2015 First Boston Symposium of Encoded Library Platforms
Robert A.
Goodnow Jr.†
a and
Christopher P.
Davie†
b
aAstraZeneca, 35 Gatehouse Drive, Waltham, MA 02451, USA. E-mail: robert.doodnow@astrazeneca.com
bGlaxoSmithKline, 830 Winter Street, Waltham, MA 02451, USA. E-mail: christopher.p.davie@gsk.com
The practice of DNA-encoded library synthesis and screening1 continues to evolve with increasing numbers of practitioners as well as reported successes and innovations. In recent years, the steady increase in publications about DNA-encoded platforms signals growing interest and enthusiasm.2 Some of the most important steps in the evolution of this technology and its applications include the development of new chemistries that are compatible with a DNA environment, advances in sequencing, computational efforts to take full advantage of this abundance of data, and customization of affinity-based selections. In addition, the application of this method for hit and lead generation in the drug discovery process foretells its potential return on investment. The International Symposium on DNA-Encoded Chemical Libraries in Zürich is now well established as a European venue for public discussion of these platforms. Based on the success of that meeting and noting a growing interest for this technology in the Boston area, a Symposium of Encoded Library Platforms was organized and convened on November 6, 2015 at AstraZeneca in Waltham, MA. World experts and passionate followers of DNA-encoded library-based platforms convened to share in a day of presentations, posters and discussions of this fast-moving field of science. The enthusiastic participation of some 150 international participants from approximately 50 institutions in this meeting signalled a desire and willingness to enhance common understanding of the latest developments of this technology and its application in drug discovery and chemical biology, and to connect scientists for knowledge sharing and possible collaborations. Through a series of concise lectures, posters, and panel commentary by world-leading experts, future trends in this exciting field have come to light.
Meeting co-organizer Robert Goodnow of AstraZeneca kicked off the meeting with a brief summary of the key aspects of this technological innovation conveyed in the seminal report by Brenner and Lerner.3 These include orthogonality of solid-phase peptide and oligonucleotide synthesis, “split-and-pool” methodology, encoding with DNA, affinity selection methods, amplification with PCR, installation of restriction sites to avoid primer dominance in selective hybridization, and off-DNA re-synthesis. This paper, a thought experiment, contained most if not all the key essential considerations of an encoded chemistry approach. In the intervening twenty-four years, the scientific community has elaborated the details around these key aspects.
The Brenner and Lerner publication appeared at a time when many scientists had been engaged in addressing the problem of encoding for split-and-pool combinatorial chemistry (Fig. 1). The concurrent evolution of combinatorial chemistry in face of an enduring encoding problem for large mixtures made the situation ripe for a good solution. Subsequent implementations of DNA-encoded library technology (often referred to as DELT or ELT) during the 1990s and 2000s made possible the development of new on-DNA chemistries and the synthesis of many libraries available for affinity selections. In the past five years, this chemistry technology has come to fruition as illustrated by the report of many high affinity hits for important drug discovery targets.2
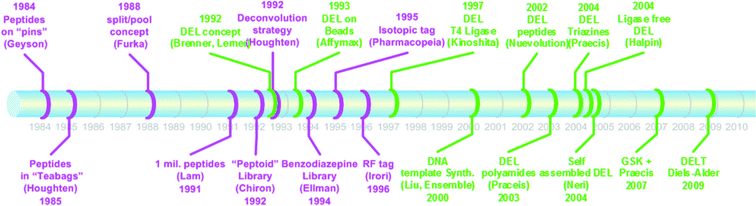 | ||
| Fig. 1 Key events in the development of combinatorial chemistry as a background to the Brenner and Lerner DNA-encoding innovation. | ||
Goodnow went on to describe needs in the pharmaceutical industry that have created so called drivers for innovation and application of this approach. An often underappreciated aspect is the driver of low cost in the prosecution of this technology. To illustrate this idea, Goodnow examined two scenarios. First, he simulated the cost for creation of a one million small molecule screening library from commercially available historical single compounds, from bespoke combinatorial libraries and from custom synthesis.4 Second, he simulated the cost to produce a published DNA-encoded library (DEL) based on publically available information. Whereas a custom library of one million compounds could cost hundreds of millions to two billion US dollars, a DEL of 800 million compounds as reported by Clark et al.5 would cost some two hundred thousand US dollars. It is important to note that the compounds existent in both approaches are unique with respect to many aspects including compound numbers and sources, structural diversity, assay formats and durations, as well as hit identification and follow-up practices. Nevertheless, a ten thousand fold cost differential is compelling and must be recognized as a driver of some force. Professors David Liu and Dario Neri noted the new opportunity that academic scientists enjoy for access to chemical diversity in great numbers due to DELT; in the past, relatively large numbers of compounds were available only through collaborations with pharmaceutical companies.
Goodnow's introductory seminar was followed by a series of excellent presentations from academic and industry leaders in the field: Prof. David Liu (Harvard University), Dr. Christopher Arico-Muendel (GlaxoSmithKline), Dr. Stephen Hale (Ensemble), Dr. Nils Jakob Vest Hansen (Vipergen), Dr. Alexander Satz (Roche), Dr. Thomas Franch (NuEvolution), Dr. Matthew Clark (X-Chem), and keynote speaker Prof. Dario Neri (ETH Zürich). These presentations summarized recent developments in the field, ranging from advances in the DNA-compatible reaction toolbox and library design to a broad spectrum of targets and target classes that have proven successful with ELT. Prof. David Liu's presentation included application of his group's DNA-templated synthesis platform to the identification of small-molecule macrocycles, most notably recent discovery of the first physiologically active inhibitor of insulin-degrading enzyme (IDE).6 Keynote speaker Prof. Dario Neri's seminar, entitled “Single-Pharmacophore and Dual-Pharmacophore DNA-Encoded Chemical Libraries: A Comparative Evaluation”, provided a perspective on the strengths and weaknesses of each of these methodologies his group has been exploring.7 Additional highlights included GSK's presentation on a soluble epoxide hydrolase (sEH) candidate GSK2256294 that has completed Phase 1 studies.8 X-Chem mentioned an autotoxin candidate derived from their ELT platform being developed by its spinoff company, X-Rx, in collaboration with Gilead.9 The symposium then wrapped up with a lively panel discussion led by Dr. Derek Lowe (Vertex Pharmaceuticals) and included experts in the field from both academic and industry settings.
Subsequent articles in this special edition of MedChemComm will provide detailed research updates and perspectives from some of the symposium speakers as well as additional researchers in the field of DNA-Encoded Library Technology (ELT). Further, those interested should also keep in mind the upcoming 5th International Symposium on DNA-Encoded Chemical Libraries coordinated by Prof. Dario Neri and Dr. Jörg Scheuermann (Zürich, Switzerland; August 26, 2016)‡ as well as the Second Boston Symposium of Encoded Library Platforms that will be held in the Boston area in 2017.§
Notes and references
- For recent platform overview articles see: T. Gura, Science, 2015, 350, 1139 CrossRef CAS PubMed; A. Mullard, Nature, 2016, 530, 367 CrossRef PubMed.
- For recent review and perspective articles see: H. Salamon, M. K. Škopić, K. Jung, O. Bugain and A. Brunschweiger, ACS Chem. Biol., 2016, 11, 296 CrossRef CAS PubMed; R. M. Franzini and C. Randolph, J. Med. Chem., 2016 DOI:10.1021/acs.jmedchem.5b01874; R. M. Franzini, D. Neri and J. Scheuermann, Acc. Chem. Res., 2014, 47, 1247 CrossRef PubMed; R. E. Kleiner, C. E. Dumelin and D. R. Liu, Chem. Soc. Rev., 2011, 40, 5707 RSC.
- S. Brenner and R. A. Lerner, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 5381 CrossRef CAS.
- R. Goodnow in A Handbook for DNA-Encoded Chemistry, ed. Robert A. Goodnow, Jr., John Wiley & Son, Inc., Hoboken, New Jersey, 1st edn, 2014, ch. 18, pp. 417–426 Search PubMed.
- M. A. Clark, R. A. Acharya, C. C. Arico-Muendel, S. L. Belyanskaya, D. R. Benjamin, N. R. Carlson, P. A. Centrella, C. H. Chiu, S. P. Creaser, J. W. Cuozzo, D. P. Davie, Y. Ding, G. J. Franklin, K. D. Franzen, M. L. Gefter, S. P. Hale, N. J. V. Hansen, D. I. Israel, J. Jiang, M. J. Kavarana, M. S. Kelley, C. S. Kollmann, F. Li, K. Lind, S. Mataruse, P. F. Medeiros, J. A. Messer, P. Myers, H. O'Keefe, M. C. Oliff, C. E. Rise, A. L. Satz, S. R. Skinner, J. L. Svendsen, L. Tang, K. van Vloten, R. W. Wagner, G. Yao, B. Zhao and B. A. Morgan, Nat. Chem. Biol., 2009, 5, 647 CrossRef CAS PubMed.
- D. Liu, presented in part at First Boston Symposium of Encoded Library Platforms, AstraZeneca, Waltham, MA, November, 2015.
- R. M. Franzini, D. Neri and J. Scheuermann, Acc. Chem. Res., 2014, 47, 1247 CrossRef CAS PubMed.
- A. L. Lazaar, L. Yang, R. L. Boardley, N. S. Goyal, J. Robertson, S. J. Baldwin, D. E. Newby, I. B. Wilkinson, R. Tal-Singer, R. J. Mayer and J. Cheriyan, Br. J. Clin. Pharmacol., 2016, 81, 971 CrossRef CAS PubMed; P. L. Podolin, B. J. Bolognese, J. F. Foley, E. Long III, B. Peck, S. Umbrecht, X. Zhang, P. Zhu, B. Schwartz, W. Xie, C. Quinn, H. Qi, S. Sweitzer, S. Chen, M. Galop, Y. Ding, S. L. Belyanskaya, D. I. Israel, B. A. Morgan, D. J. Behm, J. P. Marino Jr., E. Kurali, M. S. Barnette, R. J. Mayer, C. L. Booth-Genthe and J. F. Callahan, Prostaglandins Other Lipid Mediators, 2013, 104-105, 25 CrossRef PubMed.
- Business Wire Home Page, http://www.businesswire.com/news/home/20151111005581/en/X-Rx-Announces-Autotaxin-Inhibitor-Collaboration-Gilead-Sciences, (accessed March 22, 2016).
Footnotes |
| † Co-organizers of the First Boston Symposium of Encoded Library Platforms (November 6, 2015). |
| ‡ For additional information on the 5th International Symposium on DNA-Encoded Chemical Libraries you can send an email to <decl2016@pharma.ethz.ch>. |
| § Those interested in being contacted about the Second Boston Symposium of Encoded Library Platforms should email Robert Goodnow (robert.doodnow@astrazeneca.com) and Chris Davie (christopher.p.davie@gsk.com). |
| This journal is © The Royal Society of Chemistry 2016 |
