Design and functionalization of responsive hydrogels for photonic crystal biosensors
Sukwon
Jung
,
Joel L.
Kaar
* and
Mark P.
Stoykovich
*
Department of Chemical and Biological Engineering, University of Colorado, Boulder 80303, USA. E-mail: joel.kaar@colorado.edu; mark.stoykovich@colorado.edu
First published on 10th May 2016
Abstract
Photonic crystals embedded in analyte responsive hydrogels provide a powerful platform for biosensing and have benefitted from recent advances in polymer synthesis and functionalization, nanofabrication, and optical characterization. In this review, state-of-the-art techniques for the fabrication and functionalization of photonic crystal hydrogels are presented, along with a fundamental description of how such materials transduce detection of a target analyte into a simple optical readout. Specific biosensing applications using photonic crystal hydrogels are highlighted, including for the detection of DNA hybridization and single-site DNA modifications, enzyme activity, and small molecule metabolites. Moreover, perspectives on using molecular-level interactions to design and engineer more sensitive or more selective biosensors, as well as future directions in this expanding field, are discussed.
Design, System, ApplicationAs a novel platform for the development of high-throughput and point-of-care biosensors, photonic crystal hydrogels hold considerable promise. Such materials consist of periodic nanostructures embedded within a responsive polymer network, and undergo changes in physical and optical properties in the presence of a target molecule. Due to the visual readout, simple design, and molecular-level customization, these materials have potential widespread utility for the detection of a broad spectrum of target biological molecules, ranging from ionic species, metabolites, nucleic acids, enzymes, and small molecules that alter biological function, including pesticides and nerve agents. To date, there has been considerable advancements in the design, fabrication, and functionalization of photonic crystal hydrogels, leading to far-reaching improvements in sensitivity and selectivity. In light of such advancements, exciting directions in the molecular design and implementation of these materials have emerged, leading to new opportunities to exploit the advantages of this biosensing platform. |
1. Introduction
Biosensing is a rapidly expanding field, with commercial applications in medical diagnostics and personal healthcare, security and defense, and environmental detection.1–4 The diversity in the design, operation, and sophistication of biosensors is commensurate with applications ranging from rapid, portable, low-cost screening (e.g., handheld blood glucose meters, dipsticks for urinalysis, and lateral flow assays)5 to highly quantitative and selective measurements (e.g., enzyme-linked immunosorbent assays (ELISA), polymerase chain reaction (PCR)-based assays, and nanomaterial-based scanometric assays).6 In general, recognition of the target analyte is transduced into an optical/colorimetric, electronic, magnetic, or mechanical signal.3,7–9 In order to be useful, such a sensor should be highly-selective toward the target analyte in complex environments, interference-free, sensitive to low analyte concentrations (e.g., ppm or ppb levels), and have a wide operational range; however, in practice, there is usually tradeoff amongst these features. The complexity and specialization of each biosensor must also be considered, particularly as they are translated from the benchtop to commercial devices. Of particular interest is the development of sensing “platforms” that are broadly applicable to a wide spectrum of analytes and can readily be used in field or lab-based settings.In this review, we focus on recent advances to a biosensing platform that is based on photonic crystals embedded in analyte-sensitive hydrogels. Photonic crystals with two-dimensional (2D) or three-dimensional (3D) structures provide a periodic dielectric in a low-loss medium that is capable of manipulating or controlling light, often in the visible spectral range, such that particular colors are reflected or transmitted.10,11 Swelling or shrinking of the hydrogel, in response to the analyte of interest, induces changes in the structure of the photonic crystal that are thus transduced into an optical response.10–12 Although such sensor designs were originally demonstrated for the detection of ionic and small molecule species (e.g., Cu2+, Co2+, Ni2+, Zn2+, Hg2+, Pb2+, H+, glucose, ammonia, surfactants, creatinine, and parathion),13–20 the direct detection of biomolecules and biomolecule function, including enzyme activity, represents a new and exciting application space for this sensing approach. As summarized here, biosensors based on photonic crystal hydrogels are particularly well-suited for the detection of metabolites, biomacromolecules (such as proteins and DNA), and even cells and the secretion of extracellular products. Notably, the transduction of the detector signal into a simple optical readout by the photonic crystal may be useful for point-of-care diagnostics.12 Another attractive aspect of photonic crystal hydrogel biosensors is that they permit label-free detection of analytes, thereby eliminating the requirement of exogenous labels or reagents.21–23 This unique aspect of this sensing approach, which may be implemented in multiplexed formats for high-throughput drug or metabolite screening, may constitute a significant advantage over conventional bioassays.
Throughout this review, molecular-level aspects of the design and functionalization of the photonic crystal hydrogels are highlighted as enabling features in the development of novel biosensor assays. Such hydrogels may specifically be designed to respond to diverse stimuli and molecular interactions that allow for the detection of biological activity or biochemical interactions,24,25 as well as changes to physical structure or mechanical forces,26,27 molecular configurations,28–31 ionic environments,16,32,33 or even hydrogen bonding.34–36 One significant advantage of hydrogel-based biosensors is their ready functionalization with recognition molecules, including enzymes,37,38 antibodies,39,40 aptamers,41,42 and nucleic acids43,44 using conventional bioconjugation strategies. Additionally, the hydrogel component itself may be rationally tuned such as by altering polymer composition to respond to local changes in chemical environment without functionalization.18,34,36 The following sections summarize the design and fabrication of photonic crystal hydrogel biosensors as well as the mechanisms by which molecular interactions between the analyte and hydrogel may be exploited to elicit a detectable response.
2. Design and fabrication of photonic crystal hydrogel sensors
Biosensors with photonic crystals embedded in a hydrogel matrix may be categorized broadly by the types of periodic nanostructures that are used. The most common types of photonic crystal biosensors are based on: 1) crystalline colloidal arrays, 2) inverse opal assembly, and 3) holograms. In this section, the design and fabrication approaches for each of these photonic crystal hydrogel platforms, as well as general strategies for their functionalization with target recognition molecules, are reviewed. Additionally, emerging approaches for the preparation of block copolymer-based photonic crystal hydrogels have recently garnered attention as a sensing platform and will also be highlighted.2.1. Crystalline colloidal array hydrogels
Crystalline colloidal arrays (CCAs) are 3D photonic crystals formed via the electrostatic self-assembly of highly-charged and monodisperse nanoparticles in concentrated stabilized suspensions.45–47 Emulsion polymerization methods are typically used to synthesize crosslinked polymer beads (e.g., of polystyrene (PS) or poly(methyl methacrylate) (PMMA)) that are 70–200 nm in diameter with surface charge densities of 1–6 μC cm−2.48–50 The periodic nanostructures that result from the spontaneous formation of the CCAs can diffract incident light at specific wavelengths specified by their lattice spacing and crystal structure. Depending on the nanoparticle size, suspension concentration, and processing conditions, either face centered cubic (FCC) or body centered cubic (BCC) crystals may be self-assembled that minimize the total free energy of the system, with the FCC structure being favored in concentrated suspensions of large nanoparticles (>8 v/v% and >100 nm in diameter) and the BCC structure being favored in dilute suspensions of small nanoparticles (<3 v/v% and <80 nm in diameter).45,51–53 Moreover, the CCA structure and particle spacing may be finely tuned via control of the particle size, charge density, and solution concentration.45,54The CCAs may be encapsulated in hydrogel matrices, as shown in Fig. 1a, by self-assembly of the nanoparticle structures in the presence of hydrogel precursors and subsequent initiation of radical polymerization of the crosslinked hydrogel networks by exposure to heat or UV light.55–57 Often the CCA nanoparticles are themselves functionalized such that they are covalently linked into the hydrogel network during polymerization,25 which assists in immobilizing the CCA structure and prevents leaching of the nanoparticles from the hydrogel, including during swelling or shrinkage. Since assembly of the CCA is driven by electrostatic interactions, the selection of the monomer, photoinitiator, and environment (e.g., ionic strength) is critical. In light of this, mixtures of the CCAs and hydrogel precursors are commonly equilibrated with ion-exchange resins to remove any charged species other than those on the CCA nanoparticles (i.e., from unreacted monomer from the preparation of the colloidal nanoparticles, inhibitors, or the aqueous solvent) prior to self-assembly. Additionally, many water soluble photoinitiators or charged monomers, such as acrylic acid or azide-functionalized monomers that might serve as reactive groups for functionalization of the hydrogels post-preparation, may disrupt CCA assembly and thus are generally avoided as precursors for hydrogel synthesis. The most common monomer and crosslinker utilized for CCA hydrogel fabrication are acrylamide and N,N′-methylenebisacrylamide, respectively. As discussed below, such acrylamide-based hydrogels have been extensively used as a platform for hydrogel functionalization with target recognition moieties, either via post-polymerization modification or copolymerization of acrylamide with uncharged but reactive monomeric species (e.g., with alkynes, epoxides, or isocyanates).
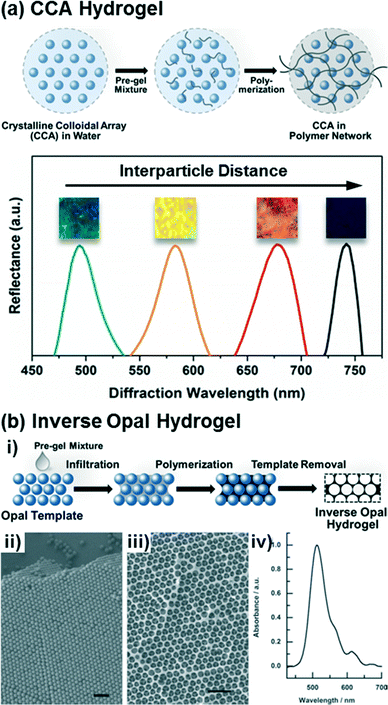 | ||
| Fig. 1 Preparation of CCA and inverse opal based photonic crystal hydrogels. (a) Scheme of the fabrication procedure for CCA hydrogels and representative diffraction spectra of the CCA hydrogels depending on the crystal lattice spacing. Adapted from ref. 25. Copyright (2014) American Chemical Society. (b) Scheme of the fabrication procedure for inverse opal hydrogels (i), SEM images of an opal template (ii) and its corresponding inverse opal structure (iii), and a diffraction spectrum of the inverse opal hydrogel (iv). Adapted from ref. 58. Copyright (2012) WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
2.2. Inverse opal hydrogels
Inverse opal hydrogels are constructed via opal templating methods59–62 as shown in Fig. 1b. Initially, in order to construct the opal template, monodisperse nanoparticles (typically silica, PS, or PMMA) are close-packed on substrates (Fig. 1b,ii) mainly via a vertical deposition method.54,61 The rate of deposition, dictated in part by the nanoparticle concentration in solution and the solvent volatility, must be slow to allow the formation of well-packed and defect-free crystal structures. Following deposition of the nanoparticles, the interstitial voids between particles of the opal templates are infiltrated with a pre-gel mixture consisting of monomers and crosslinkers by capillary action. Unlike the CCA hydrogels, the inverse opal hydrogels can be constructed with ionic monomers that can be utilized as recognition molecules60,63,64 or conjugation handles for further functionalization.65 The filled opal templates are then polymerized and the opal templates are selectively removed from the polymerized hydrogel matrices through wet-etching processes, resulting in periodic nanosized voids in the hydrogel (Fig. 1b,iii). The as-prepared inverse opal hydrogels can diffract specific wavelengths (Fig. 1b,iv) due to the periodicity and regularity of the opal structures. Similarly to the CCA hydrogels, the optical response of the inverse opal hydrogels may be reported as a relative change in the diffraction wavelength that arises due to changes in the lattice spacing. It is important to note that the optical response from inverse opal structures in hydrogels differ from inverse opals in rigid inorganics, such as TiO2 and SiO2, in which the voids become filled with the analyte solution and changes in the effective index of refraction of the material induce an optical response.66–69 Although the void structure may diminish the mechanical integrity of inverse opal hydrogels relative to CCA hydrogels, the continuous macropores of the inverse opal hydrogels may allow for less hindered diffusion of molecules (i.e., target analyte) and thus improve sensitivity and response time.2.3. Holographic hydrogels
Holography allows for the photopatterning of periodic nanostructures (i.e., one-dimensional photonic crystals consisting of multilayer Bragg diffraction gratings) in hydrogel matrices.21,70,71 In one implementation, the Bragg diffraction gratings for holographic hydrogel sensors are composed of silver nanoparticle stacks (Fig. 2a,i) created via photochemical patterning using laser interferometry (Fig. 2a,ii). Briefly, this holographic photopatterning process involves diffusion of a silver ion precursor into the polymer matrices (steps 3–4 in Fig. 2a,ii), silver halide formation (steps 5–6), laser exposure of an interference pattern using a Lloyd's mirror approach (step 7), development for silver nanoparticle formation (step 8), and post-processing steps to eliminate any unreacted precursors and to improve the diffraction efficiency (steps 9–11). Compared to other photonic crystal hydrogels, the periodic diffraction gratings of the holographic hydrogels can be accurately and easily controlled, permitting the optical properties of such hydrogels to be readily tuned.21,71,72 Two- or three-dimensional holographic polymer structures may also be defined, if desired, using multi-beam patterning techniques.73–75 In contrast to the self-assembly processes used to form CCA or inverse opal hydrogels, the multiple processing steps and patterning techniques required to generate holographic hydrogels present an inherent challenge. Recently, these challenges have largely been overcome by the emergence of laser ablation and photopolymerization methods for structuring holographic hydrogels (see the recent review ref. 21 for details of these methods).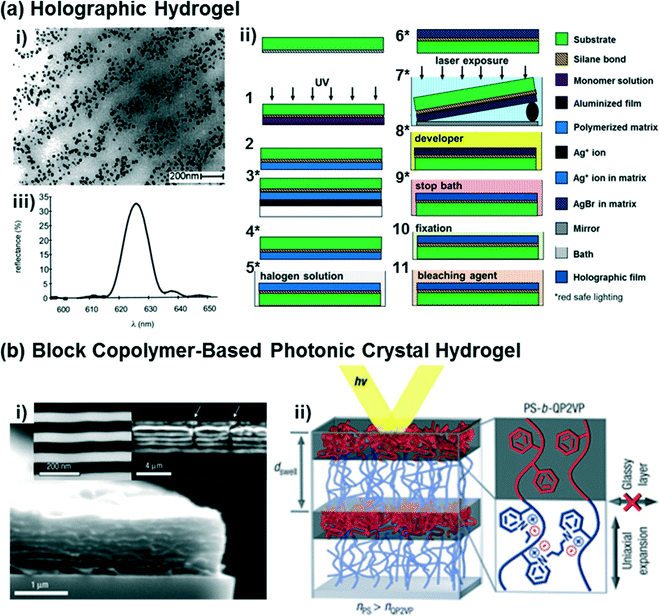 | ||
| Fig. 2 (a) A representative TEM image of holographic hydrogels consisting of periodic layers of silver nanoparticles (dark dots) embedded in a hydrogel matrix (i), as fabricated using a Lloyd's mirror interferometry approach that exposes regular grating patterns in the hydrogel swollen with silver halide precursors (ii). A representative diffraction spectrum from the holographic hydrogels is shown in (iii). Adapted from ref. 21 and 76. Copyright (2002 & 2014) American Chemical Society. (b) Representative SEM images of a diblock copolymer-based photonic crystal hydrogel that forms lamellar sheets (i), and a schematic diagram of the polystyrene-b-quaternized poly(2-vinyl pyridine) molecular structure and its optical response based on the self-assembled structure (ii). Adapted from ref. 77. Copyright (2007) Nature Publishing Group. | ||
2.4. Block copolymer-based photonic crystal hydrogels
Recently, diblock copolymers have been exploited to construct multilayer Bragg diffraction gratings.77–80 Block copolymers consist of two or more chemically distinct polymer chains that are covalently linked to each other,81 with great chemical and structural diversity being accessible at the molecular-level. Block copolymers self-assemble into well-ordered nanostructures (e.g., spheres, cylinders, or lamella)82,83 with control over the domain periodicity and refractive index contrast, such that such materials can be used for the rapid fabrication of photonic crystals.77,79,80,84 As one example, a periodic lamellar structure was constructed via self-assembly of a diblock copolymer consisting of hydrophobic and hydrophilic polyelectrolyte blocks (polystyrene-b-quaternized poly(2-vinyl pyridine), PS-b-QP2VP),77 as shown in Fig. 2b. The PS block provides the overall structural integrity to the material and prevents it from dissolving, while in aqueous solvents swelling of the QP2VP layers in the lamellar structured gel film (Fig. 2b,ii) modulates both the domain spacing and the refractive index contrast, and thus enables diffraction of the incident light. While the strategy of using the self-assembly of block copolymers enables the rapid fabrication of multilayer diffraction gratings, there are limitations that must be addressed for sensing applications, including low diffraction efficiency, structure defectivity, and variability in the domain sizes (i.e., lattice spacings).84 Another significant challenge in using block copolymers for self-assembling photonic materials is in achieving structures that are large enough in dimension to interact with the incident light, particularly at visible wavelengths. Approaches for synthesizing and self-assembling bottlebrush block copolymers85 into photonic materials are being pursued to address this challenge.86–882.5. Functionalization of photonic crystal hydrogels
To exploit photonic crystal hydrogels as platforms for biosensing, they must be sensitive and responsive to target analytes while maintaining their ability to display colors indicative of their structure. This is generally achieved by functionalizing the hydrogels with recognition molecules that interact specifically or non-specifically with the target analytes. The recognition molecules (detailed further in section 4) range from small molecules and crown ethers to complex biomolecules such as peptides, DNA, and proteins. The most common technique for functionalizing the hydrogels is direct copolymerization of recognition molecules having polymerizable groups (e.g., vinyl, acryloyl, methacryloyl, or allyl groups) into the hydrogel network.14,76,89–92 Although monomers functionalized with biomolecules may be copolymerized into the hydrogel network, copolymerization may result in structural changes (e.g., changes in secondary or tertiary of proteins or enzymes) that diminish analyte binding. Such changes may specifically be induced by radicals,93 UV light,94 and/or high temperatures95 during polymerization.An alternative approach for the incorporation of recognition molecules into photonic crystal hydrogels entails post-polymerization conjugation to the hydrogel network.24,25,96,97 This approach requires the initial hydrogels to have some reactive functional groups to which the recognition molecules may be attached. As one example, acrylamide-based hydrogels can be hydrolyzed to substitute chemically reactive carboxylic acid groups for inert amide ones.25,98 Hydrogels may also be copolymerized with well-defined concentrations of monomers containing chemically reactive functional groups (e.g., alkynes, carboxylic acids, or epoxides).24,64,97,99 Multiple and chemically orthogonal functional groups may even be incorporated into the same hydrogel. For example, amine-containing molecules may be conjugated with carboxylic acid-containing hydrogels using EDC/NHS coupling chemistry19,20,25 and, separately, azide-containing molecules can be directly conjugated with alkyne-containing hydrogels via the highly selective and rapid click cycloaddition reaction.97 Diverse combinations of conjugation chemistries for the functionalization of hydrogels after polymerization are available, which have been described in detail elsewhere.100–102
3. Optical response of photonic crystal hydrogels: theoretical basis and molecular engineering
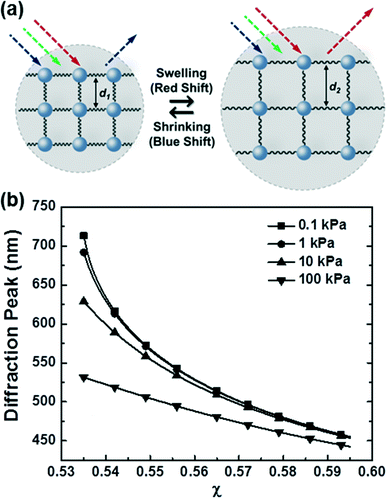
For biosensing applications, swelling or shrinking of the hydrogel in response to analyte recognition can trigger a visual change in the optical properties of the embedded photonic crystal by modifying its structure. As shown in the schematic diagram in Fig. 3a, photonic crystal hydrogels diffract certain wavelengths of incident light and adopt colors based on their structure. The wavelengths that constructively interfere and form a diffraction pattern in reflection satisfy the Bragg–Snell equation mλ = 2d(neff2 − cos2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ)1/2,103–105 where m is the order of diffraction, λ is the wavelength of the diffracted light, d is the distance between diffracting planes, neff is the mean effective refractive index of the photonic crystal hydrogel, and θ is the angle of the incident light as measured with respect to the hydrogel surface normal. Photonic crystals that undergo volumetric changes, as induced in response to interactions with target analytes or mechanical stresses, adopt different lattice spacing (d in Fig. 3a) and therefore preferentially diffract incident light at different wavelengths, with the colorimetric response red- or blue-shifted with swelling or shrinking, respectively.11 The optical response of photonic crystal hydrogels that are unconstrained and isotropic is directly related to the overall volumetric changes as follows:
θ)1/2,103–105 where m is the order of diffraction, λ is the wavelength of the diffracted light, d is the distance between diffracting planes, neff is the mean effective refractive index of the photonic crystal hydrogel, and θ is the angle of the incident light as measured with respect to the hydrogel surface normal. Photonic crystals that undergo volumetric changes, as induced in response to interactions with target analytes or mechanical stresses, adopt different lattice spacing (d in Fig. 3a) and therefore preferentially diffract incident light at different wavelengths, with the colorimetric response red- or blue-shifted with swelling or shrinking, respectively.11 The optical response of photonic crystal hydrogels that are unconstrained and isotropic is directly related to the overall volumetric changes as follows: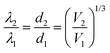 | (1) |
 | ||
| Fig. 3 Optical response of photonic crystal hydrogel sensors. (a) Schematic diagram describing the optical response of a photonic crystal hydrogel in response to changes in its volume. (b) Theoretically expected optical response of photonic crystal hydrogels as a function of the polymer-solvent interaction parameter χ and the shear modulus of the hydrogel. Adapted from ref. 97. Copyright (2015) American Chemical Society. | ||
The optical response can thus be described by Flory–Rehner theory for hydrogel swelling106 in which the hydrogel volume at equilibrium (V) is determined by balancing the osmotic pressure across the hydrogel–environment interface.
| ΠT = Πion + ΠM + ΠE = 0 | (2) |
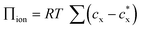 | (3) |
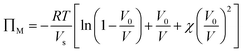 | (4) |
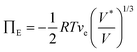 | (5) |
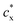 are the mobile ion concentrations for species x inside and outside the hydrogels, respectively. In addition, Vs represents the molar volume of the solvent and V0 and V* are the hydrogel volume in the dry and unstrained states, respectively.106
are the mobile ion concentrations for species x inside and outside the hydrogels, respectively. In addition, Vs represents the molar volume of the solvent and V0 and V* are the hydrogel volume in the dry and unstrained states, respectively.106
According to this theory, the Donnan potential induced by the immobilized ion concentration within the hydrogels may change upon interaction with target analytes and provides a driving force (i.e., change in osmotic pressure Πion) that elicits the hydrogel to swell or shrink. The Donnan potential may be manipulated by changes in the total concentration of the immobilized ions or the ionic strength of the solvent medium for a greater optical response. Moreover, material parameters of the hydrogels, including the polymer-solvent interaction parameter (χ) and the crosslinking density (ve), play a crucial role in determining the hydrogel volume at equilibrium. First, the χ parameter describes the thermodynamic preference of the polymer networks to interact with the solvents, with smaller values of χ indicative of more favorable polymer–solvent interactions compared to the polymer–polymer interactions and resulting in swollen gels that are more sensitive and optically responsive to changes in the polymer or solvent environment. Next, the crosslinking density is proportional to the shear modulus of the hydrogel (G′ = veRT ϕ1/3 where G′ and ϕ are the shear modulus and the polymer volume fraction, respectively)106 and the elastic restoring force that resists gel deformation away from the unstrained state. Hydrogels with a lower extent of crosslinking density (i.e., lower moduli) therefore have greater volumetric changes and are more sensitive to external stimuli.
Combined, the Flory–Rehner equation for hydrogel swelling and the Bragg–Snell equation for diffraction provide a theoretical basis for the molecular-level design of photonic crystal hydrogels with high sensitivity and desirable optical responses (e.g., responses in the visible or near-IR range) for biosensing. Tuning of the photonic crystal hydrogels may be accomplished, in part, by rational manipulation of the hydrogel properties and the sensing environment. For example, the effect of χ and shear modulus on the optical response of photonic crystal hydrogels is predicted in Fig. 3b,97 illustrating how the optical response of the photonic crystal can be readily tuned. The calculated results show a significant shift in the diffraction wavelength from the unstrained state, from blue to red colored materials, with decreasing χ or shear modulus that lower the energetic penalty for hydrogel swelling. Over the range that was examined, the optical response is more sensitive to changes in the polymer-solvent interactions than to the shear modulus, which allows even subtle modifications to the polymer or solvent quality to be detectable. In practice, these properties of the hydrogel may be modulated by the crosslink density and the composition of the monomeric constituents; the crosslink density may most directly be modulated via changes in the monomer concentrations in a pre-gel solution with a fixed crosslinker concentration and χ may be tuned by copolymerization with more or less hydrophilic monomers than the majority monomer (e.g., N-hydroxyethyl acrylamide or N-tert-butylacrylamide relative to acrylamide). Similarly, the functionalization of the polymer networks with recognition molecules for specific target detection (i.e., selectivity) has also to be considered in the design of the sensors based on photonic crystal hydrogels, as this functionalization may significantly affect the polymer-solvent interactions, the mechanical properties, or the immobilized charge in the hydrogels and thus the optical response.
4. Engineering of photonic crystal hydrogels for biosensing
In this section, we provide a focused review of the state-of-the-art application of photonic crystal hydrogels to biosensing, specifically highlighting the biological aspects of the target analytes or recognition molecules, the molecular-level design and functionalization of the sensors, and the fundamental basis for the optical response. Reviews on a broader range of sensing applications with photonic crystal hydrogels, for example for detecting temperature changes, pH, ionic strength, humidity, solvents, or pressure, are available elsewhere.10,11,21,1074.1. Detection of metabolites, intracellular molecules, and toxins
Photonic crystal hydrogel sensors have demonstrated success in the detection and quantification of small-molecule chemicals of importance in personal health,17,19,20,38,108–110 food monitoring,36,58,90 and environmental screening.15,90,111Table 1 summarizes the detection approaches and capabilities of some photonic crystal hydrogel biosensors designed to target metabolites, intracellular molecules, and toxins. In the simplest conception, photonic crystal hydrogels may quantify the concentration of organic solvents miscible in water (e.g., methanol, ethanol, 1-propanol, or 1-butanol) by utilizing the volumetric modulations that arise from non-specific changes to the polymer-solvent interaction parameter.35,36,112 Glucose, other secreted metabolites, and intracellular molecules may be detected in a similar manner that relies on non-specific interactions between the analyte and the hydrogel.| Target analyte | Recognition molecule/strategy | Mechanism of hydrogel response | Sensitivity | Dynamic range | Ref. | |
|---|---|---|---|---|---|---|
| a Dissolved gas. b Gas. c With stabilizer poly(ethylene glycol) or 15-crown-5. d With high PBA loading density, e With stabilizer (3-acrylamidopropyl)trimethylammonium chloride. f Low ionic strength. g High ionic strength. h In 130 mM and 150 mM Na+ background. i Bacterial growth monitoring. j In tear fluid. k In urine. | ||||||
| CCA sensor | Cd2+ | Thiourea | Crosslinking | 0.01 mM | <10 mM | 90 |
| Hg2+ | Aptamer | Crosslinking | 10 nM | <0.1 mM | 125 | |
| Pb2+ | Crown ether (18-crown-6) | Donnan potential | 0.1 μMf | <20 mMf | 14 | |
| Donnan potential | 0.5 μMg | <2.5 mMg | 108 | |||
| Aptamer | Crosslinking | 1 nM | <1 mM | 125 | ||
| Ammonia | Berthelot reaction | Crosslinking | 50 μM | <0.35 mM | 17 | |
| Glucose | Phenylboronic acid (PBA) | Donnan potential | 50 μMf | <40 mMf | 117 | |
| Crosslinkingc | 0.2 mMg | <10 mMg | 109 | |||
| Crosslinkingd | 0.15 mMg | <5 mMg | 121 | |||
| Glycine | Tetracycline–copper(II) complex | Donnan potential | 0.01 nM | <10 mM | 126 | |
| Inverse opal sensor | Hg2+ | Carboxylic acid | Crosslinking | 10 nM | <1 mM | 63 |
| Acetate | Thiourea | Donnan potential | 1 mM | <10 mM | 122 | |
| CO2a | Tertiary amine | Donnan potential | 0.2% | <100% | 123 | |
| Cyanide | Trifluoroacetyl | Donnan potential (major) χ parameter (minor) | 0.1 μM | <1 mM | 58 | |
| Glucose | PBA | Donnan potential | 0.1 mM | <100 mM | 118 | |
| Holographic sensor | Divalent metal ions | Iminodiacetic acid | Crosslinking | ∼5 mM | <40 mM | 13 |
| K+ | Crown ether (18-crown-6) | Donnan potential | 1 mMh | <33 mMh | 76 | |
| Pb2+ | 8-Hydroxyquinoline | Crosslinking | 0.1 mM | <10 mM | 127 | |
| Ammoniab | Sulfonate | Crosslinking | 0.19% | <12.5% | 128 | |
| Glucose | PBA | Donnan potential | 2 mMi | <11 mMi | 119 | |
| Crosslinkinge | 1 mM | <6 mM | 129 | |||
| Crosslinkinge | 5 mMj | <20 mMj | 110 | |||
| Donnan potential | 2.4 mM (pH 7) 0.9 mM (pH 8) | <10 mMk | 92 | |||
Although the lack of specificity allows for the straightforward design of a single sensor for the detection of many types of analytes, the χ parameter that describes the polymer–solvent interactions is complicated. As a free-energy parameter, it captures enthalpic and entropic contributions, and may be fairly well estimated as χ = χs + (Vsolvent/RT)(δsolvent − δpolymer)2 where χs and Vsolvent represent an entropic component of χ and the molar volume of the solvent, respectively. δsolvent and δpolymer are the solubility parameters for the solvent and polymer, respectively.81 On a per molecule basis, the detector response may therefore be optimized by maximizing the change in χ in the presence of the target analyte from the reference state. Two key observations may be made based on this theory. First, the dynamic range of hydrogel sensors that respond to changes in χ is maximized when the reference material without analyte has a χ = 0 or χ = χC that corresponds to the critical interaction parameter at which the hydrogel fully collapses (note, more sophisticated theories have been developed113 in these polymer concentrated regimes in which the interaction parameter is assumed to be concentration dependent such that χ = χ1 + χ2ϕ). Second, since most hydrogels used for sensing applications start with interaction parameters between these limits (0 < χ < χC), the presence of the analyte may either increase or decrease the favorability of the polymer-solvent interactions such that hydrogel swelling or shrinking may be observed and the corresponding optical response may either red- or blue-shift. Fortunately, the solubility parameters are available for many common solvents and polymers,114,115 allowing for the prediction of the sensor response and its relative magnitude to specific analytes with relative ease. Group contribution theory81,116 may also be used to estimate the solubility parameters of novel polymers or specific polymer modifications as may occur during hydrogel functionalization and analyte binding.
Metabolites that interact with specificity to the photonic crystal hydrogel may also be detected. As one example, glucose sensors based on responsive hydrogels have been developed based on the inclusion of phenylboronic acid (PBA) derivatives as specific recognition molecules.91,92,117–120 The PBAs copolymerized with the hydrogel polymer network can bind with the diols in glucose, such that the PBA–glucose complexes have lower pKa values than the uncomplexed PBAs leading to borate anion formation at standard pHs for detection. Based on this binding mechanism, the PBA-functionalized photonic crystal hydrogels have a greater immobilized negative charge and the volumetric response resulting from changes in the Donnan potential may be correlated to a quantifiable glucose concentration. Although functionalization of the sensors with greater concentrations of PBAs may be anticipated to yield higher sensitivities, the relative hydrophobicity of PBA increases the χ value of the hydrogel matrices resulting in a less sensitive optical response in some instances.119,120 The molecular structure of the PBAs, including the position of the boronic acid on the phenyl ring and the inclusion of electron withdrawing substituents, can also influence the optical response of the sensors by changing the pKa of the PBA–glucose complexes.98 Using such approaches, glucose concentrations down to 50 μM were detected at low ionic strength,117 yet the optical response of this sensor was significantly reduced at high ionic strengths due to the dependence of the sensing mechanism on the Donnan potential. To address this challenge, ∼10-fold higher PBA loadings have been applied with demonstrated success121 and alternative sensing mechanisms have also been developed. One promising mechanism elicited hydrogel contraction via formation of a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PBA–glucose complex stabilized with additional components (e.g., poly(ethylene glycol) or the crown ether 15-crown-5) in the hydrogels.109 This approach allowed for detection of glucose down to concentrations of ∼0.2 mM in solutions with high ionic strength.
1 PBA–glucose complex stabilized with additional components (e.g., poly(ethylene glycol) or the crown ether 15-crown-5) in the hydrogels.109 This approach allowed for detection of glucose down to concentrations of ∼0.2 mM in solutions with high ionic strength.
Inverse opal hydrogel sensors may also be designed to detect and quantify specific charged metabolites or toxins. For example, a trifluoroacetyl-functionalized inverse opal hydrogel sensor was designed in order to detect cyanide ions.58 Owing to the hydrophobicity of the functional monomer containing the trifluoroacetyl group (2-(methacryloyloxy)ethyl-4-(2,2,2-trifluoroacetamido) benzonate), the as-prepared inverse opal hydrogel was relatively hydrophobic. Upon the reaction between cyanide and the trifluoroacetyl group, negatively charged species were immobilized within the hydrogel leading to a decrease in the χ value as well as an increase in the Donnan potential. The hydrogel then swelled and a redshift in the diffraction wavelength was correlated to the cyanide concentrations. Note that, depending on the hydrogel hydrophobicity, the addition of organic solvents (e.g., 10% v/v CH3CN) to modify the solvent quality may yield improved optical responses of the inverse opal hydrogel sensors due to more favorable polymer-solvent interactions (i.e., lowered χ values). A thiourea-functionalized inverse opal hydrogel has also been designed to detect acetate.122 In this system, acetate was bound to amide groups in thiourea by hydrogen bonding and again led to negative charge immobilization within the hydrogel. This sensor design has limitations similar to others that are dependent on changes in the immobilized charge concentration and Donnan potential, namely that the sensors become ineffective at high acetate concentrations or in environments with high ionic strength.
A range of dissolved gases, including those produced during cellular metabolism, may also be detected with photonic crystal hydrogels. The diagnostic marker ammonia was quantified in human blood serum using CCA hydrogel sensors functionalized with 3-aminophenol.17 In such materials, the Berthelot reaction results in the creation of new crosslinks within the hydrogels and thus hydrogel contraction. Additionally, inverse opal sensors for the measurement of dissolved CO2 (ref. 123) and ammonia gas124 have been reported. The inverse opal CO2 gas sensor was designed by exploiting protonated tertiary amine groups in the sensor that selectively adsorbed dissolved CO2. A substantially different design has been used for the detection of ammonia gas, in which the inverse opal hydrogels were formed from conducting polymers consisting of poly(3,4-ethylenedioxythiophene) and pyrrole that exhibit changes in both volume and conductivity in the presence of ammonia gas and allow for optical and electrical signals to be measured simultaneously.
4.2. Enzymatic reaction-based biosensing with photonic crystal hydrogels
Enzymatic reactions may also be applied in photonic crystal hydrogel sensors, as highlighted in Table 2, to detect enzymatic activity, gene expression, or the presence of a particular substrate or product. Often detection schemes based on enzymatic reactions exploit the specificity of enzymes toward their target substrate and their catalytic behavior that enables signal amplification. As one example, creatinine, which is a common marker for kidney failure, has been quantified in bodily fluids using CCA hydrogels functionalized with creatinine deiminase and 2-nitrophenol.19 In this two-step approach, the free creatinine was hydrolyzed by the enzyme while producing hydroxide anions, such that the local pH in the hydrogel increased and deprotonated the 2-nitrophenol. The deprotonation of the 2-nitrophenol enhanced the interactions between the medium and the hydrogel matrix leading to hydrogel swelling and a redshift in diffraction. Creatinine concentrations may thus be measured even under high ionic strength conditions ranging from 0.01 to 0.7 mM, and therefore allows such biosensors to be applied directly to samples of human serum.| Target analyte | Recognition molecule/strategy | Mechanism of hydrogel response | Sensitivity | Dynamic range | Ref. | |
|---|---|---|---|---|---|---|
| a Suppressed gel contraction by inhibitor Hg2+. b Deoxygenated condition. c Gel contraction. | ||||||
| CCA sensor | Hg2+ | Urease–urea reaction | Donnan potentiala | 1 ppb | <20 ppb | 15 |
| Cholesterol | Cholesterol oxidase | Donnan potential | 0.4 mM | <5 mM | 38 | |
| Creatinine | Creatinine deiminase/2-nitrophenol | χ parameter | 10 μM | <1 mM | 19 | |
| Glactose | Galactosidase | Donnan potential | 0.1 mM | <0.33 mM | 37 | |
| Glucose | Glucose oxidase | Donnan potential | 10 pMb | <0.5 mM | 14 | |
| Methyl paraoxon | Organophosphorus hydrolase | χ parameter | 0.2 μM | <0.2 mM | 130 | |
| Parathion | Acetylcholinesterase (AChE) | Donnan potential | 4.2 fM | <42 pM | 20 | |
| Protein kinase A | Peptide | Donnan potential | 4 U μL−1 | <25 U μL−1 | 25 | |
| Donnan potentialc | 0.1 U μL−1 | <10 U μL−1 | 97 | |||
| Urea | Urease–urea reaction | Donnan potential | 0.05 mM | <0.5 mM | 132 | |
| Inverse opal sensor | Glucose | Glucose oxidase | Donnan potential | 2 mM | <18 mM | 96 |
| Holographic sensor | AChE | AChE–acetylcholine reaction | Donnan potential | 3.13 U mL−1 | <200 U mL−1 | 135 |
| Penicillin G | Penicillinase–penicillin reaction | Donnan potential | 1 mM | <25 mM | 131 | |
| Urea | Urease–urea reaction | Donnan potential | ∼5 mM | <50 mM | 131 | |
A similar reaction sequence was used for the detection of the organophosphorus compound methyl paraoxon, which is a pesticide and common nerve agent simulant, with the enzyme organophosphorus hydrolase (OPH) and the pH-sensitive 3-aminophenolate immobilized in CCA hydrogels.130 A similar compound, parathion, which is also a pesticide and acetylcholinesterase (AChE) inhibitor, was detected down to femtomolar concentrations using CCA hydrogels immobilized with AChE.20 The enzymatic reaction between immobilized AChE and parathion led to phosphorylation of serine residues in the AChE and thus hydrogel swelling induced through the Donnan potential. The real-time and in-field detection of organophosphorus compounds, which are potential chemical warfare agents, represents an important potential application space that may take advantage of the visual reporting provided by photonic crystal hydrogels.
A photonic crystal hydrogel sensor functionalized with cholesterol oxidase (ChOx) has been also developed to measure the concentration of cholesterol in solution.38 For this, a CCA hydrogel platform containing chemically reactive epoxide groups was prepared through copolymerization of acrylamide and glycidyl methacrylate, then conjugated with ChOx via reactions between the epoxides and nucleophilic groups on the ChOx surface (e.g., Lys, Cys, His, or Tyr residues). The enzymatic reaction between cholesterol and ChOx resulted in the reduction of the flavin moiety in ChOx, which increased the immobilized anion concentration within the hydrogel. Other metabolites and diagnostic markers, such as urea131,132 and penicillin G,131 have been detected and quantified with photonic crystal hydrogel sensors using enzyme reactions to induce the volumetric and optical responses. Similarly, glucose was detected down to 10 pM concentrations under deoxygenated conditions by a glucose oxidase (GOx)-functionalized CCA hydrogel. A redshift in diffraction peak resulted from an increase in the concentration of immobilized anion in the gel as the flavin moiety of the GOx became reduced upon enzymatic reaction with glucose.14,37 Note that such sensing systems often become ineffective in the presence of oxygen which can re-oxidize the flavin, as well as in relatively high ionic strength buffering solutions that may be required for enzyme stabilization but that diminish the hydrogel response to the Donnan potential. Fortunately, it has been demonstrated that these practical issues may be resolved by sparging N2 through the sensing media during the reaction and rinsing the biosensor in a low ionic strength solution prior to measurement of the optical response.
Due to their role in regulating cellular processes, enzymes that catalyze the post-translational modification (PTM) of proteins represent important therapeutic targets for many diseases. At the molecular level, the attachment of chemical or biological moieties provides a mechanism to modulate intracellular protein levels and activity by altering protein folding, activity, stability, and cellular localization. Notably, changes in the intracellular level and activity of proteins involved in signaling networks can alter cell fate and lead to a myriad of diseases, including cancer, neurodegenerative diseases (e.g., Alzheimer's, Parkinson's), cardiovascular diseases, and autoimmune diseases (e.g., diabetes). However, enzymes that catalyze PTMs are inherently challenging to assay due to the lack of measurable signal that is generated by PTM reactions. Conventional biochemical methods to assay enzymes that catalyze PTMs nearly all use radiolabeled, fluorescent, or luminescent substrates. Photonic crystal hydrogel sensors have recently been designed to evaluate the activity of kinases that phosphorylate proteins to modulate cellular processes and function.25,97 For this purpose, CCA hydrogels were functionalized with a short peptide substrate (LRRASLG) that is a target for protein kinase A (PKA), as shown in Fig. 4. The serine residue in the recognition peptides were phosphorylated during the enzymatic reaction, resulting in additional immobilized negative charge and a greater Donnan potential within the hydrogels and thus inducing a detectable change in the hydrogel volume. Importantly, the sensitivity of the PKA responsive CCA hydrogel sensors were improved ∼8 fold by simply eliminating background charges on the hydrogel polymer backbones with a click chemistry approach for peptide conjugation instead of a conventional method using carboxylated CCA hydrogels and the EDC/NHS coupling reaction.97 By monitoring the diffraction shift, the PKA activities in the presence of the inhibitor H-89 (ref. 25) or in a cell lysate97 were quantitatively evaluated. Additionally, the reverse reaction (i.e., dephosphorylation) catalyzed by the enzyme phosphatase was also readily monitored using this approach, demonstrating the diverse nature of potential post-translational modification reactions that can be detected. Among the potential PTM reactions that may also be detected using this approach include glycosylation, acetylation, methylation, lipidation, sulfonation, nitrosylation, ubiquitination, and sumoylation, which have important implications in development and disease pathogenesis.133,134
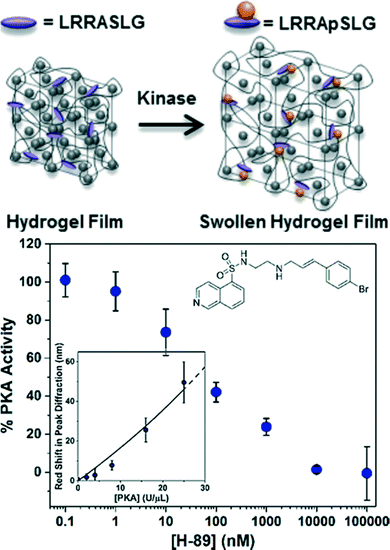 | ||
| Fig. 4 CCA hydrogel biosensor designed to monitor PKA activity. A redshift in diffraction wavelength is observed during the enzymatic reaction of PKA with recognition peptides that are conjugated in the hydrogel. Such biosensors are also able to detect the inhibition of kinase reactions by small molecules like H-89 (inset). Adapted from ref. 25. Copyright (2014) American Chemical Society. | ||
In order to detect and measure mercury concentrations in water, CCA hydrogel sensors were designed to quantify the inhibition of urease, which catalyzes the hydrolysis of urea, by Hg2+.15 Carboxylated CCA hydrogels (i.e., negatively-charged hydrogels) were initially functionalized with the urease by the EDC-mediated coupling reaction between carboxylic acids in the hydrogel and primary amines (e.g., available in lysines or the n-terminal amine) on the surface of the protein. When the enzymatic conversion of urea occurred with the available urease, free ions (NH4+ and HCO3−) were produced that caused the hydrogel to shrink by modifying the mobile ion concentration and Donnan potential. However, the inhibition of the enzyme by Hg2+ suppressed the hydrogel contraction, such that, by monitoring the relative extent of the blue-shift in diffraction, the concentration of Hg2+ in water was determined over the range of 1 to 20 ppb. The upper limit to the accurate detection of Hg2+ concentrations arises due to the Donnan potential dependent sensing mechanism; high Hg2+ concentrations yield high ionic strength solutions that inherently minimize the optical response.
4.3. Biospecific interaction-based biosensing with photonic crystal hydrogels
Photonic crystal hydrogel sensors for the detection of biomacromolecules such as DNA and proteins have been designed by exploiting the specific interactions available to biomolecules, as summarized in Table 3. DNA, in particular, provides an attractive target for detection given the high charge density provided by the phosphate backbone. In one study, CCA hydrogels were functionalized with “probe” single-stranded (ss) DNA having a sequence complementary to a “target” ssDNA consisting of an 18-mer sequence selected from the gene of the tumor suppressor protein p53 that is a marker used in early stage cancer detection (scheme in Fig. 5).24 The target ssDNA was readily distinguished from an analogous ssDNA with a sequence having a single base mismatch via sequence specific nucleic acid hybridization. The hybridization of the target DNA to the probe drastically increased the concentration of charge immobilized in the hydrogels and resulted in a visual report of the presence of the target DNA sequence, as shown in Fig. 5. The melting temperature of the hybridized DNA pairs was also characterized using the photonic crystal hydrogel system, and indicated that unsurprisingly the annealing conditions for hybridization and the ionic strength of the solution played important roles in the thermodynamic stability of the double stranded DNA.| Target analyte | Recognition molecule/strategy | Mechanism of hydrogel response | Sensitivity | Dynamic range | Ref. | |
|---|---|---|---|---|---|---|
| a Low ionic strength. b High ionic strength. c CFU: colony-forming unit. | ||||||
| CCA sensor | Single-stranded (ss) DNA | Probe ssDNA | Donnan potential | 0.5 μM | <1 mM | 24 |
| Methylated ssDNA | Probe ssDNA | χ parameter | 0.1 mM | <0.5 mM | 24 | |
| Avidin | Biotin | Crosslinking | 0.09 mg mL−1a | <1 mg mL−1a | 141 | |
| 0.5 mg mL−1b | <1 mg mL−1b | |||||
| Concanavalin A | Mannose | Crosslinking | 0.02 mg mL−1 | <2 mg mL−1 | 142 | |
| Candida albicans (bacteria) | Concanavalin A | Crosslinking | 32 CFU mL−1c | <6 × 107 CFU mL−1c | 144 | |
| Inverse opal sensor | ssDNA | Probe ssDNA | Crosslinking | 1 nM | <1 mM | 138 |
| Avidin | Biotin | Crosslinking | 80 nM | <10 μM | 89 | |
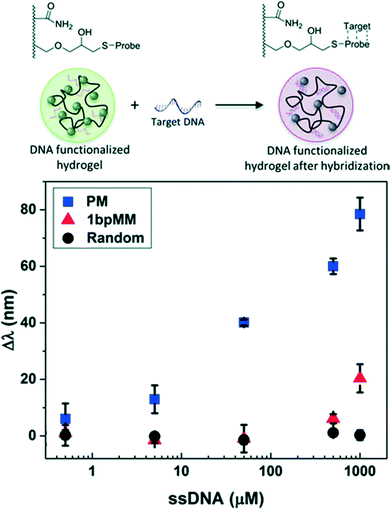 | ||
| Fig. 5 CCA hydrogel biosensor designed to detect target DNA. A concentration-dependent redshift in diffraction wavelength was observed upon sequence specific nucleic acid hybridization due to increases in the immobilized negative charge. Shown are the responses for target DNA sequences that are a perfect match (PM), a one base pair mismatch (1bpMM), and randomized relative to the probe sequence. Adapted from ref. 24. Copyright (2015) Royal Society of Chemistry. | ||
Epigenetic and environmental modifications to DNA may also be detected by photonic crystal hydrogel sensors. For example, methylated DNA was distinguished from the unmethylated DNA reference using CCA hydrogel sensors by exploiting a difference in the magnitude of χ upon hybridization.24 It was determined that, given the observed response, each methylation group yielded a ∼2–3 nm shift in the wavelength of peak diffraction and that single methylation sites on a 18-mer DNA sequence may be detected. Similarly, it may be predicted that other chemical modifications to DNA, including hydroxymethylation or functionalities that are highly hydrophobic, will also be readily detectable using this approach. The response to such modifications may be predicted by estimates of the solubility parameters;81,116 for example, inclusion of a single phenyl modification can be predicted to have approximately three times the effect on χ as a methyl modification.24 Similarly, chemical carcinogens that form DNA adducts may be detected (e.g., benzo[a]pyrene is known to be covalently linked to guanines in DNA through a series of chemical reactions136,137 and to cause transversion mutations). DNA biosensing schemes based on photonic crystal hydrogels may then even provide opportunities to screen for chemical carcinogens and DNA adducts that lead to mutations from which cancer originates.
In other implementations based on the inverse opal structure, ssDNA was incorporated directly into the hydrogel as a crosslinker through polymerizable functional groups at the 5′- and 3′-ends.138 Upon hybridization of the complementary strand, the conformation of the DNA crosslinkers were modified such that the hydrogel contracted (i.e., resulting from an increase in the elastic restoring force of the hydrogel) and thus a blue-shift in diffraction was observed.
Nucleic acid-based aptamers that can bind to specific molecules or ions while changing their configuration have also gained increased attention as recognition molecules due to their high selectivity.139 Recently, CCA hydrogels with crosslinks formed by the aptamers whose sequences lead to T (thymine)–Hg2+–T complexes and G (guanine)-quadruplexes with Pb2+ were constructed for selective detection of Hg2+ and Pb2+ in water.125 Upon interaction with Hg2+ and Pb2+, the aptamers underwent a conformational change from a linear to hairpin-like and G-quadruplex structure, respectively, and induced hydrogel contraction. With these aptamer-based CCA hydrogel sensors, Hg2+ and Pb2+ were quantified down to 10 and 1 nM, respectively, and displayed negligible interference in the optical response at 100-fold excess concentrations of other metal ions (Ag+, Ba2+, Mn2+, Mg2+, Cr3+ and Fe3+).
Protein–ligand recognition can also be utilized to construct photonic crystal hydrogel biosensors. The formation of new crosslinks in the hydrogel network upon target protein binding to multiple recognition sites (i.e., ligands) may lead to significant hydrogel contraction.140 By utilizing this volumetric change in ligand-functionalized photonic crystal hydrogels, detection of model proteins (i.e., avidin and Concanavalin A) with CCA and inverse opal hydrogels has been demonstrated.89,141,142 In addition, tumor-specific markers (i.e., glycoproteins) have been detected by shrinkage of lectin- and antibody-functionalized hydrogels in response to the formation of lectin–glycoprotein–antibody complexes that act as reversible crosslinks.143 Such designs based on photonic crystal hydrogels are promising as label-free and reusable immunoassays.
5. Additional design considerations and emerging directions
Photonic crystal sensors embedded in responsive hydrogels have significant potential, as reviewed in prior sections, for detecting the presence or activity of biological analytes including metabolites, enzymes, and DNA. Although improving the sensitivity and selectivity toward such analytes remain challenges of importance, this biosensor design benefits from the simplicity of the materials and the ability to visually quantify the response. Whereas we have thus far emphasized the role played by the hydrogel materials, the physical mechanisms by which the hydrogels are equilibrated and respond to stimuli, and the conjugation methods used to functionalize the biosensors with recognition molecules, in this concluding section we will discuss other considerations in the design and engineering of photonic crystal hydrogel biosensors, as well as emerging directions for their application.When designing hydrogel responsive sensors for biomacromolecules or even microorganisms with sizes from 10 nm–10 μm, it is critical to consider the diffusion of such bulky components into the hydrogel matrix, especially when their direct bioactivity on the hydrogel is critical to the response mechanism. In such cases, the mesh size of the hydrogel network may sterically hinder the analyte penetration into the hydrogels and limit their function to the hydrogel surface.145–147 The Renkin equation148 can be used to estimate the hindered diffusion of large particles or macromolecules in the hydrogel matrix as a function of the network mesh size: Dm/D0 = (1 − ds/dm)2 × [1 − 2.10(ds/dm) + 2.09(ds/dm)2 − 0.95(ds/dm)5] where Dm and D0 represent the diffusion coefficients of targets in the hydrogel and solution, respectively, and ds and dm represent the hydrodynamic diameter of the target and the mesh size of the hydrogels, respectively. It is therefore necessary to design the hydrogel networks with dm ≫ ds, which may be achieved by reducing the crosslinking density, to assure rapid and complete penetration of the target analytes into the detector volume.
Uniformity of the 3D structure of the photonic crystal hydrogels, particularly over large volumes, is also desirable. A single-crystalline structure would in theory yield diffraction spectra with peaks that are monochromatic or at least much narrower, thereby providing greater detector sensitivity and allowing for smaller sampling volumes. In practice, however, variations in the crystalline structure arise during the self-assembly processes used to fabricate the photonic crystals, e.g., the arrays of colloidal spherical particles in the CCA and inverse opal designs, or the lamellar structures formed by block copolymers. Defects analogous to those in atomic crystals or liquid crystalline systems arise, which lead to crystalline grains of finite size.149 Although the free-energy of self-assembled systems would be minimized by annihilating defects to achieve a single-crystalline configuration, kinetic and thermodynamic barriers limit such processes particularly over large volumes, even when performed slowly (e.g., by dip coating54,61). Techniques may be applied to direct the self-assembly of crystalline structures with enhanced long-range order and perfection, particularly in thin films, using topographic guiding structures (i.e., graphoepitaxy) or external guiding fields (e.g., electric fields). Moreover, since the hydrogel and crystalline structures are often prepared simultaneously, the components influence each other and the resulting structure. For example, there is a competition between the relative rates of hydrogel polymerization and self-assembly that must necessarily favor slow polymerization rates; more subtle is that the mechanical stress upon hydrogel crosslinking may in turn induce reconstruction of the self-assembled morphology. Fabrication approaches that effectively decouple self-assembly from hydrogel preparation, or that avoid self-assembly altogether as in the holographic hydrogel materials, may therefore find greater utility in the future.
Characterization of the photonic crystal response is likewise an area that demands further development. One of the primary advantages of photonic crystals, as commonly implemented for sensing, is that they can be tuned to diffract in the visible range of wavelengths. This allows for detection by eye of the sensor response as a color change, albeit insensitively given the angular dependence of the response and the relatively poor ability of human vision to distinguish quantitatively between similar colors (e.g., greenish-blue vs. green vs. yellowish-green). Fortunately sensors based on photonic crystal hydrogels are also compatible with existing techniques for optical characterization including spectrophotometry and optical microscopy. In spite of the diverse tools that are readily available for characterizing such materials, the majority of the literature reports the use of reflectance spectroscopy in the visible spectral range from the surface of films of photonic crystal hydrogels. In some cases, however, it may be desirable to purposefully design the biosensors to have diffraction spectra with peaks outside the visible range. Given the strong absorbance of water, the hydrogel materials, and biological tissue in the UV, the most obvious alternative would be to design the sensors to operate in the near-IR window. Implantable photonic crystal hydrogel biosensors for continuous diagnostics (e.g., monitoring of glucose or cholesterol levels) or medical imaging may thus be envisioned. Alternatively, it may be desirable to tune the biosensor to operate in select ranges of the visible spectrum, for example in cases in which samples are simultaneously characterized by fluorescence microscopy or some of the biological components strongly autofluoresce.
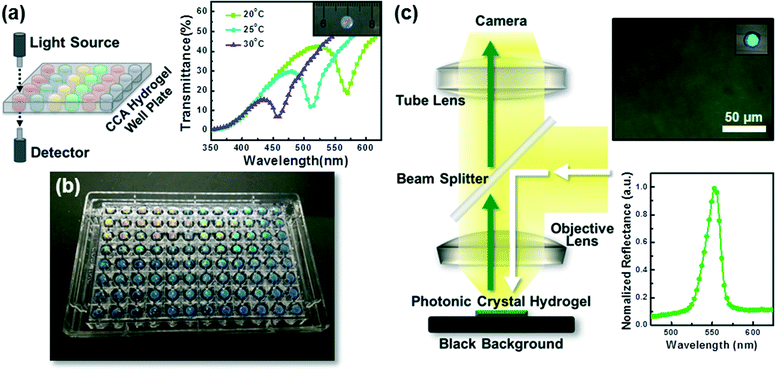
Photonic crystal hydrogels may also be characterized in transmission, rather than reflection, by fabricating them in thin film formats, as shown in Fig. 6a, such that the diffraction wavelength is reported as a minimum in transmittance (%T) or a maximum in attenuance150 defined as D = −log10(%T/100). Performing spectroscopy in transmission, when combined with the readily adaptable fabrication schemes of CCA or inverse opal hydrogels (see sections 2.1 and 2.2), allows for the implementation of photonic crystal hydrogel biosensors in standard microtiter plates (e.g., 96- and 384-well microplates), as we have demonstrated in Fig. 6b. Microplate formats of photonic crystal hydrogel sensors may enable high-throughput characterization of target analytes using conventional microplate readers and allow for rapid drug discovery151 or screening enzymatic and cellular activity.152,153 These microplates may also be utilized for simultaneous multiple target sensing (i.e., multiplexing) upon functionalization of the hydrogel matrices in each well with specific recognition molecules as described in section 2.5.
Furthermore, it is possible to image local areas of photonic crystal hydrogel biosensors using optical microscopy, as we demonstrate in Fig. 6c. When combined with microscopic techniques (e.g., reflected light and dark field microscopy) and imaging analysis methods that may allow for collection of optical response spectra on a pixel-by-pixel basis, there are powerful tools to quantitatively sense localized changes to the hydrogel. The ability to map the local response at the microscale may find application for systems that have spatial variations in activity or that locally modify the hydrogel structure. Cellular systems may be of particular interest. Notably, the presence of Candida albicans, which causes fungal infections in humans, has been detected with CCA hydrogels functionalized with protein Concanavalin A that can specifically bind to mannan epitopes displayed on the surface of C. albicans.144 Microscopy techniques would allow detection at a single cell level, including distinguishing specific cell types amongst a diverse colony. More broadly, photonic crystal hydrogel sensors would allow for selection of cell sub-populations, for example when plated and cultured from large libraries generated by cellular or metabolic engineering techniques, that yield with vigor extracellular products of particular interest.
Similarly, the in vitro characterization of the interaction of cells with their substrate and environment in cell culture and tissue engineering is a field of active investigation due to its importance in cell growth and migration. Hydrogels of polyacrylamide or poly(ethylene glycol) embedded with photonic crystals are compatible with tissue culture as two-dimensional substrates and would allow for quantification and mapping of the local mechanical forces applied by adherent cells on the substrate.154 These forces occur primarily at dynamic positions known as focal adhesions and include, in part, pulling forces that arise due to contractile macromolecular assemblies.155 The ability of cells to remodel their substrates, either by degradation or deposition of additional extracellular matrix material, may also be explored using photonic crystal hydrogel sensors. For example, hydrogels with oligopeptide crosslinks sensitive to cleavage by matrix metalloproteinases (MMPs) could be used to visually display local changes to the mechanical modulus that occur upon matrix remodeling.156
In conclusion, responsive hydrogels embedded with photonic crystals function successfully as biosensors by transducing physical changes in structure to optical readouts. Such systems are sensitive to mechanical stresses, chemical interactions, and biological activity, and may be exploited to identify small molecule chemicals and metabolites, large biomacromolecules and their activity, and even cellular behavior. Significant effort has been put forth in terms of the molecular-level design, fabrication, and functionalization of photonic crystal hydrogels to yield biosensors that are sensitive, selective, label-free, reversible and reliable, and capable of providing quantitative information. However, opportunities remain to further engineer the materials and characterization methods prior to implementation in commercial bioassays, particularly for applications that can exploit the compatibility with existing tools for high-throughput and multiplexed screening.
Acknowledgements
This work was supported in part by funding from the National Science Foundation (DMR1411320) and the University of Colorado Liquid Crystal Material Research Center (NSF DMR0820579). We also thank Dr. Kelsey MacConaghy for helpful discussion and contributions towards the development of the theoretical model of hydrogel swelling.References
- S. Song, H. Xu and C. H. Fan, Int. J. Nanomed., 2006, 1, 433–440 CrossRef CAS PubMed.
- A. P. F. Turner, Chem. Soc. Rev., 2013, 42, 3184–3196 RSC.
- J. Kirsch, C. Siltanen, Q. Zhou, A. Revzin and A. Simonian, Chem. Soc. Rev., 2013, 42, 8733–8768 RSC.
- S. Rodriguez-Mozaz, M. J. L. de Alda, M. P. Marco and D. Barcelo, Talanta, 2005, 65, 291–297 CAS.
- A. K. Yetisen, M. S. Akram and C. R. Lowe, Lab Chip, 2013, 13, 2210–2251 RSC.
- D. A. Giljohann and C. A. Mirkin, Nature, 2009, 462, 461–464 CrossRef CAS PubMed.
- S. M. Yoo and S. Y. Lee, Trends Biotechnol., 2016, 34, 7–25 CrossRef CAS PubMed.
- H. Lee, T. H. Shin, J. Cheon and R. Weissleder, Chem. Rev., 2015, 115, 10690–10724 CrossRef CAS PubMed.
- J. L. Arlett, E. B. Myers and M. L. Roukes, Nat. Nanotechnol., 2011, 6, 203–215 CrossRef CAS PubMed.
- J. P. Ge and Y. D. Yin, Angew. Chem., Int. Ed., 2011, 50, 1492–1522 CrossRef CAS PubMed.
- C. Fenzl, T. Hirsch and O. S. Wolfbeis, Angew. Chem., Int. Ed., 2014, 53, 3318–3335 CrossRef CAS PubMed.
- A. K. Yetisen, H. Butt, L. R. Volpatti, I. Pavlichenko, M. Humar, S. J. Kwok, H. Koo, K. S. Kim, I. Naydenova, A. Khademhosseini, S. K. Hahn and S. H. Yun, Biotechnol. Adv., 2016, 34, 250–271 CrossRef CAS PubMed.
- B. M. Gonzalez, G. Christie, C. A. B. Davidson, J. Blyth and C. R. Lowe, Anal. Chim. Acta, 2005, 528, 219–228 CrossRef.
- J. H. Holtz and S. A. Asher, Nature, 1997, 389, 829–832 CrossRef CAS PubMed.
- D. Arunbabu, A. Sannigrahi and T. Jana, Soft Matter, 2011, 7, 2592–2599 RSC.
- K. Lee and S. A. Asher, J. Am. Chem. Soc., 2000, 122, 9534–9537 CrossRef CAS.
- K. W. Kimble, J. P. Walker, D. N. Finegold and S. A. Asher, Anal. Bioanal. Chem., 2006, 385, 678–685 CrossRef CAS PubMed.
- J. T. Zhang, N. Smith and S. A. Asher, Anal. Chem., 2012, 84, 6416–6420 CrossRef CAS PubMed.
- A. C. Sharma, T. Jana, R. Kesavamoorthy, L. J. Shi, M. A. Virji, D. N. Finegold and S. A. Asher, J. Am. Chem. Soc., 2004, 126, 2971–2977 CrossRef CAS PubMed.
- J. P. Walker and S. A. Asher, Anal. Chem., 2005, 77, 1596–1600 CrossRef CAS PubMed.
- A. K. Yetisen, I. Naydenova, F. D. Vasconcellos, J. Blyth and C. R. Lowe, Chem. Rev., 2014, 114, 10654–10696 CrossRef CAS PubMed.
- Y. J. Zhao, X. W. Zhao and Z. Z. Gu, Adv. Funct. Mater., 2010, 20, 2970–2988 CrossRef CAS.
- X. D. Fan, I. M. White, S. I. Shopova, H. Y. Zhu, J. D. Suter and Y. Z. Sun, Anal. Chim. Acta, 2008, 620, 8–26 CrossRef CAS PubMed.
- K. I. MacConaghy, D. M. Chadly, M. P. Stoykovich and J. L. Kaar, Analyst, 2015, 140, 6354–6362 RSC.
- K. I. MacConaghy, C. I. Geary, J. L. Kaar and M. P. Stoykovich, J. Am. Chem. Soc., 2014, 136, 6896–6899 CrossRef CAS PubMed.
- X. L. Jia, J. Y. Wang, K. Wang and J. T. Zhu, Langmuir, 2015, 31, 8732–8737 CrossRef CAS PubMed.
- Y. F. Yue, T. Kurokawa, M. A. Haque, T. Nakajima, T. Nonoyama, X. F. Li, I. Kajiwara and J. P. Gong, Nat. Commun., 2014, 5, 4659 CAS.
- Z. K. Yang, D. J. Shi, M. Q. Chen and S. R. Liu, Anal. Methods, 2015, 7, 8352–8359 RSC.
- C. Guo, C. H. Zhou, N. Sai, B. A. Ning, M. Liu, H. S. Chen and Z. X. Gao, Sens. Actuators, B, 2012, 166, 17–23 CrossRef.
- Y. Fuchs, O. Soppera, A. G. Mayes and K. Haupt, Adv. Mater., 2013, 25, 566–570 CrossRef CAS PubMed.
- N. Griffete, H. Frederich, A. Maitre, S. Ravaine, M. M. Chehimi and C. Mangeney, Langmuir, 2012, 28, 1005–1012 CrossRef CAS PubMed.
- A. J. Marshall, D. S. Young, S. Kabilan, A. Hussain, J. Blyth and C. R. Lowe, Anal. Chim. Acta, 2004, 527, 13–20 CrossRef CAS.
- F. Xue, Z. H. Meng, F. L. Qi, M. Xue, F. Y. Wang, W. Chen and Z. Q. Yan, Analyst, 2014, 139, 6192–6196 RSC.
- M. Xu, A. V. Goponenko and S. A. Asher, J. Am. Chem. Soc., 2008, 130, 3113–3119 CrossRef PubMed.
- Z. Pan, J. K. Ma, J. Yan, M. Zhou and J. P. Gao, J. Mater. Chem., 2012, 22, 2018–2025 RSC.
- A. G. Mayes, J. Blyth, M. Kyrolainen-Reay, R. B. Millington and C. R. Lowe, Anal. Chem., 1999, 71, 3390–3396 CrossRef CAS.
- J. H. Holtz, J. S. W. Holtz, C. H. Munro and S. A. Asher, Anal. Chem., 1998, 70, 780–791 CrossRef CAS.
- M. K. Maurer, S. E. Gould and P. J. Scott, Sens. Actuators, B, 2008, 134, 736–742 CrossRef CAS.
- D. C. Appleyard, S. C. Chapin and P. S. Doyle, Anal. Chem., 2011, 83, 193–199 CrossRef CAS PubMed.
- S. Jung and H. Yi, Biomacromolecules, 2013, 14, 3892–3902 CrossRef CAS PubMed.
- J. Liu, H. X. Liu, H. Z. Kang, M. Donovan, Z. Zhu and W. H. Tan, Anal. Bioanal. Chem., 2012, 402, 187–194 CrossRef CAS PubMed.
- B. F. Ye, H. B. Ding, Y. Cheng, H. C. Gu, Y. J. Zhao, Z. Y. Xie and Z. Z. Gu, Adv. Mater., 2014, 26, 3270–3274 CrossRef CAS PubMed.
- S. Jung and H. Yi, Langmuir, 2012, 28, 17061–17070 CrossRef CAS PubMed.
- H. M. Yi, L. Q. Wu, R. Ghodssi, G. W. Rubloff, G. F. Payne and W. E. Bentley, Anal. Chem., 2004, 76, 365–372 CrossRef CAS PubMed.
- R. J. Carlson and S. A. Asher, Appl. Spectrosc., 1984, 38, 297–304 CrossRef CAS.
- N. A. Clark, A. J. Hurd and B. J. Ackerson, Nature, 1979, 281, 57–60 CrossRef CAS.
- V. M. Rotello, Nanoparticles: Building Blocks for Nanotechnology, Springer US, New York, 2004 Search PubMed.
- C. E. Reese, C. D. Guerrero, J. M. Weissman, K. Lee and S. A. Asher, J. Colloid Interface Sci., 2000, 232, 76–80 CrossRef CAS PubMed.
- A. S. Tse, Z. J. Wu and S. A. Asher, Macromolecules, 1995, 28, 6533–6538 CrossRef CAS.
- Y. N. Xia, B. Gates, Y. D. Yin and Y. Lu, Adv. Mater., 2000, 12, 693–713 CrossRef CAS.
- J. J. Bohn, A. Tikhonov and S. A. Asher, J. Colloid Interface Sci., 2010, 350, 381–386 CrossRef CAS PubMed.
- V. I. Khrupa, I. R. Entin and L. I. Datsenko, Zh. Tekh. Fiz., 1991, 61, 196–198 CAS.
- R. Williams and R. S. Crandall, Phys. Lett. A, 1974, 48, 225–226 CrossRef CAS.
- S. Furumi, H. Fudouzi and T. Sawada, Laser Photonics Rev., 2010, 4, 205–220 CrossRef CAS.
- S. A. Asher, J. Holtz, L. Liu and Z. J. Wu, J. Am. Chem. Soc., 1994, 116, 4997–4998 CrossRef CAS.
- J. M. Weissman, H. B. Sunkara, A. S. Tse and S. A. Asher, Science, 1996, 274, 959–960 CrossRef CAS PubMed.
- T. Kanai, D. Lee, H. C. Shum, R. K. Shah and D. A. Weitz, Adv. Mater., 2010, 22, 4998–5002 CrossRef CAS PubMed.
- X. S. Li, L. H. Peng, J. C. Cui, W. N. Li, C. X. Lin, D. Xu, T. Tian, G. X. Zhang, D. Q. Zhang and G. T. Li, Small, 2012, 8, 612–618 CrossRef CAS PubMed.
- C. I. Aguirre, E. Reguera and A. Stein, Adv. Funct. Mater., 2010, 20, 2565–2578 CrossRef CAS.
- Y. J. Lee and P. V. Braun, Adv. Mater., 2003, 15, 563–566 CrossRef CAS.
- A. Stein, B. E. Wilson and S. G. Rudisill, Chem. Soc. Rev., 2013, 42, 2763–2803 RSC.
- K. Ueno, K. Matsubara, M. Watanabe and Y. Takeoka, Adv. Mater., 2007, 19, 2807–2812 CrossRef CAS.
- M. L. Zhang, F. Jin, M. L. Zheng and X. M. Duan, RSC Adv., 2014, 4, 20567–20572 RSC.
- J. Y. Wang, Y. Cao, Y. Feng, F. Yin and J. P. Gao, Adv. Mater., 2007, 19, 3865–3871 CrossRef CAS.
- C. D. Sorrell and M. J. Serpe, Anal. Bioanal. Chem., 2012, 402, 2385–2393 CrossRef CAS PubMed.
- I. B. Burgess, L. Mishchenko, B. D. Hatton, M. Kolle, M. Loncar and J. Aizenberg, J. Am. Chem. Soc., 2011, 133, 12430–12432 CrossRef CAS PubMed.
- C. Y. Kuo, S. Y. Lu, S. Chen, M. Bernards and S. Jiang, Sens. Actuators, B, 2007, 124, 452–458 CrossRef CAS.
- J. L. Li, X. W. Zhao, H. M. Wei, Z. Z. Gu and Z. H. Lu, Anal. Chim. Acta, 2008, 625, 63–69 CrossRef CAS PubMed.
- J. L. Li and T. S. Zheng, Sens. Actuators, B, 2008, 131, 190–195 CrossRef CAS.
- J. Blyth, R. B. Millington, A. G. Mayes and C. R. Lowe, Imaging Sci. J., 1999, 47, 87–91 CAS.
- A. K. Yetisen, Y. Montelongo, N. M. Farandos, I. Naydenova, C. R. Lowe and S. H. Yun, Appl. Phys. Lett., 2014, 105, 261106 Search PubMed.
- C. P. Tsangarides, A. K. Yetisen, F. D. Vasconcellos, Y. Montelongo, M. M. Qasim, T. D. Wilkinson, C. R. Lowe and H. Butt, RSC Adv., 2014, 4, 10454–10461 RSC.
- M. Campbell, D. N. Sharp, M. T. Harrison, R. G. Denning and A. J. Turberfield, Nature, 2000, 404, 53–56 CrossRef CAS PubMed.
- J. H. Moon, J. Ford and S. Yang, Polym. Adv. Technol., 2006, 17, 83–93 CrossRef CAS.
- L. J. Wu, Y. C. Zhong, C. T. Chan, K. S. Wong and G. P. Wang, Appl. Phys. Lett., 2005, 86, 241102 CrossRef.
- A. G. Mayes, J. Blyth, R. B. Millington and C. R. Lowe, Anal. Chem., 2002, 74, 3649–3657 CrossRef CAS PubMed.
- Y. Kang, J. J. Walish, T. Gorishnyy and E. L. Thomas, Nat. Mater., 2007, 6, 957–960 CrossRef CAS PubMed.
- O. B. Ayyub, M. B. Ibrahim, R. M. Briber and P. Kofinas, Biosens. Bioelectron., 2013, 46, 124–129 CrossRef CAS PubMed.
- M. Bockstaller, R. Kolb and E. L. Thomas, Adv. Mater., 2001, 13, 1783–1786 CrossRef CAS.
- H. S. Lim, J. H. Lee, J. J. Walish and E. L. Thomas, ACS Nano, 2012, 6, 8933–8939 CrossRef CAS PubMed.
- R. J. Young and P. A. Lovell, Introduction to Polymers, CRC Press, New York, 2011 Search PubMed.
- H. C. Kim, S. M. Park and W. D. Hinsberg, Chem. Rev., 2010, 110, 146–177 CrossRef CAS PubMed.
- N. A. Lynd, A. J. Meuler and M. A. Hillmyer, Prog. Polym. Sci., 2008, 33, 875–893 CrossRef CAS.
- A. C. Edrington, A. M. Urbas, P. DeRege, C. X. Chen, T. M. Swager, N. Hadjichristidis, M. Xenidou, L. J. Fetters, J. D. Joannopoulos, Y. Fink and E. L. Thomas, Adv. Mater., 2001, 13, 421–425 CrossRef CAS.
- R. Verduzco, X. Y. Li, S. L. Pesek and G. E. Stein, Chem. Soc. Rev., 2015, 44, 2405–2420 RSC.
- G. M. Miyake, V. A. Piunova, R. A. Weitekamp and R. H. Grubbs, Angew. Chem., Int. Ed., 2012, 51, 11246–11248 CrossRef CAS PubMed.
- G. M. Miyake, R. A. Weitekamp, V. A. Piunova and R. H. Grubbs, J. Am. Chem. Soc., 2012, 134, 14249–14254 CrossRef CAS PubMed.
- B. R. Sveinbjornsson, R. A. Weitekamp, G. M. Miyake, Y. Xia, H. A. Atwater and R. H. Grubbs, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14332–14336 CrossRef PubMed.
- J. P. Couturier, M. Sutterlin, A. Laschewsky, C. Hettrich and E. Wischerhoff, Angew. Chem., Int. Ed., 2015, 54, 6641–6644 CrossRef CAS PubMed.
- F. Y. Lin and L. P. Yu, Anal. Methods, 2012, 4, 2838–2845 RSC.
- D. Nakayama, Y. Takeoka, M. Watanabe and K. Kataoka, Angew. Chem., Int. Ed., 2003, 42, 4197–4200 CrossRef CAS PubMed.
- A. K. Yetisen, Y. Montelongo, F. D. Vasconcellos, J. L. Martinez-Hurtado, S. Neupane, H. Butt, M. M. Qasim, J. Blyth, K. Burling, J. B. Carmody, M. Evans, T. D. Wilkinson, L. T. Kubota, M. J. Monteiro and C. R. Lowe, Nano Lett., 2014, 14, 3587–3593 CrossRef CAS PubMed.
- R. A. Floyd and J. M. Carney, Ann. Neurol., 1992, 32, S22–S27 CrossRef CAS PubMed.
- N. Kobayashi, S. Katsumi, K. Imoto, A. Nakagawa, S. Miyagawa, M. Furumura and T. Mori, Pigm. Cell Res., 2001, 14, 94–102 CAS.
- A. W. P. Vermeer and W. Norde, Biophys. J., 2000, 78, 394–404 CrossRef CAS PubMed.
- L. Jin, Y. J. Zhao, X. Liu, Y. L. Wang, B. F. Ye, Z. Y. Xie and Z. Z. Gu, Soft Matter, 2012, 8, 4911–4917 RSC.
- K. I. MacConaghy, D. M. Chadly, M. P. Stoykovich and J. L. Kaar, Anal. Chem., 2015, 87, 3467–3475 CrossRef CAS PubMed.
- C. J. Zhang, M. D. Losego and P. V. Braun, Chem. Mater., 2013, 25, 3239–3250 CrossRef CAS.
- E. T. Tian, Y. Ma, L. Y. Cui, J. X. Wang, Y. L. Song and L. Jiang, Macromol. Rapid Commun., 2009, 30, 1719–1724 CrossRef CAS PubMed.
- G. T. Hermanson, Bioconjugate Techniques, Elsevier Science, Burlington, 2008 Search PubMed.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974–6998 CrossRef CAS PubMed.
- W. X. Xi, T. F. Scott, C. J. Kloxin and C. N. Bowman, Adv. Funct. Mater., 2014, 24, 2572–2590 CrossRef CAS.
- H. Fudouzi, in Nanomaterials and Nanoarchitectures: A Complex Review of Current Hot Topics and their Applications, ed. M. Bardosova and T. Wagner, Springer Netherlands, Dordrecht, 2015, ch. 1, pp. 1–19 Search PubMed.
- C. Paquet and E. Kumacheva, Mater. Today, 2008, 11, 48–56 CrossRef CAS.
- Y. J. Zhao, Z. Y. Xie, H. C. Gu, C. Zhu and Z. Z. Gu, Chem. Soc. Rev., 2012, 41, 3297–3317 RSC.
- P. J. Flory, Principles of Polymer Chemistry, Cornell University Press, Ithaca, 1953 Search PubMed.
- H. Wang and K. Q. Zhang, Sens. Actuators, B, 2013, 13, 4192–4213 CAS.
- S. A. Asher, S. F. Peteu, C. E. Reese, M. X. Lin and D. Finegold, Anal. Bioanal. Chem., 2002, 373, 632–638 CrossRef CAS PubMed.
- V. L. Alexeev, A. C. Sharma, A. V. Goponenko, S. Das, I. K. Lednev, C. S. Wilcox, D. N. Finegold and S. A. Asher, Anal. Chem., 2003, 75, 2316–2323 CrossRef CAS PubMed.
- X. P. Yang, X. H. Pan, J. Blyth and C. R. Lowe, Biosens. Bioelectron., 2008, 23, 899–905 CrossRef CAS PubMed.
- C. E. Reese and S. A. Asher, Anal. Chem., 2003, 75, 3915–3918 CrossRef CAS PubMed.
- A. K. Yetisen, M. M. Qasim, S. Nosheen, T. D. Wilkinson and C. R. Lowe, J. Mater. Chem. C, 2014, 2, 3569–3576 RSC.
- B. Erman and P. J. Flory, Macromolecules, 1986, 19, 2342–2353 CrossRef CAS.
- C. M. Hansen, Hansen Solubility Parameters: A User's Handbook, CRC Press, New York, 2007 Search PubMed.
- A. F. M. Barton, CRC Handbook of Solubility Parameters and Other Cohesion Parameters, CRC Press, New York, 1991 Search PubMed.
- J. M. G. Cowie and V. Arrighi, Polymers: Chemistry and Physics of Modern Materials, CRC Press, New York, 2007 Search PubMed.
- S. A. Asher, V. L. Alexeev, A. V. Goponenko, A. C. Sharma, I. K. Lednev, C. S. Wilcox and D. N. Finegold, J. Am. Chem. Soc., 2003, 125, 3322–3329 CrossRef CAS PubMed.
- Y. J. Lee, S. A. Pruzinsky and P. V. Braun, Langmuir, 2004, 20, 3096–3106 CrossRef CAS PubMed.
- M. C. Lee, S. Kabilan, A. Hussain, X. P. Yang, J. Blyth and C. R. Lowe, Anal. Chem., 2004, 76, 5748–5755 CrossRef CAS PubMed.
- S. Kabilan, A. J. Marshall, F. K. Sartain, M. C. Lee, A. Hussain, X. P. Yang, J. Blyth, N. Karangu, K. James, J. Zeng, D. Smith, A. Domschke and C. R. Lowe, Biosens. Bioelectron., 2005, 20, 1602–1610 CrossRef CAS PubMed.
- M. Ben-Moshe, V. L. Alexeev and S. A. Asher, Anal. Chem., 2006, 78, 5149–5157 CrossRef CAS PubMed.
- S. Kado, H. Otani, Y. Nakahara and K. Kimura, Chem. Commun., 2013, 49, 886–888 RSC.
- W. Hong, Y. Chen, X. Feng, Y. Yan, X. B. Hu, B. Y. Zhao, F. Zhang, D. Zhang, Z. Xu and Y. J. Lai, Chem. Commun., 2013, 49, 8229–8231 RSC.
- Q. F. Zhong, H. Xu, H. B. Ding, L. Bai, Z. D. Mu, Z. Y. Xie, Y. J. Zhao and Z. Z. Gu, Colloids Surf., A, 2013, 433, 59–63 CrossRef CAS.
- B. F. Ye, Y. J. Zhao, Y. Cheng, T. T. Li, Z. Y. Xie, X. W. Zhao and Z. Z. Gu, Nanoscale, 2012, 4, 5998–6003 RSC.
- M. Liu and L. P. Yu, Analyst, 2013, 138, 3376–3379 RSC.
- A. K. Yetisen, Y. Montelongo, M. M. Qasim, H. Butt, T. D. Wilkinson, M. J. Monteiro and S. H. Yun, Anal. Chem., 2015, 87, 5101–5108 CrossRef CAS PubMed.
- J. L. M. Hurtado and C. R. Lowe, ACS Appl. Mater. Interfaces, 2014, 6, 8903–8908 CAS.
- A. M. Horgan, A. J. Marshall, S. J. Kew, K. E. S. Dean, C. D. Creasey and S. Kabilan, Biosens. Bioelectron., 2006, 21, 1838–1845 CrossRef CAS PubMed.
- J. P. Walker, K. W. Kimble and S. A. Asher, Anal. Bioanal. Chem., 2007, 389, 2115–2124 CrossRef CAS PubMed.
- A. J. Marshall, D. S. Young, J. Blyth, S. Kabilan and C. R. Lowe, Anal. Chem., 2004, 76, 1518–1523 CrossRef CAS PubMed.
- F. Zeng, S. Z. Wu, Z. W. Sun, H. Y. Xi, R. F. Li and Z. L. Hou, Sens. Actuators, B, 2002, 81, 273–276 CrossRef CAS.
- S. R. Bhaumik, E. Smith and A. Shilatifard, Nat. Struct. Mol. Biol., 2007, 14, 1008–1016 CAS.
- M. Sethuraman, N. Clavreul, H. Huang, M. E. McComb, C. E. Costello and R. A. Cohen, Free Radical Biol. Med., 2007, 42, 823–829 CrossRef CAS PubMed.
- E. V. Tan and C. R. Lowe, Anal. Chem., 2009, 81, 7579–7589 CrossRef CAS PubMed.
- A. M. Jeffrey, K. W. Jennette, S. H. Blobstein, T. B. Weinstein, F. A. Beland, R. G. Harvey, H. Kasai, I. Miura and K. Nakanishi, J. Am. Chem. Soc., 1976, 98, 5714–5715 CrossRef CAS PubMed.
- E. Eisenstadt, A. J. Warren, J. Porter, D. Atkins and J. H. Miller, Proc. Natl. Acad. Sci. U. S. A., 1982, 79, 1945–1949 CrossRef CAS.
- Y. J. Zhao, X. W. Zhao, B. C. Tang, W. Y. Xu, J. Li, L. Hu and Z. Z. Gu, Adv. Funct. Mater., 2010, 20, 976–982 CrossRef CAS.
- W. Z. Zhou, P. J. J. Huang, J. S. Ding and J. Liu, Analyst, 2014, 139, 2627–2640 RSC.
- K. Gawel, D. Barriet, M. Sletmoen and B. T. Stokke, Sensors, 2010, 10, 4381–4409 CrossRef CAS PubMed.
- J. T. Zhang, X. Chao, X. Y. Liu and S. A. Asher, Chem. Commun., 2013, 49, 6337–6339 RSC.
- J. T. Zhang, Z. Y. Cai, D. H. Kwak, X. Y. Liu and S. A. Asher, Anal. Chem., 2014, 86, 9036–9041 CrossRef CAS PubMed.
- T. Miyata, M. Jige, T. Nakaminami and T. Uragami, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 1190–1193 CrossRef CAS PubMed.
- Z. Y. Cai, D. H. Kwak, D. Punihaole, Z. M. Hong, S. S. Velankar, X. Y. Liu and S. A. Asher, Angew. Chem., Int. Ed., 2015, 54, 13036–13040 CrossRef CAS PubMed.
- S. Jung and H. Yi, Chem. Mater., 2015, 27, 3988–3998 CrossRef CAS.
- N. W. Choi, J. Kim, S. C. Chapin, T. Duong, E. Donohue, P. Pandey, W. Broom, W. A. Hill and P. S. Doyle, Anal. Chem., 2012, 84, 9370–9378 CrossRef CAS PubMed.
- A. G. Lee, C. P. Arena, D. J. Beebe and S. P. Palecek, Biomacromolecules, 2010, 11, 3316–3324 CrossRef CAS PubMed.
- R. E. Beck and J. S. Schultz, Science, 1970, 170, 1302–1305 CAS.
- W. Bollmann, Crystal Defects and Crystalline Interfaces, Springer, Berlin, 2012 Search PubMed.
- S. E. Braslavsky, Pure Appl. Chem., 2007, 79, 293–465 CrossRef CAS.
- H. Wesche, S. H. Xiao and S. W. Young, Comb. Chem. High Throughput Screening, 2005, 8, 181–195 CrossRef CAS PubMed.
- J. P. Goddard and J. L. Reymond, Curr. Opin. Biotechnol., 2004, 15, 314–322 CrossRef CAS PubMed.
- J. A. Dietrich, A. E. McKee and J. D. Keasling, Annu. Rev. Biochem., 2010, 79, 563–590 CrossRef CAS PubMed.
- T. Iskratsch, H. Wolfenson and M. P. Sheetz, Nat. Rev. Mol. Cell Biol., 2014, 15, 825–833 CrossRef CAS PubMed.
- U. S. Schwarz and S. A. Safran, Rev. Mod. Phys., 2013, 85, 1327–1381 CrossRef CAS.
- J. L. Leight, D. L. Alge, A. J. Maier and K. S. Anseth, Biomaterials, 2013, 34, 7344–7352 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |