Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
High-resolution complementary chemical imaging of bio-elements in Caenorhabditis elegans†
Dominic J.
Hare‡
 ab,
Michael W. M.
Jones‡
cd,
Verena C.
Wimmer
b,
Nicole L.
Jenkins
b,
Martin D.
de Jonge
c,
Ashley I.
Bush
ab,
Michael W. M.
Jones‡
cd,
Verena C.
Wimmer
b,
Nicole L.
Jenkins
b,
Martin D.
de Jonge
c,
Ashley I.
Bush
 b and
Gawain
McColl
*b
b and
Gawain
McColl
*b
aElemental Bio-imaging Facility, University of Technology Sydney, Broadway, New South Wales 2007, Australia
bThe Florey Institute of Neuroscience and Mental Health, University of Melbourne, Parkville, Victoria 3010, Australia. E-mail: gmccoll@florey.edu.au; Tel: +61 3 9035 6608
cAustralian Synchrotron, Clayton, Victoria 3168, Australia
dARC Centre of Excellence in Advanced Molecular Imaging, La Trobe Institute for Molecular Sciences, La Trobe University, Melbourne, 3086, Australia
First published on 9th November 2015
Abstract
Here, we present a sub-μm multimodal approach to image essential elements in Caenorhabditis elegans. A combination of chemical imaging technologies reveals total metal concentration, chemical state and the protein to which an element is associated. This application of distinct yet complementary chemical imaging techniques provided unique insight into essential and trace elements at the subcellular level.
Caenorhabditis elegans is a model system that displays highly compartmentalised elemental distribution ranging from high abundance species to ultra-trace elements. When imaging these elements, abundance within the sample does not necessarily equate to high sensitivity or the capacity for high spatial resolution.1 Application of synchrotron-based X-ray fluorescence microscopy (XFM) is no exception; a number of factors determine whether an analyte can be detected and spatially mapped at subcellular resolution. These factors relate to both technical limitations and the nature of the sample itself. Considerations for imaging elements in C. elegans include: the energy of the incident beam, duration of exposure, the atomic mass of the element, the energy of the fluorescence, the characteristics of the detector used, the composition and thickness of the sample, and the environmental conditions in which the analysis takes place.2
Fluorescence profiles of specific elements must be discerned from elastic (Rayleigh) and inelastic (Compton) scattering;3i.e. element-specific fluorescence must not be obscured by scatter peak tails. The incident X-ray energy determines which excitation events can occur, but also positions the scattering peaks within the X-ray spectrum. Lower mass (Z) elements have reduced cross-sections, fluorescent yields and are easily absorbed by the sample matrix, all limiting sensitivity. While self-absorption may be negligible for heavier elements in thinner specimens, it places real limitations on low-Z elements, even within single cells.4,5 In addition, for specimens measured in air, argon (Ar) fluorescence causes a major interfering peak in the collected spectra (with associated tail) that can overwhelm the signal derived from lighter bio-elements, making their detection impossible.6 Previously, within whole animals biologically important mid-Z elements, such as calcium (Ca), zinc (Zn) and redox-active metals have often been analysed without corresponding data on low-Z elements (e.g. phosphorus (P) and sulfur (S)).
C. elegans have been a successful test bed for pushing the boundaries of microscopy,7,8 and are particularly well suited for whole-organism imaging of fundamental biochemistry. Examples span from Raman vibrational spectroscopy for imaging lipid metabolism9 to scanning electron microscopy for profiling the C. elegans connectome.10,11 We have used this nematode to extensively study metal metabolism via XFM, conducting population studies,12 tomography13 and X-ray absorption near-edge structure (XANES) spectroscopy14 to appraise the complex biochemistry of metals in vivo. C. elegans are highly resistant to ionising radiation,15 which permits analysis of hydrated and anesthetised samples for mid-Z elements using hard X-rays (>10 keV).
While hydrated imaging of anesthetised samples is preferred in principle, the water content increases the absorption of low energy fluorescence, and therefore dehydration aids detection of elements with atomic masses below potassium (K; Z < 19). Preserving subcellular distribution of elements is challenging, particularly following chemical fixation. Even brief (<30 s) formalin fixation of thin tissue sections can cause redistribution and leaching of transition metals and electrolytes.16 However, we previously demonstrated that cryofixation of C. elegans in liquid N2-cooled propane followed by lyophilisation does not cause significant variation in elemental content or subcellular distribution.12
Evolving detector capabilities, such as those of the 384-channel Maia reduces sampling overheads by collecting spectra ‘on-the-fly’ (i.e. continuously during a transit across the specimen).18 The reduced overheads allow scanning of larger samples including whole C. elegans in a practical timeframe. Furthermore, this enables spatial oversampling, where data is collected at intervals less than the full width at half maximum (FWHM) of the beam profile. Although the modulation transfer function (MTF) supports the utility of sampling at half of the beam size, in most practical cases this oversampling is not performed due to detector time limitations. Recent upgrades to the Maia detector have improved low-energy sensitivity, thus mitigating this limitation.
Here, we have combined the Maia detector (Rev C), a N2 environment for reduced Ar fluorescence, sample dehydration and incident energy selections to simultaneously image a broad range of endogenous biological elements from P to strontium (Sr) in wild type C. elegans. Specimens were mapped via XFM using 12.9 keV and 18.5 keV incident beams. Fluorescence emission was collected by the Maia detector mounted in the backscatter geometry (see ESI† for Experimental Methods). We present highly spatially resolved and correlated images of low-Z elements previously considered as ‘difficult’ analytes with respect to XFM in biology, whilst also using different modes of imaging to demonstrate the high complementarity possible using a unified imaging approach.1
Mapping biological samples in a frozen hydrated state can reduce incident beam ‘damage’, permitting radiation exposure of up to 1010 Gy, allowing the extended dwell time necessary for the collection of multiple spectra as in XANES analysis19 or oversampling. However, cryogenic conditions can be prone to artifacts4 and retain problematic water content.20 At room temperature, cryofixed and lyophilised Vicia faba (fava bean) chromosomes showed no morphological change at radiation doses of up to 107 Gy,21 suggesting cryogenic measurements may not always be necessary. Our dehydrated samples received a combined radiation dose of ∼106 Gy with no apparent morphological changes in Compton scattering used to identify microstructural features,22 or in known elemental distributions previously described.12
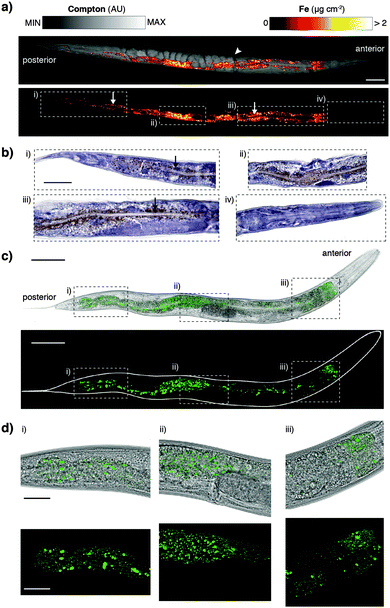
Derived Compton maps were used to relate C. elegans anatomy to all other measured elements in the hyperspectral image stack (Fig. 1a; full resolution maps available as ESI†). Sulfur was ubiquitously distributed and best recapitulated the majority of anatomical structures (Fig. 1b). Phosphorus also reproduced structural definition of the specimen, though was found at comparatively lower concentrations in the head and full extent of the tail. Consistent with expectations, both elements showed high concentrations within embryos known to be rich in lipids, phospholipids and yolk proteins required for development.
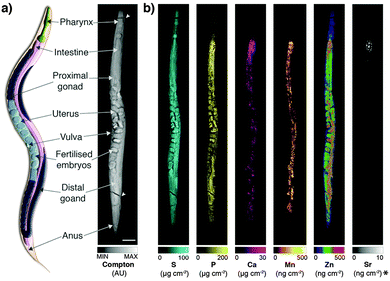 | ||
| Fig. 1 (a) Schematic representation of major anatomical structures and features of an adult hermaphrodite C. elegans (adapted from WormAtlas17) matched to corresponding features in the XFM map of Compton inelastic scattering. Two fractures in the specimen occurred during cryofixation and lyophilisation (white arrowheads). Scale bar = 50 μm. (b) Corresponding quantitative maps of S, P, Ca, Mn, Zn (all using an incident energy of 12.9 keV) and Sr (18.5 keV; marked with asterisk). | ||
As we have previously shown, Ca and manganese (Mn) were highly compartmentalised along the intestinal lumen,13 as was Zn, in addition to being rich within the gonad and embryos, consistent with Zn finger transcription factors necessary for early development. Using the higher incident energy of 18.5 keV, we also found subcellular concentrations of Sr in the most anterior intestine. Strontium commonly substitutes for Ca in biological systems at a greatly reduced concentration;23 higher sensitivity of XFM is needed to map to quantify distribution further along the intestinal tract.
Due to minimised time penalties from oversampling, the spatial resolution achieved approached that of light microscopy (approximately 200 nm24), allowing application of statistical approaches to objectively determine if true co-localisation was occurring between specific elements that are both low-Z and highly mobile in the previously hydrated specimen. Potassium and chlorine (presumably as K+ and Cl−) appeared to have highly correlated cytoplasmic distribution (Fig. 2a). Pearson and Mander's correlation measures of the merged images showed strong association (Pearson's ρ = 0.708; Mander's R = 0.926). Using Li's method for intensity correlation analysis (ICA),25 which overcomes several limitations of both Pearson and Mander's overlay comparisons, we determined the ICA quotient (Q) for the entire organism was 0.234 (+0.5 = perfect correlation; −0.5 = no correlation). Examination of both merged elements and the mapped product of difference from the mean (PDM) used to calculate the overall ICA Q showed distinct regions of K-enrichment within the gonad, as well as marked positive correlation in embryos. Interrogation of the pixel (concentration) histograms for each element showed a bimodal distribution indicative of highly enriched K in the gonad. Frequency distribution and PDM versus signal intensity (areal concentration) plots further demonstrated a skew towards areas of high K.
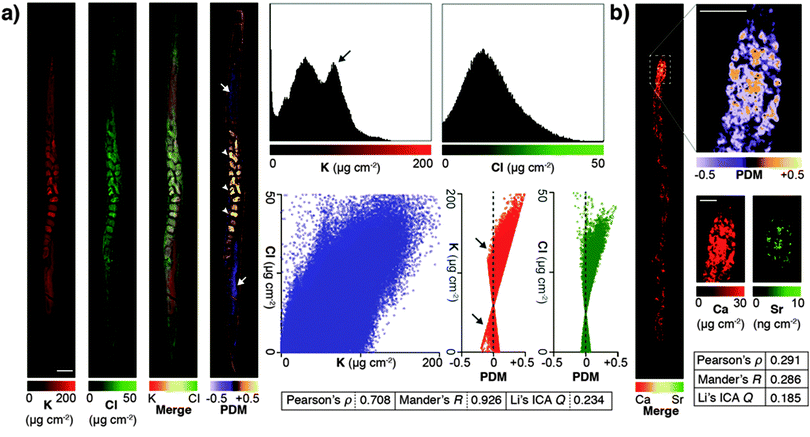 | ||
| Fig. 2 (a) High abundance and low-Z elements K and Cl were analyzed for co-localisation in an individual C. elegans specimen. Images were merged and underwent whole organism correlation analysis using Pearson's, Mander's and Li's intensity correlation analysis (ICA; inset table).25 Visualisation of the product of difference from the mean (PDM; presented on a black background) at high resolution improved interpretation by presenting the ICA quotient (Q) on a pixel-by-pixel basis. Using this method, both elements showed marked correlation in embryos (0 > Q > +0.5; white arrowheads), as well as distinct potassium-rich regions within the distal and proximal gonad (−0.5 < Q < 0; white arrows). Scale bar = 50 μm. Histograms of pixel values revealed a bimodal distribution for K, consistent with K enrichment in the gonad (black arrow). (b) The advantages of visualising correlation are clear when comparing co-localisation of high abundance Ca with low abundance Sr, which shares similar biochemistry but is close to the XFM limit of detection. Co-localisation is less robust (lower Pearson's ρ, Mander's R and ICA Q) across the whole organism (inset table); though in the anterior intestine where both Ca and Sr are most concentrated shows high spatial correlation (0 < Q < +0.5). | ||
Li's ICA Q measure is particularly useful for visualising the degree of spatial co-localisation; to demonstrate this we assessed the correlation between Ca and Sr, which within the whole organism was less distinct due to low Sr concentration (ICA Q = 0.185). In the anterior intestinal cells where Sr was detectable, we observed high correlation between Ca and Sr in the resulting PDM image (Fig. 2b), consistent with intestinal co-localisation observed in other taxa.26 These results illustrate that PDM imaging allows both spatial correlation at a sub μm level of detail, as well as the within the whole organism.
The XFM methods used here significantly improved spatial resolution for in vivo mapping. Previously we have shown a distribution of iron (Fe) about the intestine at approximately 2 μm resolution.12 Our sub-μm imaging approach permitted assessment of Fe revealing a level of detail comparable to histological staining and light microscopy. We found that punctate Fe deposits (Fig. 3a) resembled Fe distributions in formalin-fixed, paraffin-embedded sections stained using the Perls method for non-heme Fe (Fig. 3b).27 In addition to localised Fe deposits, XFM mapping also showed a more generalised Fe distribution not seen in Perls staining, indicative of heme. The correspondence between these two diverse imaging modalities suggests that both approaches accurately report on in vivo Fe. Although Perls staining is not quantitative, these results differ from those reported by Hackett et al.,16 who suggested formalin fixation alone alters Fe distribution in biological tissue. We suggest that neither radiation damage from XFM, nor extensive chemical processing for histological staining necessarily disturbs the distribution of non-heme Fe in C. elegans.
To further explore consistency between complementary imaging methods, we examined the in vivo localisation of the dominant Fe storage protein ferritin.28 Using a green fluorescent protein (GFP) fusion to ferritin we compared high-resolution confocal fluorescence in vivo microscopy to XFM mapping and Perls staining (Fig. 3c and d). Distribution of GFP fluorescence, and thereby ferritin localisation, was again remarkably similar to the Fe puncta previously imaged (see Movie, ESI†). Ferritin accounts for almost half of the Fe content of C. elegans and therefore represents a good proxy for non-heme Fe. The multiple imaging methods used serve as validation of each respective technique, providing the first consistent representation of subcellular Fe within a whole organism. Future directions of this complementary imaging approach could employ C. elegans with mutated genes that affect Fe metabolism, as well as ageing studies (such as those described in James et al.14) to exploit the higher resolution mapping protocol described here. Here we focused on Fe to demonstrate complementary imaging of metal distribution; similar studies could employ the genetically encoded fluorescent calcium sensor GCaMP,29 which has been used in the C. elegans model system,30 to compare total body Ca concentration with cell-specific Ca2+ content.
In summary, we have demonstrated sub-μm XFM mapping of bio-elements, both rare and ubiquitous, ranging from low-Z to highly abundant transition metals in a model organism ideally suited for studying metal metabolism. Mapping low-Z elements with confidence will facilitate new experimental paradigms. With image resolution approaching that of light microscopy, implementation of high-level correlative image analysis methods previously reserved for standard fluorescence microscopy are now within reach of contemporary XFM. We also demonstrated how multiple modality imaging provides a greater appreciation of subcellular metal distribution. Using both well-established histochemical analysis and confocal fluorescence imaging provides a more comprehensive picture of Fe distribution within C. elegans, an essential element for development and health. A unified approach to imaging using multiple methods accessible to chemists and biologists alike will permit further advances in understanding metal biochemistry.
Acknowledgements
An Australian Research Council Discovery Project (DP130100357) to A. I. B., G. M. and M. D. J. and a UTS Chancellor's Postdoctoral Fellowship to D. J. H. supported this work. We would like to thank the XFM beamline at the Australian Synchrotron, Kirsten Grant, Fransisca Sumardy and Ian Birchall (Florey) for technical assistance, the Caenorhabditis Genetics Center (CGC) supported by the US National Institutes of Health – Office of Research Infrastructure Programs (P40 OD010440) for providing strains, and the Victorian Government's Operational Infrastructure Support Program.Notes and references
- D. J. Hare, E. J. New, M. D. de Jonge and G. McColl, Chem. Soc. Rev., 2015, 44, 5941–5958 RSC.
- E. Lombi, M. D. Jonge, E. Donner, C. G. Ryan and D. Paterson, Anal. Bioanal. Chem., 2011, 400, 1637–1644 CrossRef CAS PubMed.
- M. J. Pushie, I. J. Pickering, M. Korbas, M. J. Hackett and G. N. George, Chem. Rev., 2014, 114, 8499–8541 CrossRef CAS PubMed.
- J. Deng, D. J. Vine, S. Chen, Y. S. G. Nashed, Q. Jin, N. W. Phillips, T. Peterka, R. Ross, S. Vogt and C. J. Jacobsen, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2314–2319 CrossRef CAS PubMed.
- S. Majumdar, J. R. Peralta-Videa, H. Castillo-Michel, J. Hong, C. M. Rico and J. L. Gardea-Torresdey, Anal. Chim. Acta, 2012, 755, 1–16 CrossRef CAS PubMed.
- L. Finney, Y. Chishti, T. Khare, C. Giometti, A. Levina, P. A. Lay and S. Vogt, ACS Chem. Biol., 2010, 5, 577–587 CrossRef CAS PubMed.
- B.-C. Chen, W. R. Legant, K. Wang, L. Shao, D. E. Milkie, M. W. Davidson, C. Janetopoulos, X. S. Wu, J. A. Hammer III, Z. Liu, B. P. English, Y. Mimori-Kiyosue, D. P. Romero, A. T. Ritter, J. Lippincott-Schwartz, L. Fritz-Laylin, R. D. Mullins, D. M. Mitchell, J. N. Bembenek, A.-C. Reymann, R. Böhme, S. W. Grill, J. T. Wang, G. Seydoux, U. S. Tulu, D. P. Kiehart and E. Betzig, Science, 2014, 346, 1257998 CrossRef PubMed.
- M. Chalfie, Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, Science, 1994, 263, 802–805 CAS.
- P. Wang, B. Liu, D. Zhang, M. Y. Belew, H. A. Tissenbaum and J. X. Cheng, Angew. Chem., Int. Ed., 2014, 53, 11787–11792 CrossRef CAS PubMed.
- D. Hall, E. Hartwieg and K. Nguyen, in Caenorhabditis elegans: Cell Biology and Physiology, ed. J. Rothman and A. Singson, Academic Press, New York, 2012 Search PubMed.
- M. Xu, T. A. Jarrell, Y. Wang, S. J. Cook, D. H. Hall and S. W. Emmons, PLoS One, 2013, 8, e54050 CAS.
- S. A. James, M. D. de Jonge, D. L. Howard, A. I. Bush, D. Paterson and G. McColl, Metallomics, 2013, 5, 627–635 RSC.
- G. McColl, S. A. James, S. Mayo, D. L. Howard, G. F. Moorhead, D. Paterson, M. D. de Jonge and A. I. Bush, PLoS One, 2012, 7, e32685 CAS.
- S. A. James, B. R. Roberts, D. J. Hare, M. D. de Jonge, I. E. Birchall, N. L. Jenkins, R. A. Cherny, A. I. Bush and G. McColl, Chem. Sci., 2015, 6, 2952–2962 RSC.
- J. B. Weidhaas, D. M. Eisenmann, J. M. Holub and S. V. Nallur, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9946–9951 CrossRef CAS PubMed.
- M. J. Hackett, J. A. McQuillan, F. El-Assaad, J. B. Aitken, A. Levina, D. D. Cohen, R. Siegele, E. A. Carter, G. E. Grau, N. H. Hunt and P. A. Lay, Analyst, 2011, 136, 2941–2952 RSC.
- Z. Altun and D. Hall, in WormAtlas, http://www.wormatlas.org/hermaphrodite/hermaphroditehomepage.htm, 2015.
- C. G. Ryan, D. P. Siddons, G. Moorhead, R. Kirkham, G. De Geronimo, B. E. Etschmann, A. Dragone, P. A. Dunn, A. Kuczewski, P. Davey, M. Jensen, J. M. Ablett, J. Kuczewski, R. Hough and D. Paterson, J. Phys.: Conf. Ser., 2009, 186, 012013 CrossRef.
- T. Bacquart, G. Devès, A. Carmona, R. Tucoulou, S. Bohic and R. Ortega, Anal. Chem., 2007, 79, 7353–7359 CrossRef CAS PubMed.
- R. Tjallingii, U. Röhl, M. Kölling and T. Bickert, Geochem., Geophys., Geosyst., 2007, 8, Q02004 CrossRef.
- S. Williams, X. Zhang, C. Jacobsen, J. Kirz, S. Lindaas, J. Hof and S. S. Lamm, J. Microsc., 1993, 170, 155–165 CrossRef.
- M. D. de Jonge and S. Vogt, Curr. Opin. Struct. Biol., 2010, 20, 606–614 CrossRef CAS PubMed.
- R. Wasserman, The Transfer of Calcium and Strontium Across Biological Membranes, Academic Press, New York, 2012 Search PubMed.
- S. Goodman, Medical Cell Biology, Academic Press, New York, 2007 Search PubMed.
- Q. Li, A. Lau, T. J. Morris, L. Guo, C. B. Fordyce and E. F. Stanley, J. Neurosci., 2004, 24, 4070–4081 CrossRef CAS PubMed.
- N. Sugihira, E. Kobayashi and K. T. Suzuki, Biol. Trace Elem. Res., 1990, 25, 79–88 CrossRef CAS PubMed.
- G. Orchard and B. Nation, Histopathology, Oxford University Press, Oxford, 2011 Search PubMed.
- K. Honarmand Ebrahimi, P.-L. Hagedoorn and W. R. Hagen, Chem. Rev., 2015, 115, 295–326 CrossRef CAS PubMed.
- J. Nakai, M. Ohkura and K. Imoto, Nat. Biotechnol., 2001, 19, 137–141 CrossRef CAS PubMed.
- P. T. McGrath, M. V. Rockman, M. Zimmer, H. Jang, E. Z. Macosko, L. Kruglyak and C. I. Bargmann, Neuron, 2009, 61, 692–699 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental methods, figures and movie. See DOI: 10.1039/c5mt00288e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2016 |