Faa, V. Nurchi, L. Demelia, R. Ambu, G. Parodo, T. Congiu, R. Sciot, P. Vaneyken, R. Silvagni and G. Crisponi, J. Hepatol., 1995, 22, 303–308 CrossRef CAS PubMed.
- H. Hayashi, A. Hattori, Y. Tatsumi, K. Hayashi, Y. Katano, J. Ueyama, S. Wakusawa, M. Yano and H. Goto, Med. Mol. Morphol., 2013, 46, 133–140 CrossRef PubMed.
- B. Wu and J. S. Becker, Metallomics, 2012, 4, 403–416 RSC.
- R. Lobinski, C. Moulin and R. Ortega, Biochimie, 2006, 88(11), 1591–1604 CrossRef CAS PubMed.
- C. J. Fahrni, Curr. Opin. Chem. Biol., 2007, 11, 121–127 CrossRef CAS PubMed.
- T. Paunesku, S. Vogt, J. Maser, B. Lai and G. Woloschak, J. Cell. Biochem., 2006, 99, 1489–1502 CrossRef CAS PubMed.
- H. Kinoshita, Y. Hori, T. Fukumoto, T. Ohigashi, K. Shinohara, Y- Hayashi and Y. Ku, Med. Mol. Morphol., 2010, 43, 19–25 CrossRef PubMed.
- M. Ralle, N. J. Blackburn and S. Lutsenko, AIP Conference Proceedings 882, 13th International Conference on X-ray Absorption Fine Structure, XAFS13, 2006, 328-330.
- Z. Y. Qin, J. A. Caruso, B. Lai, A. Matusch and J. S. Becker, Metallomics, 2011, 3, 28–37 RSC.
- A. A. Dias, M. Carvalho, M. L. Carvalho and S. Pessanha, J. Anal. At. Spectrom., 2015, 30, 2488–2495 RSC.
- F. Blaske, O. Reifschneider, G. Gosheger, C. A. Wehe, M. Sperling, U. Karst, G. Hauschild and S. Höll, Anal. Chem., 2014, 86, 615–620 CrossRef CAS PubMed.
- Y. Pushkar, G. Robison, B. Sullivan, S. X. Fu, M. Kohne, W. D. Jiang, S. Rohr, B. Lai, M. A. Marcus, T. Zakharova and W. Zheng, Aging Cell, 2013, 12, 823–832 CrossRef CAS PubMed.
- Z. Savoly, G. Pepponi, P. I Nagy, C. Streli, G. Buzanich, E. Chinea-Cano and G. Zaray, X-Ray Spectrom., 2013, 42, 321–329 CrossRef CAS.
- A. Al-Ebraheem, J. Goettlicher, K. Geraki, S. Ralph and M. J. Farquharson, X-Ray Spectrom., 2010, 39, 332–337 CrossRef CAS.
- C. M. Weekley, A. Shanu, J. B. Aitken, S. Vogt, P. K. Witting and H. H. Harris, Metallomics, 2014, 6, 1602–1615 RSC.
- H. Riesemeier, K. Ecker, W. Gorner, B. R. Müller, M. Radtke and M. Krumrey, X-Ray Spectrom., 2005, 34, 160–163 CrossRef CAS.
- P. van Espen, K. Janssens and J. Nobels, Chemom. Intell. Lab. Syst., 1986, 1, 109–114 CrossRef CAS.
- B. Ravel and M. Newville, Synchrotron Radiat., 2005, 12, 537–541 CrossRef CAS PubMed.
- Y. Shiono, S. Wakusawa, H. Hayashi, T. Takikawa, M. Yano, T. Okada, H. Mabuchi, S. Kono and H. Miyajima, Am. J. Gastroenterol., 2001, 96, 3147–3151 CrossRef CAS PubMed.
- P. Ferenci, P. Steindl-Munda, W. Vogel, W. Jessner, M. Gschwantler, R. Stauber, C. Datz, F. Hackl, F. Wrba, P. Bauer and O. Lorenz, Clin. Gastroenterol. Hepatol., 2005, 3, 811–818 CrossRef CAS PubMed.
- U. Merle, M. Schaefer, P. Ferenci and W. Stremmel, Gut, 2007, 56, 115–120 CrossRef CAS PubMed.
- S. G. Boaru, U. Merle, R. Uerlings, A. Zimmermann, C. Flechtenmacher, C. Willheim, E. Eder, P. Ferenci, W. Stremmel and R. Weiskirchen, J. Cell. Mol. Med., 2015, 19, 806–814 CrossRef CAS PubMed.
- B. Sarkar and E. A. Roberts, Metallomics, 2011, 3, 20–27 RSC.
| This journal is © The Royal Society of Chemistry 2016 |