Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Discrimination between bacterial species by ratiometric analysis of their carbohydrate binding profile†
Lucienne
Otten
a,
Elizabeth
Fullam
b and
Matthew I.
Gibson
 *a
*a
aDepartment of Chemistry, University of Warwick, Gibbet Hill Road, Coventry, CV4 7AL, UK. E-mail: m.i.gibson@warwick.ac.uk
bSchool of Life Sciences, University of Warwick, Gibbet Hill Road, Coventry, UK
First published on 1st December 2015
Abstract
Antibiotic resistance is a global health concern meaning there is an urgent need for new treatments and diagnostics. Here glycosylated surfaces are used to profile the binding patterns of a range of Gram-negative, Gram-positive and mycobacteria. This enables the creation of ‘barcodes’ to enable identification and discrimination between the strains, which could not be achieved by single-point glycan binding and offers a new concept in bacteria detection.
Bacteria cause diseases including meningitis, pneumonia, diarrhea and many hospital acquired infections such as Staphylococcus aureus1 or Clostridium difficile2 and are responsible for millions of deaths every year. ‘Old’ diseases such as tuberculosis, caused by the bacterium Mycobacterium tuberculosis, for which antibiotics had previous proven to be effective, are once again on the rise3–5 with an estimated 2 billion carriers globally. Since penicillin was discovered in 1929 it revolutionized the treatment of bacterial infections6 but the current drug pipeline has dried up alongside development of resistance.7 The spread of resistance is very rapid; for example methicillin was clinically used in 1960 with the first case of methicillin resistant S. aureus reported in 1961.8 These factors have contributed to the prediction that by 2050 the number of deaths associated with antimicrobial resistance will outnumber those caused by cancer.9,10 The problem of antibiotic resistance is propagated by the lack of rapid point-of-care diagnostics tools to ensure correct use of the remaining drugs. Most of the current Food and Drug Administration (FDA) approved detection methods rely on a culturing step; the bacteria are isolated and grown for visual, or other examination methods, which can take several days11 and some strains are intrinsically challenging to culture. Since Robert Koch first described culturing for identification of bacteria over 140 years ago the techniques used has hardly changed since.12,13 However biofilm based infections are now becoming the most common problem, with these bacteria proving incredibly challenging to culture.14 It is estimated that only 1% of bacteria are culturable on standard culture media and several of the species that commonly cause hospital acquired infections can enter a viable but not culturable phase where they are not detectable through culturing techniques.12,14,15 The emerging alternative is the use of polymerase chain reaction (PCR) or sequencing methods. PCR based processes include; (i) the analysis of the full genome of microbial species and identification through comparison with a database16,17 and (ii) Ibis, a method which involves selective PCR of a species-specific element of DNA which is flanked by highly conserved regions. Whilst PCR based methods are very rapid (with Ibis reportedly only taking 6 hours) they are still relatively expensive (Ibis needs a mass spectrometer) and rely on extraction of nucleic acids from within bacteria, which can be problematic in Gram-positive species due to their thick peptidoglycan wall, which is difficult to lyse.14
Many bacteria infections require an initial adhesion step, for example, the high mannose-binding adhesin FimH is a crucial virulence factor found in uropathogenic Escherichia coli and among enterobacteriacae.18,19 Biofilm based infections are responsible for many chronic bacteria associated conditions such as pneumonia20 and tuberculosis21 and also the most difficult to culture.22–24 Considering the above, adhesins are potential targets for novel prophylactic anti-adhesion therapies25,26 or as diagnostics.27–30 Several nanoparticle-based detections systems exploiting adhesins have been reported including galabiose particles for the detection of Streptococcus suis.20,31 Mannose conjugated gold nanoparticles have been shown to detect FimH positive strains of E. coli in a colorimetric assay.27 These particle-based systems highlight the potential for carbohydrates in detection but are intrinsically limited by the promiscuity of glycan–lectin (carbohydrate binding proteins) interactions. Therefore, specific identification of lectins (or bacteria) is complicated by the non-specific binding which can give false positives. To overcome this, we have reported the use of a powerful statistical analysis technique for the identification of carbohydrate binding toxins with similar binding specificity using linear discriminant analysis (LDA) – essentially we created a ‘bar code’ to described each lectin.32 Here we demonstrate a multiplexed bacteria assay based on their relative binding affinity to a small and accessible array of glycans. The relative binding affinity of each bacteria was determined to generate a training matrix, which can subsequently be used for discrimination/identification. A range of Gram-negative, Gram-positive including mycobacteria are studied, including surrogates for M. tuberculosis.
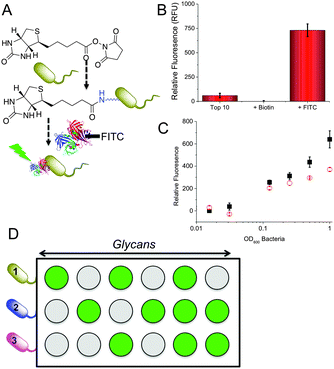
The key aim here was to assess if the relative binding profiles of different bacterial strains to a range of glycans could be used to generate a training matrix as an easy method to enable multiplexed diagnostics based on each strains relative binding to different sugars. To generate glycosylated surfaces we employed hydrazide functional 96 well plates (Carbo-BIND™). Glycosylation with reducing carbohydrates was enabled by simply heating at 50 °C with an aniline catalyst. This method gives a mixture between ring-closed (approximately 60%) and ring opened sugars. Whilst generating some heterogeneity, the discrimination method we employ (see later) is not affected by this, and in theory any other glycan immobilisation method could be used. To enable visualisation of bacterial binding for this proof-of-concept study, a two-step labelling procedure was employed. Bacteria were first biotinylated with Biotin-NHS, followed by reaction with FITC-labelled streptavidin (Fig. 1A). We choose this method to enable facile binding measurement rather than as a true diagnostic tool. Alternative detection methods include microscopy, SPR, bilayer interferometry or nanoparticle binding, but are outside the scope of this manuscript. Successful labelling of the bacteria was showed by measuring the fluorescence (Fig. 1B) of the bacterial cultures and visual examination.
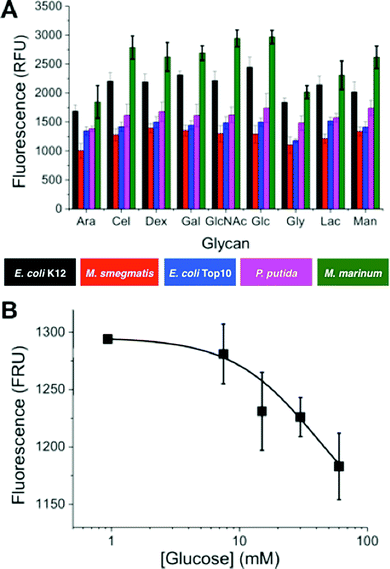
To evaluate glycan binding, Top10 and K12 (E. coli) were incubated with a glucose functional surface as a function of bacterial concentration. A clear dose-dependant response was observed with the K12's showing more adherence at higher ODs (defined as the absorbance at 600 nm), which we attribute to their expression of FimH (Fig. 1C). This experiment also serves to highlight the challenge of single-channel glycan-based sensors – both bacteria would give a positive response. Fig. 1D shows the multiplexed concept to be used here. Looking ‘across’ from any given glycan, it would not be possible to identify the bacteria. However, using multiple glycan ‘inputs’ enables a barcode to be created which is unique to the bacteria. It is therefore clear that several glycans are required to gain sufficient resolution. 9 different glycans were immobilised onto the plates and 5 different bacteria strains (labelled as described above) were interrogated. The bacterial species examined here are; two strains of E. coli Top10 and K12, Pseudomonas putida, Mycobacterium smegmatis and Mycobacterium marinum. Of the two strains of E. coli: Top10 is FimH negative and K12 is FimH positive and thus a mimic of pathogenic strains. P. putida is similar in nature to Pseudomonas aeruginosa which causes life threatening infections that can affect multiple organs but commonly causes pneumonia in cystic fibrosis patients.33,34M. marinum and M. smegmatis are both model organisms for M. tuberculosis, the causative agent of tuberculosis, with M. marinum sharing around 3000 orthologs with over 85% amino acid identity with M. tuberculosis.35 The species analysed included Gram-positive and Gram-negative species, pathogenic and non-pathogenic samples and different strains of the same species providing a focussed, but diverse set of bacteria. The relative binding of these bacteria to each surface was evaluated by fluorescence and the profiles are show in Fig. 2A.
Fig. 2A shows that individual strains exhibited distinct levels of adhesion. For example M. marinum showed higher binding to most glycans than the others. It is beyond the scope of this study to identify the individual components responsible for the binding, and it should be highlighted that this knowledge is not essential for this biosensory approach (vide infra). However, it was important to confirm the interactions were due to glycan–adhesion, and not just non-specific binding. A competition experiment was conducted whereby OD = 1 M. smegmatis was incubated with the surfaces, but with increasing glucose concentrations in the buffer (1–50 mM). As the glucose concentration was increased, there was a clear decrease in the amount of binding (Fig. 2B).
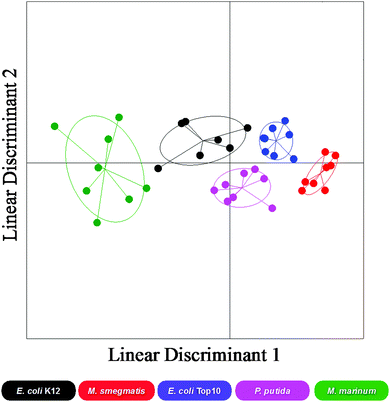
There are many putative carbohydrate binding proteins (including transporters) in the strains, and any given adhesin can also bind to other glycans, even if it has relatively lower affinity. Considering this, any single sugar cannot identify a specific bacteria, within the error of the measurement. However, each bacteria has a distinct set (‘barcode’) of binding across the range of sugars. This large set of data was thus used as a training matrix for LDA.32,36 LDA is a powerful classification tool, which for every class (bacterial species) within the training matrix (binding profiles) it minimises the variation within each class and maximises the variation between classes by transforming the original data in order to get maximum separation between groups. For the bacterial binding data, the LDA model produced showed good resolution between bacterial species and when validated using a ‘leave-one-out’ approach (where each sample is left out in an iterative process and the model generated before classification of the missing sample) the model was able to re-classify all data points to their original class with 92% accuracy. Fig. 3 shows a 2-dimensional representation of the LDA analysis and confidence boundaries for each bacterial strain. This clearly shows that each bacteria has a distinct ‘region’ enabling discrimination. In particular, the two mycobacterial species (M. smegmatis and M. marinum) had excellent separation.
Finally a blind test was done. A culture was prepared by an independent operator and provided. This was fluorescently labelled and incubated with each of the glycan surfaces in triplicate. The average binding response was classified using the LDA model and was correctly identified as E. coli Top10 with 96% certainty. Future work will involve the extension of this method to ‘label-free’ analytical techniques and also focussing on clinical strains. The rapid and simple nature of this makes it ideal for translation and compares favourable with sequencing/PCR based methods such as Ibis which takes at least 6 hours for identification,14 and in some cases expansion and culture of the bacteria.
To conclude, we have demonstrated a rapid technique for the identification of bacterial strains based on profiling their differential binding to carbohydrate functionalised surfaces coupled with a powerful, but simple to use, classification algorithm. The procedure was shown to be valid for Gram-negative and Gram-positive/mycobacteria, indicating it has potential to diagnose or identify a broad range of pathogenic bacteria, either on its own, or in combination with other established analytical methods. In particular, the potential for rapid and low-cost diagnostics is shown including for mycobacteria. M. tuberculosis is a re-emerging global healthcare threat with no suitable detection methods.
We would like to thank Dr Alasdair Hubbard for technical assistance. Equipment was supported by the Birmingham Science City (SC) Advanced Materials project, with support from Advantage West Midlands and part funded by the European Regional Development Fund. LO is funded by the BBSRC via the Systems Biology DTC. EF is a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 104193/Z/14/Z). MIG thanks the ERC for a Starting Grant, CRYOMAT 638661.
Notes and references
- M. T. Holden, L. Y. Hsu, K. Kurt, L. A. Weinert, A. E. Mather, S. R. Harris, B. Strommenger, F. Layer, W. Witte, H. de Lencastre, R. Skov, H. Westh, H. Zemlickova, G. Coombs, A. M. Kearns, R. L. Hill, J. Edgeworth, I. Gould, V. Gant, J. Cooke, G. F. Edwards, P. R. McAdam, K. E. Templeton, A. McCann, Z. Zhou, S. Castillo-Ramirez, E. J. Feil, L. O. Hudson, M. C. Enright, F. Balloux, D. M. Aanensen, B. G. Spratt, J. R. Fitzgerald, J. Parkhill, M. Achtman, S. D. Bentley and U. Nubel, Genome Res., 2013, 23, 653–664 CrossRef CAS PubMed.
- C. M. Surawicz, L. J. Brandt, D. G. Binion, A. N. Ananthakrishnan, S. R. Curry, P. H. Gilligan, L. V. McFarland, M. Mellow and B. S. Zuckerbraun, Am. J. Gastroenterol., 2013, 108, 478–498 CrossRef CAS PubMed.
- A. Zaidi, W. Huskins, D. Thaver, Z. Bhutta, Z. Abbas and D. Goldmann, Lancet, 2005, 365, 1175–1188 CrossRef.
- J. M. Q. Hiram, F. Ricardo, C. Miguel, A. Jose, C. B. Maria, C. M. S. Juan, R. Pedro Torellas and C. Veronica, B48. case reports on Tuberculosis, American Thoracic Society, 2015, pp. A3275–A3275 Search PubMed.
- N. Rezaei, A. Aghamohammadi, D. Mansouri, N. Parvaneh and J. L. Casanova, Expert Rev. Clin. Immunol., 2011, 7, 129–131 CrossRef PubMed.
- C. M. Grossman, Ann. Intern. Med., 2008, 149, 135–136 CrossRef PubMed.
- M. Cully, Nature, 2014, 509, S16–S17 CrossRef CAS PubMed.
- D. Bethune, R. Blowers, M. Parker, E. Pask, O. Jessen, K. Rosendal, P. Bulow, V. Faber, K. Eriksen and K. Hiramatsu, Br. Med. J., 1961, 124, 124–125 Search PubMed.
- J. O'Neil, Antimicrobial resistance: Tackling a crisis for the health and wealth of nations, http://amr-review.org/home.
- B. Spellberg, R. Guidos, D. Gilbert, J. Bradley, H. W. Boucher, W. M. Scheld, J. G. Bartlett, J. Edwards and T. I. D. S. O. America, Clin. Infect. Dis., 2008, 46, 155–164 CrossRef PubMed.
- P. Tissari, A. Zumla, E. Tarkka, S. Mero, L. Savolainen, M. Vaara, A. Aittakorpi, S. Laakso, M. Lindfors, H. Piiparinen, M. Mäki, C. Carder, J. Huggett and V. Gant, Lancet, 2010, 375, 224–230 CrossRef CAS.
- S. Tan, S. Chew, S. Tan, M. Givskov and L. Yang, Curr. Opin. Biotechnol., 2014, 26, 1–6 CrossRef CAS PubMed.
- R. Koch and T. Brock, Milestones in microbiology: 1556 to 1940, 1884 Search PubMed.
- J. W. Costerton, J. C. Post, G. D. Ehrlich, F. Z. Hu, R. Kreft, L. Nistico, S. Kathju, P. Stoodley, L. Hall-Stoodley, G. Maale, G. James, N. Sotereanos and P. DeMeo, FEMS Immunol. Med. Microbiol., 2011, 61, 133–140 CrossRef CAS PubMed.
- S. Pasquaroli, G. Zandri, C. Vignaroli, C. Vuotto, G. Donelli and F. Biavasco, J. Antimicrob. Chemother., 2013, 68, 1812–1817 CrossRef CAS PubMed.
- E. L. Doughty, M. J. Sergeant, I. Adetifa, M. Antonio and M. J. Pallen, PeerJ, 2014, 2, e585 Search PubMed.
- M. R. Halachev, J. Z. Chan, C. I. Constantinidou, N. Cumley, C. Bradley, M. Smith-Banks, B. Oppenheim and M. J. Pallen, Genome Med., 2014, 6, 70 CrossRef PubMed.
- M. Hartmann, H. Papavlassopoulos, V. Chandrasekaran, C. Grabosch, F. Beiroth, T. K. Lindhorst and C. Röhl, FEBS Lett., 2012, 586, 1459–1465 CrossRef CAS PubMed.
- S. Sakarya and S. Oncu, Med. Sci. Monit., 2003, 9, RA76–82 CAS.
- N. P. Pera and R. J. Pieters, Med. Chem. Commun., 2014, 5, 1027–1035 RSC.
- A. K. Ojha, A. D. Baughn, D. Sambandan, T. Hsu, X. Trivelli, Y. Guerardel, A. Alahari, L. Kremer, W. R. Jacobs and G. F. Hatfull, Mol. Microbiol., 2008, 69, 164–174 CrossRef CAS PubMed.
- L.-C. Xu and C. A. Siedlecki, Acta Biomater., 2012, 8, 72–81 CrossRef CAS PubMed.
- J. W. Costerton, P. S. Stewart and E. P. Greenberg, Science, 1999, 284, 1318–1322 CrossRef CAS PubMed.
- B. Hoyle and J. W. Costerton, in Prog. Drug Res., ed. E. Jucker, Birkhäuser Basel, 1991, vol. 37, ch. 2, pp. 91–105 Search PubMed.
- M. Jones, L. Otten, S.-J. Richards, R. Lowery, D. Phillips, D. Haddleton and M. Gibson, Chem. Sci., 2014, 5, 1611–1616 RSC.
- S. J. Richards, M. W. Jones, M. Hunaban, D. M. Haddleton and M. I. Gibson, Angew. Chem., Int. Ed., 2012, 51, 7812–7816 CrossRef CAS PubMed.
- S.-J. Richards, E. Fullam, G. S. Besra and M. I. Gibson, J. Mater. Chem. B, 2014, 2, 1490–1498 RSC.
- R. L. Phillips, O. R. Miranda, C. C. You, V. M. Rotello and U. H. Bunz, Angew. Chem., Int. Ed., 2008, 47, 2590–2594 CrossRef CAS PubMed.
- M. D. Disney and P. H. Seeberger, Chem. Biol., 2004, 11, 1701–1707 CrossRef CAS PubMed.
- E. Bulard, A. Bouchet-Spinelli, P. Chaud, A. Roget, R. Calemczuk, S. Fort and T. Livache, Anal. Chem., 2015, 87, 1804–1811 CrossRef CAS PubMed.
- N. P. Pera, A. Kouki, S. Haataja, H. M. Branderhorst, R. M. J. Liskamp, G. M. Visser, J. Finne and R. J. Pieters, Org. Biomol. Chem., 2010, 8, 2425–2429 CAS.
- L. Otten and M. I. Gibson, RSC Adv., 2015, 5, 53911–53914 RSC.
- A. Wittgens, T. Tiso, T. T. Arndt, P. Wenk, J. Hemmerich, C. Muller, R. Wichmann, B. Kupper, M. Zwick, S. Wilhelm, R. Hausmann, C. Syldatk, F. Rosenau and L. M. Blank, Microb. Cell Fact., 2011, 10, 80 CrossRef CAS PubMed.
- N. Hoiby, E. W. Flensborg, B. Beck, B. Friis, S. V. Jacobsen and L. Jacobsen, Scand. J. Respir. Dis., 1977, 58, 65–79 CAS.
- T. P. Stinear, T. Seemann, P. F. Harrison, G. A. Jenkin, J. K. Davies, P. D. Johnson, Z. Abdellah, C. Arrowsmith, T. Chillingworth, C. Churcher, K. Clarke, A. Cronin, P. Davis, I. Goodhead, N. Holroyd, K. Jagels, A. Lord, S. Moule, K. Mungall, H. Norbertczak, M. A. Quail, E. Rabbinowitsch, D. Walker, B. White, S. Whitehead, P. L. Small, R. Brosch, L. Ramakrishnan, M. A. Fischbach, J. Parkhill and S. T. Cole, Genome Res., 2008, 18, 729–741 CrossRef CAS PubMed.
- A. Izenman, Modern multivariate statistical techniques, Springer, 2008 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures. See DOI: 10.1039/c5mb00720h |
| This journal is © The Royal Society of Chemistry 2016 |