Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biological processing of dinuclear ruthenium complexes in eukaryotic cells
Xin
Li
a,
Kirsten
Heimann
*bc,
Xuyen Thi
Dinh
d,
F. Richard
Keene
*cef and
J. Grant
Collins
*a
aSchool of Physical, Environmental and Mathematical Sciences, University of New South Wales, Australian Defence Force Academy, Canberra, ACT 2600, Australia. E-mail: g.collins@adfa.edu.au
bCollege of Science & Engineering, James Cook University, Townsville, QLD 4811, Australia. E-mail: kirsten.heimann@jcu.edu.au
cCentre for Biodiscovery and Molecular Development of Therapeutics, James Cook University, Townsville, QLD 4811, Australia
dComparative Genomics Centre, James Cook University, Townsville, QLD 4811, Australia
eCollege of Science, Technology & Engineering, James Cook University, Townsville, QLD 4811, Australia
fSchool of Physical Sciences, University of Adelaide, Adelaide, SA 5005, Australia. E-mail: richard.keene@adelaide.edu.au
First published on 25th July 2016
Abstract
The biological processing – mechanism of cellular uptake, effects on the cytoplasmic and mitochondrial membranes, intracellular sites of localisation and induction of reactive oxygen species – of two dinuclear polypyridylruthenium(II) complexes has been examined in three eukaryotic cells lines. Flow cytometry was used to determine the uptake of [{Ru(phen)2}2{μ-bb12}]4+ (Rubb12) and [Ru(phen)2(μ-bb7)Ru(tpy)Cl]3+ {Rubb7-Cl, where phen = 1,10-phenanthroline, tpy = 2,2′:6′,2′′-terpyridine and bbn = bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane} in baby hamster kidney (BHK), human embryonic kidney (HEK-293) and liver carcinoma (HepG2) cell lines. The results demonstrated that the major uptake mechanism for Rubb12 and Rubb7-Cl was active transport, although with a significant contribution from carrier-assisted diffusion for Rubb12 and passive diffusion for Rubb7-Cl. Flow cytometry coupled with Annexin V/TO-PRO-3 double-staining was used to compare cell death by membrane damage or apoptosis. Rubb12 induced significant direct membrane damage, particularly with HepG2 cells, while Rubb7-Cl caused considerably less membrane damage but induced greater levels of apoptosis. Confocal microscopy, coupled with JC-1 assays, demonstrated that Rubb12 depolarises the mitochondrial membrane, whereas Rubb7-Cl had a much smaller affect. Cellular localisation experiments indicated that Rubb12 did not accumulate in the mitochondria, whereas significant mitochondrial accumulation was observed for Rubb7-Cl. The effect of Rubb12 and Rubb7-Cl on intracellular superoxide dismutase activity showed that the ruthenium complexes could induce cell death via a reactive oxygen species-mediated pathway. The results of this study demonstrate that Rubb12 predominantly kills eukaryotic cells by damaging the cytoplasmic membrane. As this dinuclear ruthenium complex has been previously shown to exhibit greater toxicity towards bacteria than eukaryotic cells, the results of the present study suggest that metal-based cationic oligomers can achieve selective toxicity against bacteria, despite exhibiting a non-specific membrane damage mechanism of action.
Introduction
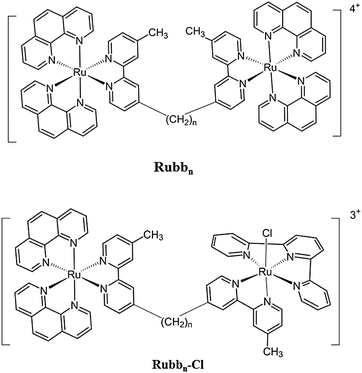
There is considerable interest in developing ruthenium complexes as therapeutic agents, and a variety of coordinatively-labile mononuclear ruthenium(II) and ruthenium(III) complexes have shown significant promise as anticancer drugs.1–6 Furthermore, a range of inert mononuclear ruthenium(II) complexes have shown promising in vitro anticancer and/or antimicrobial properties.7–15 More recently, there has been increasing interest in “tuning” the structure of the ruthenium complexes to increase their activity and selectivity for bacterial or cancer cells. One such strategy, the synthesis of cationic oligomers,16–19 takes advantage of the greater presence of negatively-charged components in the bacterial membrane compared to eukaryotic cells.20,21 The cell wall and outer membrane of bacteria contain a higher proportion of negatively-charged phospholipids – e.g. phosphatidyl-glycerol, as well as a variety of negatively-charged macromolecules such as teichoic acids and lipopolysaccharides – than do eukaryotic cells.20,21We have synthesised and examined the antibacterial activities of a series of dinuclear polypyridylruthenium(II) complexes [{Ru(phen)2}2{μ-bbn}]4+ (Rubbn, see Fig. 1) where (phen = 1,10-phenanthroline and bbn = bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane).22–25 The Rubbn complexes with a short linking chain (bb2, bb5 and bb7) showed little or no antibacterial activity, whereas Rubb12 showed excellent activity and was 40–100 fold less toxic towards eukaryotic cells.22,25 However, it was noted that Rubb12 showed very similar 24-hour IC50 values towards three significantly different eukaryotic cell lines – a baby hamster kidney (BHK), a human embryonic kidney (HEK-293) and a human liver carcinoma (HepG2).25 The similar cytotoxicity data between the eukaryotic cell lines suggest a common, non-specific, mechanism of action. Previous studies with bacteria demonstrated that although Rubb12 preferentially localises at ribosomes within a bacterial cell,26 it also significantly permeabilises the cytoplasmic membrane.27 Non-specific membrane damage would likely result in the ruthenium complex exhibiting very similar IC50 values against the three eukaryotic cell lines. However, a previous study suggested that the Rubbn complexes enter L1210 murine leukaemia cells largely by passive diffusion, accumulate in the mitochondria and induce cell death by apoptosis.28 By contrast, the results of a confocal microscopy study with the BHK, HEK-293 and HepG2 cell lines demonstrated that Rubb12 did not accumulate in the mitochondria, but predominantly localised in the nucleolus of the cell.25 Taken together, the combined results suggest that the biological processing of the Rubbn complexes in white blood cells (L1210) is significantly different to that in kidney and liver cells (BHK, HEK-293 and HepG2). In order to gain a deeper understanding of the effect of the Rubbn complexes on organ cells, and in particular the organs responsible for processing drugs, it was of interest to determine the cellular uptake mechanism and mechanism of cell death of the Rubb12 complex in the BHK, HEK-293 and HepG2 cells.
Although this study was aimed at understanding the biological processing of the Rubbn complexes in eukaryotic cells, it also provides important information for the development of cationic antimicrobial or anticancer oligomers in general. As previously noted, there is considerable current interest in the use of cationic oligomers/polymers for combating bacterial resistance to current drugs.16,19 However, because many of these potential drugs disrupt the bacterial membrane, their clinical application could be limited as they would also interact with eukaryotic membranes in a similar manner. As alkyl chains are commonly used to link the monomeric units of antimicrobial (and anticancer) cationic oligomers/polymers,16–19 the results of this study could well be applicable to a wider range of drugs than the Rubbn complexes.
In addition to the Rubbn complexes, we have also synthesised and examined the biological activities of a series of dinuclear complexes that contain one inert metal centre and one coordinatively-labile metal centre (the Rubbn-Cl complexes, see Fig. 1).29 Interestingly, while each of the Rubb12-Cl and Rubb16-Cl complexes showed similar toxicities across the BHK, HEK-293 and HepG2 cell lines, Rubb7-Cl showed surprising differences in cytotoxicities.29 Against the BHK cells, Rubb7-Cl was by far the least toxic of the Rubbn-Cl and Rubbn complexes (24 h-IC50 = 238 μM); whereas by contrast, against the HepG2 cell line it was by far the most toxic (24 h-IC50 = 3.7 μM).29 The significantly different response exhibited by the BHK, HEK-293 and HepG2 cells to Rubb7-Cl suggests that the ruthenium complex does not act in a non-specific manner (e.g. membrane damage), but through a specific pathway that leads to apoptosis. Consequently, the cellular uptake mechanism and mechanism of cell death for Rubb7-Cl against the BHK, HEK-293 and HepG2 cells was also examined and compared to that obtained for Rubb12.
Results
Cellular uptake mechanism of the ruthenium complexes
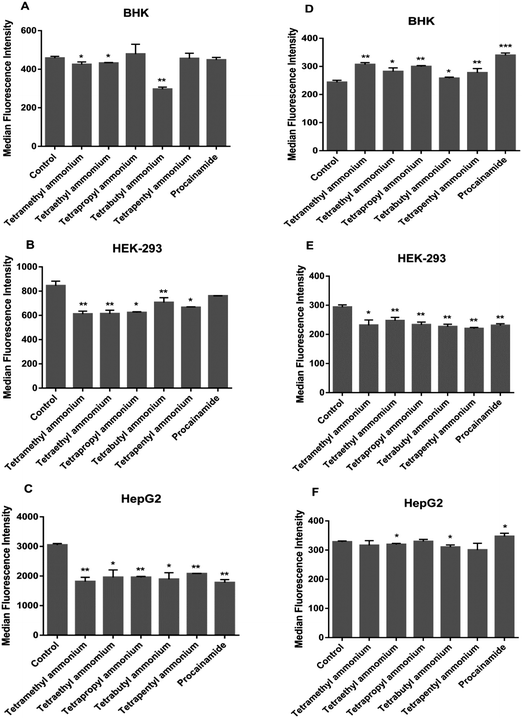
Cellular uptake studies {described in (a)–(d) below} were carried out using flow cytometry for the Rubb12 and Rubb7-Cl complexes in the kidney and liver cell lines. These studies aimed to differentiate between three major uptake pathways: (1) active transport, which is energy-dependent; (2) passive diffusion, an energy-independent form of molecular transport; and (3) carrier-assisted diffusion, an energy-independent pathway, facilitated by membrane protein carriers such as organic cation transporters (OCT).The Rubb12 and Rubb7-Cl complexes have different luminescence signal strengths. Therefore, the median fluorescence intensity (MFI) values cannot be used to compare cellular uptake of different complexes, but only for analysing changes in the uptake of one complex under different conditions in different cells.
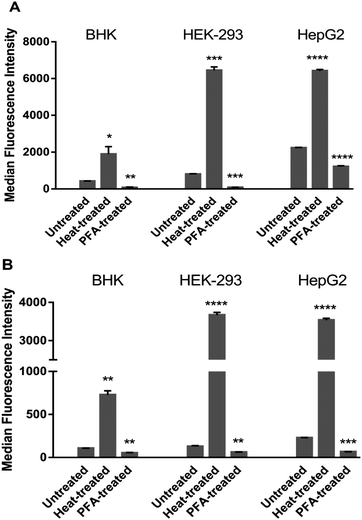
Both complexes accumulated significantly more in heat-treated cells than in untreated and PFA-fixed cells irrespective of the cell line. In control and PFA-fixed samples, the uptake generally followed HepG2 ≥ HEK-293 > BHK for both complexes. However, in heat-treated cells, MFIs in HEK-293 cells were slightly higher than HepG2, and the MFIs in both human cell lines were more than 3-fold higher than for the baby hamster kidney BHK cells. After PFA-fixation, the cellular uptake of Rubb12 and Rubb7-Cl was strongly impeded by 50–90% compared with the untreated controls in the three cell lines.
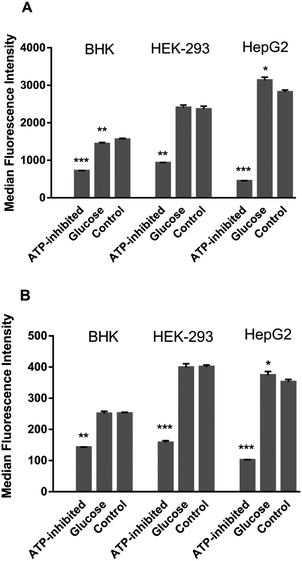
The ATP-production stimulator D-glucose did not enhance uptake of the ruthenium complexes in BHK and HEK-293 cells, with uptake of Rubb12 in BHK even slightly lower. This suggested that the cellular ATP quota was optimal, preventing further accumulation by the treatment. However, for HepG2 cells, D-glucose treatment did induce slightly greater accumulation for Rubb12 and Rubb7-Cl. The inhibition of ATP production caused a significant decrease in the uptake of both ruthenium complexes in all three cell lines, indicating that uptake was predominantly through an energy-dependent (active) process.
Detection of apoptosis and membrane damage
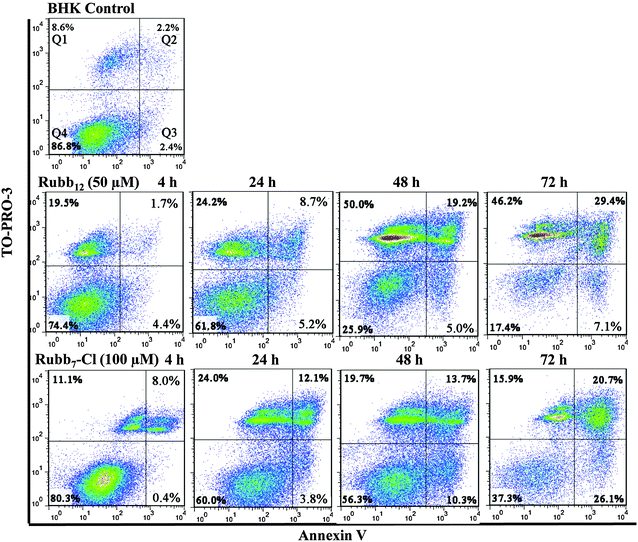
Annexin V/TO-PRO-3 double-staining assays were carried out with BHK and HepG2 cells to compare cell death by membrane damage or apoptosis for Rubb12 and Rubb7-Cl. Early stages of apoptosis are characterised by the translocation of phosphatidylserine from the inner leaflet of the plasma membrane to the outer leaflet, which can be visualised by binding to the Annexin V fluorescent conjugate.33 As cell death progresses, TO-PRO-3 (a cell-impermeant dye) diffuses into the cells with compromised membrane integrity and binds to DNA.33 Alternatively, if the cytoplasmic membrane is directly damaged by the ruthenium complex, TO-PRO-3 will enter the cell and bind to DNA without any translocation of phosphatidylserine from the inner leaflet of the plasma membrane to the outer leaflet. Accordingly, in the flow cytometry assays, apoptosis is demonstrated by cells “moving” from Q4 (bottom-left quadrant) to Q3 (bottom-right) and then finally to Q2 (top-right). On the other hand, direct membrane damage is detected by cells moving from Q4 to Q1 (top-left), and then to Q2 as the cell becomes totally permeabilised (allowing the large Annexin V fluorescent conjugate into the cell) and then possibly to Q3 as TO-PRO-3 is displaced from the DNA by the ruthenium complex.Time-course (4-, 24-, 48- and 72-hours) flow cytometry was conducted using BHK cells with Rubb12 and Rubb7-Cl at half of their 24 h-IC50 concentrations (50 and 100 μM, respectively). A small percentage of the BHK cells (5–10%) were always damaged (observed in Q1) during the trypsin treatment required for cell harvesting; however, little membrane damage was observed for HepG2 cells. After incubation with Rubb12, viable BHK cells (bottom-left quadrant, Q4) decreased gradually with incubation time (see Fig. 6). Noticeably, there was a sharp increase in the top-left quadrant, indicating that half the cells had significant membrane damage, while the early apoptotic cells (bottom-right quadrant, Q3) only increased slowly and to a lesser extent. BHK cells treated with Rubb7-Cl showed a different cell population distribution to that observed with Rubb12. The percentage of cells that exhibited direct membrane damage was considerably smaller (Q1), but the percentage of cells exhibiting apoptosis (Q2 and Q3) was significantly higher than was observed with Rubb12.
Time-course Annexin V/TO-PRO-3 double-staining assays were also carried out with HepG2 cells treated with either Rubb12 or Rubb7-Cl (see Fig. 7). For Rubb12, extensive membrane damage was observed after a 4-hour incubation, with over 50% of the cell population observed in Q1 after 24 hours. Interestingly, after a 72-hour incubation a significant percentage of the cells were Annexin positive and TO-PRO-3 negative (Q3). Given that most of the cells were non-viable due to membrane damage at 4 and 24 hours, the significant percentage of cells in Q3 after a 72-hour incubation suggests the DNA was saturated with Rubb12, rather being indicative of early apoptosis. Saturation with Rubb12 would inhibit TO-PRO-3 from binding to DNA, leading to the observed low levels of TO-PRO-3 fluorescence.
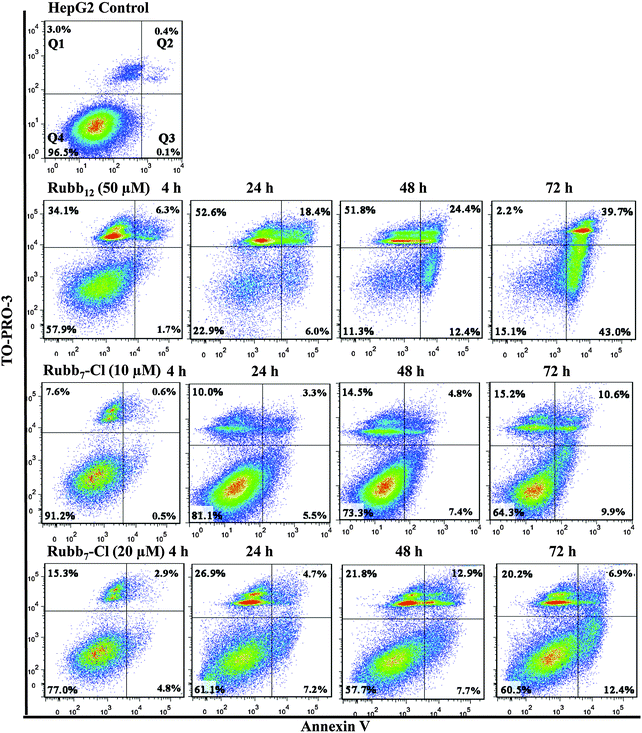 | ||
| Fig. 7 Annexin V/TO-PRO-3 staining of HepG2 cells (control) and with treatment by the ruthenium complexes treatment (from top to bottom: 50 μM Rubb12, 10 μM Rubb7-Cl and 20 μM Rubb7-Cl). | ||
HepG2 cells treated with 10 μM Rubb7-Cl appeared to induce relatively weak responses, with few cells exhibiting significant membrane damage (Q1) and even fewer cells being early-apoptotic (Q3). To further amplify cell responses, the assays were repeated with a Rubb7-Cl concentration of 20 μM. The increased concentration of Rubb7-Cl did slightly enhance the responses, with the percentage of cells exhibiting direct membrane damage doubling (Q1); however there was only a very slight increase in the number of early apoptotic cells (Q3). As the 24 h-IC50 is 3.7 μM, the flow cytometry results suggest that Rubb7-Cl is not cytotoxic against HepG2 cells due to direct membrane damage or any process leading to apoptosis.
Mitochondrial membrane potential
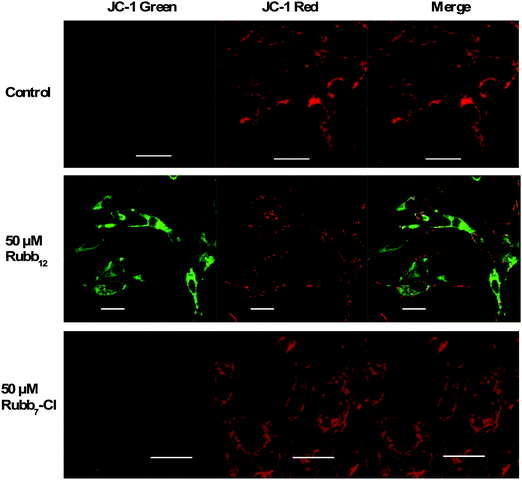
Mitochondrial dysfunction plays an important role in different cell death mechanisms, and the depolarisation of the mitochondrial membrane potential (MMP) due to changes in the mitochondrial permeability has been demonstrated to be an early stage of mitochondrial dysfunction.34 To evaluate the disruption of mitochondrial function induced by Rubb12 and Rubb7-Cl, the change in the mitochondrial membrane potential was determined by confocal microscopy using JC-1 assays.35 Functional polarised mitochondria have a high MMP promoting the formation of red fluorescent JC-1 aggregates, while depolarised mitochondria with low MMP show a diffuse green fluorescence from JC-1 monomers.BHK cells incubated with 50 μM of Rubb12 and Rubb7-Cl showed different changing patterns in MMP (Fig. 8). After a 4-hour incubation, Rubb12 treatment affected the distribution of mitochondria in BHK cells, with low MMP mitochondria spreading throughout the cytoplasm. Mitochondria with high MMP appeared to be confined to the cell periphery. Moreover, some polarised mitochondria appeared elongated. By contrast, in Rubb7-Cl treated BHK cells very few low MMP mitochondria were observed, with the high MMP mitochondria red fluorescence being maintained.
HepG2 cells incubated with 20 μM of Rubb12 and Rubb7-Cl displayed some differences in the staining pattern (Fig. 9). Treatment with Rubb12 led to the localisation of depolarised mitochondria in the perinuclear region. The presence of several condensed green hotspots could be indicative of condensed mitochondria with low MMP in response to Rubb12 treatment. On the other hand, mitochondria with high MMP displayed condensed red fluorescence. After incubation with Rubb7-Cl at the concentration of 20 μM, HepG2 cells displayed only slightly decreased red fluorescence of high MMP mitochondria, with only a few low MMP mitochondria observed in the perinuclear region.
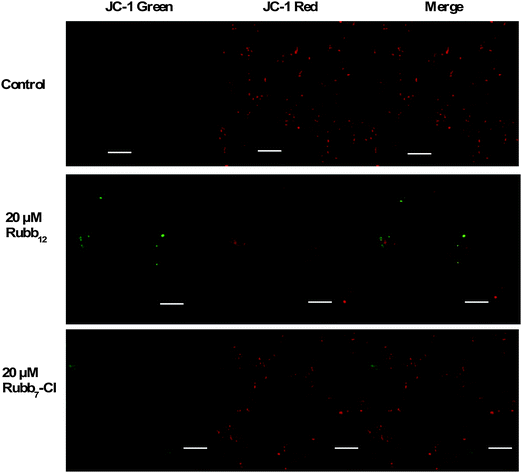 | ||
| Fig. 9 JC-1 fluorescence imaging of mitochondria in HepG2 cells after a 4-hour incubation with 20 μM of Rubb12 (middle) and Rubb7-Cl (bottom). Scale bar = 20 μm. | ||
Additionally, time-course MMP changes (0-, 4- and 20-hour incubations) were monitored in BHK cells with Rubb12 and Rubb7-Cl. As expected, longer incubation time led to increased depolarisation of mitochondria (data not shown).
Cellular localisation
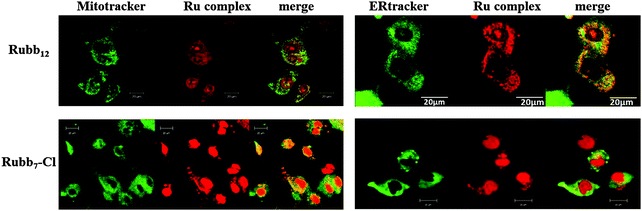
As the JC-1 assays demonstrated that Rubb12 and Rubb7-Cl affected the mitochondrial membrane potential, it was of interest to determine if the ruthenium complexes localised to any extent in the mitochondria. The cellular localisation of high and low concentrations (relative to 24 h-IC50 values) of Rubb12 and Rubb7-Cl in BHK cells has been previously reported.29 Both Rubb12 and Rubb7-Cl preferentially accumulated in the nucleolus at low concentrations, but exhibited significant DNA binding at higher concentrations. Furthermore, mitochondrial localisation was observed for Rubb7-Cl at high concentration, but not for Rubb12.29 The localisation of both ruthenium complexes at low concentrations (5 μM) in HepG2 cells has also been reported.29 However, the localisation of higher concentrations of Rubb12 and Rubb7-Cl in HepG2 cells has not been reported. Although considerable Rubb12 phosphorescence was observed in the cytoplasm of HepG2 cells, no co-localisation was observed in the mitochondria (see Fig. 10). Co-localisation experiments with ERtracker indicated that a significant proportion of the cytoplasmic Rubb12 phosphorescence was due to accumulation in the ER (Fig. 10). By contrast, Rubb7-Cl showed intense co-localisation with mitochondria (yellow overlay), but little in the ER (Fig. 10).Reactive oxygen superoxide dismutase assays
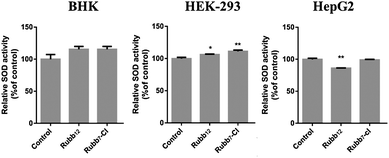
The primary superoxide scavenger, superoxide dismutase (SOD), is an attractive target for pharmacological intervention.36–38 To investigate whether the treatment with ruthenium complexes affected intracellular SOD activity, SOD assays were conducted on BHK, HEK-293 and HepG2 cells after a 20-hour incubation with Rubb12 and Rubb7-Cl (Fig. 11). Compared with the untreated controls, BHK cells appeared to slightly increase SOD activity in response to treatment with the ruthenium complexes, with a non-significant increase for Rubb12 and Rubb7-Cl being observed. Similarly, SOD activity in HEK-293 cells also showed a slight increase for both ruthenium complexes. In contrast, the cancer cell line HepG2 showed a reverse trend in SOD activity, with a significant down-regulation for Rubb12, but no obvious change for Rubb7-Cl.Discussion
Cationic oligomers, such as cationic peptides and di- and oligo-nuclear ruthenium complexes, have considerable potential as selective antibacterial agents due to their preferential interactions with the negatively-charged bacterial membrane and cell wall. One approach to the preparation of di- and oligo-nuclear ruthenium complexes is to link the individual metal centres with a ligand that contains an alkyl chain. The length of the alkyl chain could affect cellular uptake, membrane damage and intracellular localisation in bacterial and eukaryotic cells. It has been previously demonstrated that the alkyl chain length controls the uptake in bacterial and L1210 cells.23,28 However, for both bacterial and L1210 cells, the dominant mechanism of cellular uptake was passive diffusion.23,28 The aims of this study were to determine if the Rubb12 and Rubb7-Cl complexes were taken up by passive diffusion in organ cells, and to examine the effect of the ruthenium complexes on the cytoplasmic and mitochondrial membranes.Heat treatment at 60 °C destroys membrane integrity completely39 and compounds freely pass into the cells and associate with all available biological ligands. Following 60 °C treatment, both Rubb12 and Rubb7-Cl uptake was greater in the human cell lines than in the baby hamster cells (HEK-293 ≥ HepG2 > BHK), suggesting more intracellular binding sites in the two human cell lines than in BHK cells. As chromosomal DNA was one of the main intracellular targets, these results could be partly explained by the different chromosome numbers in HEK-293, HepG2 and BHK cells – with 64, 55 and 44 chromosomes, respectively.40 PFA-fixation causes cross-linking of membrane proteins (including protein carriers),41 leading to diminished uptake, with compounds predominantly entering cells via passive diffusion. The observed large loss of Rubb12 and Rubb7-Cl uptake in PFA-fixed cells may suggest that the inhibited active/carrier-assisted transportation routes contribute more than 50% of the total uptake in all cases.
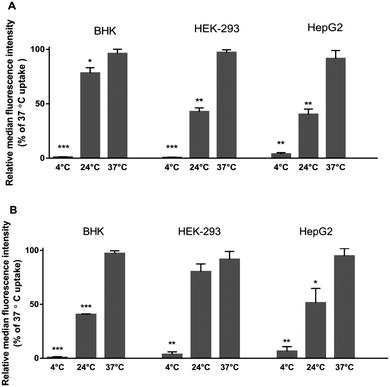
As carrier proteins are inactivated and ATP-production is limited at 4 °C,42 the small uptake of Rubb12 and Rubb7-Cl at 4 °C compared with the greater uptake at higher temperatures demonstrated the strong temperature-dependence of the uptake in the eukaryotic cells. This is consistent with the previously reported energy-dependent uptake of the rigidly linked dinuclear complex [{Ru(phen)2}2{μ-tpphz}]4+ in MCF-7 cells.43 However, other ruthenium complexes are transported via passive diffusion; for instance the mononuclear complex [Ru(DIP)2dppz]2+ showed much higher uptake at 4 °C.44 Due to the decreased membrane fluidity at 4 °C (increased rigidity of the phospholipids chains), the cell membrane would be less permeable than it would be at 37 °C.45,46 Consequently, and particularly as the interactions between the phospholipid alkyl chains and the alkyl chains of the bbn ligand are likely to be important, it is possible that the ruthenium complexes might not be able to enter the cells by passive diffusion at 4 °C. On the other hand, at 24 °C, ATP-dependent transport is mostly inhibited, while passive transport and carrier-facilitated diffusion function normally.47,48 Other studies have demonstrated that strictly ATP-required uptake of peptides was reduced by ∼75% at 24 °C compared to 37 °C,49 and Na–K-ATPase activity is inhibited by ∼70% in mammalian erythrocytes at 24 °C.50 These findings indicate that ∼25–30% of active uptake occurs at 24 °C. Accordingly, the actual active uptake can be estimated. Corrected estimations suggest active transport generally appears to be the dominant cellular uptake mechanism, contributing to more than half of the total uptake. However, for Rubb12 in BHK cells and Rubb7-Cl in HEK-239 cells slightly lower contributions of active uptake (∼30%) are estimated, indicating carrier-assisted diffusion could also be involved in the uptake.
To further ascertain the contribution of active uptake, the energy dependence of cellular uptake of Rubb12 and Rubb7-Cl was determined by pretreating cells with the ATP-production stimulator D-glucose or the metabolic inhibitors 2-deoxy-glucose and oligomycin. The unchanged uptake in the D-glucose-treated BHK and HEK-293 cells indicate an optimal energy condition, which could not be further improved. However, the increased uptake in D-glucose-treated HepG2 cells was indicative of glucose deprivation in HepG2 cells. This may be a result of a heavy reliance on glycolysis for ATP production in cancer cells instead of mitochondrial oxidative phosphorylation,51 the latter being the primary pathway to produce ATP in healthy cells.52 Therefore, it is possible that the external supply of glucose enhanced energy production in HepG2 cells, consequently leading to greater active uptake of ruthenium complexes. In contrast, the significant decrease in uptake in ATP-inhibited cells for both Rubb12 and Rubb7-Cl confirmed a significantly large contribution of active uptake of the ruthenium complexes.
The actual contribution of active uptake could be even larger than the observed proportions, considering that ATP-production cannot be completely blocked by the metabolic inhibitors.53,54 These findings, in conjunction with the lower temperature study results, strongly support the conclusion that active uptake is the major mechanism of cell entry for the ruthenium complexes studied in kidney and liver cell lines.
Based on the 24 °C uptake and the ATP-inhibited uptake results, the possible role of polyspecific organic cation transporters (OCT)-assisted diffusion of Rubb12 and Rubb7-Cl was explored. The lower uptake of Rubb12 following OCTs inhibition in the three cell lines, supports the conclusion that the uptake of Rubb12 was partially facilitated by OCTs. In contrast, Rubb7-Cl displayed no OCTs-facilitated diffusion in BHK and HepG2 cells, but a considerable contribution of OCTs-facilitated uptake was noted in HEK-293 cells. It is possible that the labile complex Rubb7-Cl interacts with the carrier proteins in a covalent mode, making dissociation harder from the membrane protein, and transport by carriers like OCTs less likely. On the other hand, a correlation between OCT binding with molecules that contain both a positive charge and high lipophilicity has been observed.55 This may explain the greater OCT-assisted uptake of the more positively-charged and possibly more lipophilic Rubb12.
In summary, the major uptake mechanism for Rubb12 and Rubb7-Cl is active transport in kidney and liver cells – with a significant contribution from carrier-assisted diffusion for Rubb12 but mainly passive diffusion for Rubb7-Cl. However, previous studies from our research group on Rubbn complexes demonstrated uptake by passive diffusion in L1210 leukaemia cells and bacteria cells.23,28 As the function of kidney and liver are filtering and detoxification,56 uptake of molecules can be expected to be more selective and finely managed, which is only possible by active uptake.
To determine the extent of the damage to the cytoplasmic membrane, and to compare the cellular consequences of membrane damage with cell death through an apoptotic pathway, Annexin V/TO-PRO-3 assays were carried out. The results clearly demonstrated that Rubb12 caused significant direct membrane damage to both BHK and HepG2 cells; however, membrane damage was considerably more rapid and extensive with the HepG2 cells. By contrast, Rubb7-Cl caused significantly less membrane damage than Rubb12 in both cell lines. Of note however, was the relatively low level of apoptotic cells up to 48 hours for both cell lines.
As an indicator of mitochondrial health, mitochondrial membrane potential (MMP) was monitored using the JC-1 assays. The results clearly demonstrate that Rubb12 depolarises the mitochondrial membrane in both BHK and HepG2 cells; whereas, Rubb7-Cl had a much smaller affect. The cellular localisation experiments indicated that although Rubb12 did not accumulate in the mitochondria in either cell line, significant mitochondrial accumulation was observed for Rubb7-Cl in both BHK and HepG2 lines. While DNA damage and ER stress beyond a certain repair-threshold could reduce MMP,55,57,58 the combined results (lack of localisation and reduction in MMP) are consistent with Rubb12 damaging the mitochondrial membrane. Given the previously-reported membrane damage induced by Rubbn in bacterial cells,27 the lipophilic ruthenium complexes could lead to depolarisation of membranes in eukaryotic cells, including the cell membrane and intracellular membrane compartments (e.g. ER, mitochondria, Golgi, endosome and lysosome), resulting in irreparable membrane damage.
Reactive oxygen species (ROS) have long been known as inducers of programmed cell death.59 ER/mitochondrial localisation of the ruthenium complexes makes generation of ROS likely, as these two organelles are intricately involved in the overall ROS regulation.60 Many ruthenium complexes have been demonstrated to increase ROS generation and induce oxidative stress and apoptosis.61–64 To estimate the oxidative perturbation caused by the ruthenium complexes Rubb12 and Rubb7-Cl, their effects on the intracellular activity of superoxide dismutase (SOD), the essential enzyme eliminating ROS, was studied. The observed enhanced SOD activity in BHK and HEK-293 cells can be indicative of increased ROS stress induced by the ruthenium complexes. This hypothesis is supported by the reported increased SOD activity in response to oxidative stress in plants65 and other animal cells.66 However, in HepG2 cells, SOD activity decreased with treatment by Rubb12 but remained unchanged for Rubb7-Cl. Since HepG2 cells are more susceptible to the ruthenium complexes than the other two cell lines, a decrease in SOD activity rather than increased activity was to be expected, due to the direct oxidative damage of SOD enzymes and/or ROS-altered SOD gene expression.67–69 Similar reduction of SOD activity has been reported in cisplatin treated rat ovarian cells70 and in vitamin-treated breast cancer cells.71 The unchanged SOD activity in Rubb7-Cl treated HepG2 could be explained by a balance between up-regulation and the inhibition of the enzyme activity/expression. These results suggest that the ruthenium complexes could induce cell death via a ROS-mediated pathway.
The present study demonstrates that the dinuclear ruthenium complex linked by the bb12 ligand (Rubb12) extensively damages the cytoplasmic membrane and probably the mitochondrial membrane. Although it has been established that Rubb12 selectively localises with RNA in eukaryotic cells,25 it is likely that membrane damage is the dominant mechanism of cell death. By contrast, Rubb7-Cl has a considerably smaller affect on the cytoplasmic and mitochondrial membranes. As a consequence, Rubb7-Cl shows greater variability in its 24 h-IC50s towards eukaryotic cells. After activation by aquation, Rubb7-Cl can form covalent bonds with biomolecules. Although Rubb7-Cl only requires 3.5 hours to be 50% aquated in aqueous solution at 25 °C,29 covalent bond formation with nucleotides and amino acids is very slow.29 This suggests that Rubb7-Cl predominantly induces its biological effect through non-covalent interactions within eukaryotic cells. HepG2 cells treated with Rubb7-Cl at a concentration more than six-fold the 72 h-IC50 exhibited only a slight cell response, with weak apoptosis and weak membrane damage. These weak cellular responses could be partly due to the low incidence of drug-induced apoptosis reported for some cancer cells.72,73 However, Rubb7-Cl targets nucleic DNA in HepG2 cells,29 which may suggest another predominant cell death pathway. The DNA binding caused by Rubb7-Cl may induce DNA damage (for example condensation), therefore halting the cell cycle.
The results of this study indicate that Rubb12 causes significant membrane damage – both cytoplasmic and mitochondrial – whereas little membrane damage was observed for Rubb7-Cl. These conclusions are consistent with the IC50 values observed for Rubb12 and Rubb7-Cl against the three eukaryotic cell lines – very similar values for Rubb12, and highly divergent values for Rubb7-Cl. Furthermore, the results of this study suggest that cationic oligomers joined by alkyl chain lengths of twelve carbons or longer will likely cause significant membrane damage to eukaryotic cells. Alternatively, cationic oligomers joined by alkyl chain lengths of seven carbons are unlikely to cause significant membrane damage, and hence may provide a better scaffold for oligomeric or polymeric antibacterial agents. The mechanism by which Rubb12 damages membranes is unknown, but it could be related to the lipophilicity of the dinuclear ruthenium complex or the actual chain length. Given that the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for Rubb12 is −2.7,22 it seems unlikely that differences in lipophilicity between Rubb12 and Rubb7-Cl is the dominant determinant of membrane damage. It is noted that the highly non-polar section of a membrane is 25–30 Å in width,74 which is approximately the same distance as that between the ruthenium centres (26 Å) in Rubb12. As the ruthenium metal centres can favourably interact (through π–π interactions of the phenanthroline ligands) and the distance between the ruthenium centres is compatible with the non-polar region of a membrane, it is possible that membrane damage is caused by aggregated Rubb12 molecules inserting perpendicularly into the membrane to form a pore. This could then lead to greater levels of membrane disruption, eventually leading to the complete permeabilisation of the cell.
P for Rubb12 is −2.7,22 it seems unlikely that differences in lipophilicity between Rubb12 and Rubb7-Cl is the dominant determinant of membrane damage. It is noted that the highly non-polar section of a membrane is 25–30 Å in width,74 which is approximately the same distance as that between the ruthenium centres (26 Å) in Rubb12. As the ruthenium metal centres can favourably interact (through π–π interactions of the phenanthroline ligands) and the distance between the ruthenium centres is compatible with the non-polar region of a membrane, it is possible that membrane damage is caused by aggregated Rubb12 molecules inserting perpendicularly into the membrane to form a pore. This could then lead to greater levels of membrane disruption, eventually leading to the complete permeabilisation of the cell.
Conclusions
Dinuclear ruthenium complexes have shown considerable potential as antimicrobial agents; however, in order to rationally design more active and more selective compounds, it is necessary to gain an understanding of the biological processing of the dinuclear complexes in bacterial and eukaryotic cells. This study provides a detailed analysis of the biological processing of the dinuclear complexes in eukaryotic cells, and in particular, an analysis of the effects of these compounds on the membranes of eukaryotic cells. The results indicate that Rubb12 damages the cytoplasmic membrane, and probably the membranes of internal organelles like the mitochondria. However, despite the fact that Rubb12 kills eukaryotic cells in a non-specific manner, it can still be 40–100 times more active against bacteria than eukaryotic cells.29 By contrast, Rubb7-Cl does not significantly damage membranes and is cytotoxic through other specific mechanism(s). As a consequence of this, Rubb7-Cl exhibits very variable cytotoxicities against various cell lines. It is commonly believed that potential new antimicrobial drugs should not be active through non-specific mechanisms (as generally evidenced by similar IC50 values against eukaryotic cells), and in particular, should not just damage cell membranes. The results of this study suggest there are sufficient differences, particularly in the charge density, between bacterial and eukaryotic membranes that cationic oligomers (like Rubb12) could form the basis of new types of antimicrobial agents.Experimental procedures
Material
The ruthenium complexes Rubb12 and Rubb7-Cl were synthesised as previously described.29,75Cell culture
Three mammalian cell lines BHK (baby hamster kidney), HEK-293 (embryonic kidney) and HepG2 (liver carcinoma) were used in this study. All cell lines were generously supplied by the Australian Army Malaria Institute (AMI, Enoggera, QLD, Australia), and originated from the American Type Culture Collection (ATCC, Manassas, USA). Cells were cultured as previously described.25 Cells used in the study were in the logarithmic growth phase and were grown to 70% confluence, and then trypsinised with 0.25% trypsin–0.02% EDTA (Sigma-Aldrich) for detachment and used in the assays described below.Cellular localisation of the ruthenium complexes
The trypsinised cells were seeded in Lumox® multiwell plates (Sarstedt, Numbrecht, Germany), and incubated with 20 to 50 μM ruthenium complexes at 37 °C with 5% CO2 for 4 or 20 h as described. Following incubation, cells were stained with 100 nM Mitotracker® Green FM (Invitrogen, Carlsbad, USA) for mitochondrial staining and 1 μM ER-Tracker™ Green (Invitrogen) for endoplasmic reticulum (ER) staining. Staining was carried out in RPMI-1640 medium under standard cultivation conditions as per the manufacturer's instructions. Following staining, cells were gently rinsed with phosphate buffer solution (PBS; pH = 7.1) prior to confocal laser scanning microscopy.The cellular localisation of the ruthenium complexes was determined using a Zeiss laser scanning confocal microscope (LSM 700, Carl Zeiss, Göttingen, Germany). Samples were viewed under 63× oil immersion using the following excitation (λex) and emission (λem) wavelengths. Metal complexes (λex = 450 nm, λem = 610 nm), Mitotracker Green FM (λex = 490 nm, λem = 516 nm) and ER-Tracker™ Green (λex = 504 nm, λem = 511 nm) were excited using a blue argon laser (λex = 488 nm), and emissions were collected at λem = 570–650 nm for the ruthenium complexes, λem = 470–550 nm for Mitotracker and ER-tracker. Image data acquisition and processing was performed using Zen software 2009 (Carl Zeiss).
Cellular uptake of ruthenium complexes
Effect of cell membrane permeability on the uptake of ruthenium complexes
Cell membrane permeability was modified by heat treatment or fixation with paraformaldehyde (PFA, Sigma-Aldrich, St. Louis, USA). The cells were cultured in three aliquots. Given the upper temperature limit for eukaryotic cells near 60 °C (due to their inability to form membranes that are both thermostable and functional),39 one aliquot was pre-incubated at 60 °C for 30 min before the ruthenium complexes were added. The heat treatment permeabilised and killed the cells, allowing for the maximal interaction between the ruthenium complexes and their intracellular targets. The second aliquot was incubated in fresh 4% PFA solution for 10 min at room temperature. The PFA solution was removed and the cell layer was rinsed with PBS before incubation with the ruthenium complexes. The third aliquot served as the control group following standard staining procedures. Ruthenium complexes (20 μM) were added to pretreated and control cells, followed by incubation at 37 °C for 4 h for all samples. After the incubation, cells were gently trypsinised, centrifuged and washed twice with PBS, and re-suspended in PBS for flow cytometry. Statistical analysis was performed using a t-test in GraphPad Prism 6.0 (GraphPad Software, San Diego, USA). A P value of <0.05 was considered significant.Dependence of cellular uptake of the ruthenium complexes on temperature
Cells were incubated with the ruthenium complexes (50 μM for BHK, 20 μM for HEK-293 and 10 μM for HepG2, reflecting the trend in the IC50 values for Rubb7-Cl) for 4 h at 4, 24 or 37 °C. Following incubation, the cells were trypsinised, centrifuged and rinsed with PBS before analysis by flow cytometry. t-Tests were performed to evaluate the significance of temperature on the uptake with 37 °C treatments serving as controls.Dependence of cellular uptake of the ruthenium complexes on ATP production
Cells were pre-incubated with 2-deoxy-glucose (30 mM, Sigma-Aldrich) and oligomycin (10 μM, Sigma-Aldrich), or the ATP stimulator D-glucose (30 mM; Sigma-Aldrich) for 2 h at 37 °C. Ruthenium complexes (20 μM) were added to pretreated cells and untreated cells (controls) and incubated at 37 °C for 4 h. After incubation, the cells were typsinised, centrifuged and rinsed twice with PBS for flow cytometry analysis. Results were statistically analysed using a t-test.Effect of organic cation transport inhibitors on the uptake of the ruthenium complexes
Cells were pre-incubated for 1 h at 37 °C with six organic cation transport inhibitors32 (Table 1; tetramethylammonium chloride, tetraethylammonium chloride monohydrate, tetrapropylammonium chloride, tetra-n-butylammonium chloride, tetra-n-pentylammonium chloride and procainamide hydrochloride; Sigma-Aldrich) at 2 mM in RPMI-1640 complete medium. Thereafter, the ruthenium complexes (20 μM) were added and incubations continued for 4 h at 37 °C. The cells were trypsinised, centrifuged and washed for analysis by flow cytometry. To compare the treated groups with controls, the data were analyzed by multiple t-tests.| OCT inhibitors | OCTs inhibited |
|---|---|
| Tetramethylammonium chloride | OCT1, OCT2 |
| Tetraethylammonium chloride | OCT1, OCT2, OCT3, OCTN1, OCTN2 |
| Tetrapropylammonium chloride | OCT1, OCT2 |
| Tetra-n-butylammonium chloride | OCT1, OCT2, OCTN1 |
| Tetra-n-pentylammonium chloride | OCT1, OCT2, OCT3, OCTN1 |
| Procainamide | OCT1, OCT2, OCT3, OCTN1, OCTN2 |
Determination of membrane damage and apoptosis induced by the ruthenium complexes
Apoptosis induced by the ruthenium complexes was assessed using the Alexa Fluor® 488 Annexin V assay (Sigma-Aldrich) for detecting phosphatidyl-serine in the external plasma membrane leaflet and TO-PRO-3 (Molecular Probes®, Invitrogen) for assessing cell membrane permeability. BHK and HepG2 cells were incubated with the ruthenium complexes for the desired times. Cells were gently trypsinised, washed and collected by centrifugation. Cell pellets were resuspended to 1 × 105 to 1 × 106 cells mL−1 in 100 μL Annexin V binding buffer. Alexa Fluor® 488 Annexin V (5 μL) was added and the samples were incubated at room temperature for 15 min in darkness. Annexin V binding buffer (400 μL) and TO-PRO-3 (5 μL) were added to each sample immediately prior to analysis on a BD LSR Fortessa flow cytometer (Franklin Lakes, USA), with a 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per sample recorded for each experiment. Data were analysed using FlowJo 7.6.1 (FlowJo Software, Ashland, USA).
000 events per sample recorded for each experiment. Data were analysed using FlowJo 7.6.1 (FlowJo Software, Ashland, USA).
Detection of mitochondrial membrane potential
Detection of mitochondrial membrane potential (MMP) was carried out using a Zeiss laser scanning confocal microscope (LSM 700, Carl Zeiss, Göttingen, Germany). BHK and HepG2 cells were treated with Rubb12 and Rubb7-Cl for 4 h in Lumox® multiwell plates (Sarstedt). After rinsing, treated cells were incubated with 5 μM JC-1 dye (mitochondrial membrane potential probe, Invitrogen) in complete RPMI-1640 medium for 30 min at 37 °C. Cells were washed with PBS before confocal microscopy. JC-1 monomers were excited with a λex = 488 nm argon-ion laser and emissions collected over the range of λem = 515–545 nm. J-aggregate forms were excited with a 568 nm argon–krypton laser and the emission signals collected over the range of λem = 575–625 nm. Time-course MMP tests were conducted using the BHK cell line. The cells were incubated with 20 μM of Rubb12 or 50 μM of Rubb7-Cl (approximately 25% of the respective 24 h-IC50 values) for 4 or 20 h, and the treated cells were processed as above.Reactive oxygen superoxide dismutase (SOD) assays
The SOD activities in the cell lines BHK, HEK-293 and HepG2 after treatment with Rubb12 and Rubb7-Cl were determined using the SOD assay kit (Sigma-Aldrich). The cell lines were incubated with the ruthenium complexes for 20 h at different concentrations (50 μM for BHK, 20 μM for HEK-293 and 10 μM for HepG2, respectively). Cells were detached from the tissue culture flask by trypsinisation and collected by centrifugation. The cell pellets were resuspended in PBS buffer solution to 5 × 106 cells mL−1. Glass beads (150–212 μm; Sigma-Aldrich) were added to the cell suspension and agitated using a bead beater (BBY24M-CE; Next Advance, Averill Park, USA) for 10 min at 4 °C to break the cell membrane. Supernatants were collected after centrifugation, and the SOD activity was determined as per the procedure provided by the manufacturer of the assay kit. Data were statistically analysed using t-tests, with the SOD activity in untreated cells serving as controls.Acknowledgements
The authors thank Professor Alan Baxter for open access to the tissue culture and specialist infrastructure in the Comparative Genomics Centre at James Cook University.References
- S. Kapitza, M. A. Jakupec, M. Uhla, B. K. Keppler and B. Marian, Cancer Lett., 2005, 226, 115 CrossRef CAS PubMed
.
-
E. Alessio, G. Mestroni, A. Bergamo and G. Sava, in Metal Ions Biological Systems, ed. A. Sigel, H. Sigel and M. Dekker, CRC Press, New York, 2004, pp. 323–351 Search PubMed
.
- L. Ronconi and P. J. Sadler, Coord. Chem. Rev., 2007, 251, 1633 CrossRef CAS
.
- Y. K. Yan, M. Melchart, A. Habtemariam and P. J. Sadler, Chem. Commun., 2005, 4764 RSC
.
- C. A. Vock, W. H. Ang, C. Scolaro, A. D. Phillips, L. Lagopoulos, L. Juillerat-Jeanneret, G. Sava, R. Scopelliti and P. J. Dyson, J. Med. Chem., 2007, 50, 2166 CrossRef CAS PubMed
.
-
G. Sava, E. Alessio, A. Bergamo and G. Mestroni, in Biological Inorganic Chemistry, ed. M. J. Clarke and P. J. Sadler, Springer, Berlin, 1999, vol. 1, pp. 143–169 Search PubMed
.
- M. R. Gill and J. A. Thomas, Chem. Soc. Rev., 2012, 41, 3179 RSC
.
- C. S. Devi, D. A. Kumar, S. S. Singh, N. Gabra, N. Deepika, Y. P. Kumar and S. Satyanarayana, Eur. J. Med. Chem., 2013, 64, 410 CrossRef PubMed
.
- F. P. Dwyer, E. C. Gyarfas, W. P. Rogers and J. H. Koch, Nature, 1952, 170, 190 CrossRef CAS PubMed
.
- A. Bolhuis, L. Hand, J. E. Marshall, A. D. Richards, A. Rodger and J. Aldrich-Wright, Eur. J. Pharm. Sci., 2011, 42, 313 CrossRef CAS PubMed
.
- S. V. Kumar, W. K. C. Lo, H. J. L. Brooks and J. D. Crowley, Inorg. Chim. Acta, 2015, 425, 1 CrossRef CAS
.
- V. Pierroz, T. Joshi, A. Leonidova, C. Mari, J. Schur, I. Ott, L. Spiccia, S. Ferrari and G. Gasser, J. Am. Chem. Soc., 2012, 134, 20376 CrossRef CAS PubMed
.
- H. Huang, P. Zhang, B. Yu, Y. Chen, J. Wang, L. Ji and H. Chao, J. Med. Chem., 2014, 57, 8971 CrossRef CAS PubMed
.
- H. Huang, B. Yu, P. Zhang, J. Huang, Y. Chen, G. Gasser, L. Ji and H. Chao, Angew. Chem., Int. Ed., 2015, 54, 14049 CrossRef CAS PubMed
.
- H. Huang, P. Zhang, H. Chen, L. Ji and H. Chao, Chem. – Eur. J., 2015, 21, 715 CrossRef CAS PubMed
.
- A. S. Abd-El-Aziz, C. Agatemor, N. Etkin, D. P. Overy, M. Lanteigne, K. McQuillan and R. G. Kerr, Biomacromolecules, 2015, 16, 3694 CrossRef CAS PubMed
.
- A. Martin, A. Byrne, C. S. Burke, R. J. Forster and T. E. Keyes, J. Am. Chem. Soc., 2014, 136, 15300 CrossRef CAS PubMed
.
- P. Liu, B.-Y. Wu, J. Liu, Y.-C. Dai, Y.-J. Wang and K.-Z. Wang, Inorg. Chem., 2016, 55, 1412 CrossRef CAS PubMed
.
- F. Li, J. G. Collins and F. R. Keene, Chem. Soc. Rev., 2015, 44, 2529 RSC
.
- H. Jenssen, P. Hamill and R. E. W. Hancock, Clin. Microbiol. Rev., 2006, 19, 491 CrossRef CAS PubMed
.
- S. K. Vooturi, C. M. Cheung, M. J. Rybak and S. M. Firestine, J. Med. Chem., 2009, 52, 5020 CrossRef CAS PubMed
.
- F. Li, Y. Mulyana, M. Feterl, J. M. Warner, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 5032 RSC
.
- F. Li, M. Feterl, Y. Mulyana, J. M. Warner, J. G. Collins and F. R. Keene, J. Antimicrob. Chemother., 2012, 67, 2686 CrossRef CAS PubMed
.
- F. Li, M. Feterl, J. M. Warner, A. I. Day, F. R. Keene and J. G. Collins, Dalton Trans., 2013, 42, 8868 RSC
.
- X. Li, A. K. Gorle, T. D. Ainsworth, K. Heimann, C. E. Woodward, J. G. Collins and F. R. Keene, Dalton Trans., 2015, 44, 3594 RSC
.
- F. Li, E. J. Harry, A. L. Bottomley, M. D. Edstein, G. W. Birrell, C. E. Woodward, F. R. Keene and J. G. Collins, Chem. Sci., 2014, 5, 685 RSC
.
- F. Li, M. Feterl, J. M. Warner, F. R. Keene and J. G. Collins, J. Antimicrob. Chemother., 2013, 68, 2825 CrossRef CAS PubMed
.
- M. J. Pisani, P. D. Fromm, Y. Mulyana, R. J. Clarke, H. Körner, K. Heimann, J. G. Collins and F. R. Keene, ChemMedChem, 2011, 6, 848 CrossRef CAS PubMed
.
- X. Li, K. Heimann, F. Li, J. M. Warner, F. R. Keene and J. G. Collins, Dalton Trans., 2016, 45, 4017 RSC
.
- E. C. Slater, Methods Enzymol., 1967, 10, 48 CAS
.
- H. Koepsell and H. Endou, Eur. J. Appl. Physiol., 2004, 447, 666 CrossRef CAS PubMed
.
- S. Barban and H. O. Schulze, J. Biol. Chem., 1961, 236, 1887 CAS
.
- I. Vermes, C. Haanen, H. Steffens-Nakken and C. Reutelingsperger, J. Immunol. Methods, 1995, 184, 39 CrossRef CAS PubMed
.
- V. Borutaite, Environ. Mol. Mutagen., 2010, 51, 406 CAS
.
- J. D. Ly, D. R. Grubb and A. Lawen, Apoptosis, 2003, 8, 115 CrossRef CAS PubMed
.
-
B. Halliwell and J. M. C. Gutteridge, Free radicals in biology and medicine, Oxford University Press, New York, 2007, pp. 449–450 Search PubMed
.
- L. W. Oberley, G. R. Buettner and G. R. Bueftner, Cancer Res., 1979, 39, 1141 CAS
.
- I. Fridovich, Acc. Chem. Res., 1972, 5, 321 CrossRef CAS
.
- M. R. Tansey and T. D. Brock, Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 2426 CrossRef CAS
.
- American Type Culture Collection, http://www.atcc.org/.
- J. A. Kiernan, Microsc. Today, 2000, 1, 8 Search PubMed
.
- H. Ginsburg and S. Yeroushalmy, J. Appl. Physiol., 1978, 282, 399 CAS
.
- M. R. Gill, J. Garcia-Lara, S. J. Foster, C. Smythe, G. Battaglia and J. A. Thomas, Nat. Chem., 2009, 1, 662 CrossRef CAS PubMed
.
- C. A. Puckett and J. K. Barton, Biochemistry, 2008, 47, 11711 CrossRef CAS PubMed
.
- M. Rossignol, T. Uso and P. Thomas, J. Membr. Biol., 1985, 87, 269 CrossRef CAS PubMed
.
- M. B. Lande, J. M. Donovan and M. L. Zeidel, J. Gen. Physiol., 1995, 106, 67 CrossRef CAS PubMed
.
- U. D. Kulkarni, R. Mahalingam, X. Li, I. Pather and B. Jasti, AAPS PharmSciTech, 2011, 12, 579 CrossRef CAS PubMed
.
- C. A. Lesch, C. A. Squier, A. Cruchley, D. M. Williams and P. Speight, J. Dent. Res., 1989, 68, 1345 CrossRef CAS PubMed
.
- J. C. Wolters, R. Abele and R. Tampé, J. Biol. Chem., 2005, 280, 23631 CrossRef CAS PubMed
.
- S. L. Kimzey and J. S. Willis, J. Gen. Physiol., 1971, 58, 634 CrossRef CAS PubMed
.
- O. Warburg, Science, 1956, 123, 309 CAS
.
-
I. M. Ahmad and M. Y. Abdalla, in Oxidative stress in cancer biology and therapy, ed. D. R. Spitz, K. J. Dornfeld, K. Krishnan and D. Gius, Humana Press, New York, 2012, pp. 1–19 Search PubMed
.
- A. N. Wick, D. R. Drury, H. I. Nakada and J. B. Wolfe, J. Biol. Chem., 1957, 224, 963 CAS
.
- M. Jastroch, A. S. Divakaruni, S. Mookerjee, J. R. Treberg and M. D. Brand, Essays Biochem., 2010, 47, 53 CrossRef CAS PubMed
.
- G. Kroemer, L. Galluzzi and C. Brenner, Physiol. Rev., 2007, 87, 99 CrossRef CAS PubMed
.
-
N. Ahmed, D. Maureen, C. Smith and E. Wood, Biology of Disease, Taylor & Francis, 2006, p. 600 Search PubMed
.
- W. X. Zong, C. Li, G. Hatzivassiliou, T. Lindsten, Q. C. Yu, J. Yuan and C. B. Thompson, J. Cell Biol., 2003, 162, 59 CrossRef CAS PubMed
.
- L. Filippin, P. J. Magalhães, G. Di Benedetto, M. Colella and T. Pozzan, J. Biol. Chem., 2003, 278, 39224 CrossRef CAS PubMed
.
- S. C. Gupta, D. Hevia, S. Patchva, B. Park, W. Koh and B. B. Aggarwal, Antioxid. Redox Signaling, 2012, 16, 1295 CrossRef CAS PubMed
.
- A. A. Alfadda and R. M. Sallam, J. Biomed. Biotechnol., 2012, 936486 Search PubMed
.
- W. Li, B.-J. Han, J.-H. Yao, G.-B. Jiang and Y.-J. Liu, RSC Adv., 2015, 5, 24534 RSC
.
- Z. Zhao, Z. Luo, Q. Wu, W. Zheng, Y. Feng and T. Chen, Dalton Trans., 2014, 43, 17017 RSC
.
- Z. Luo, L. Yu, F. Yang, Z. Zhao, B. Yu, H. Lai, K.-H. Wong, S.-M. Ngai, W. Zheng and T. Chen, Metallomics, 2014, 6, 1480 RSC
.
- C. Tan, S. Lai, S. Wu, S. Hu, L. Zhou, Y. Chen, M. Wang, Y. Zhu, W. Lian, W. Peng, J. Liangnian and A. Xu, J. Med. Chem., 2010, 53, 7613 CrossRef CAS PubMed
.
- A. S. Gupta, J. L. Heinen, A. S. Holaday, J. J. Burke and R. D. Allen, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 1629 CrossRef CAS
.
- P. States, T. Promotion and N. Series, Science, 1985, 227, 375 Search PubMed
.
- M. S. Cooke, M. D. Evans, M. Dizdaroglu and J. Lunec, FASEB J., 2003, 17, 1195 CrossRef CAS PubMed
.
- P. V. Limaye, N. Raghuram and S. Sivakami, Mol. Cell. Biochem., 2003, 243, 147 CrossRef CAS PubMed
.
-
C. Gemma, J. Vila, A. Bachstetter and P. Bickford, in Brain aging: models, methods and mechanisms, ed. D. R. Riddle, CRC Press/Taylor & Francis, 2007, pp. 353–374 Search PubMed
.
- X. Li, S. Yang, X. Lv, H. Sun, J. Weng, Y. Liang and D. Zhou, J. Gynecol. Oncol., 2013, 24, 177 CrossRef CAS PubMed
.
-
D. Feldman, J. W. Pike and F. H. Glorieux, Vitamin D, Elsevier Academic Press, 2005, p. 1952 Search PubMed
.
- D. Hanahan and R. A. Weinberg, Cell, 2000, 100, 57 CrossRef CAS PubMed
.
- S. Fulda, Int. J. Cancer, 2009, 124, 511 CrossRef CAS PubMed
.
- B. A. Lewis and D. M. Engelman, J. Mol. Biol., 1983, 166, 211 CrossRef CAS PubMed
.
- Y. Mulyana, D. K. Weber, D. P. Buck, C. A. Motti, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 1510 RSC
.
| This journal is © The Royal Society of Chemistry 2016 |