tRNA – the golden standard in molecular biology†
Mirosława Z.
Barciszewska
,
Patrick M.
Perrigue
and
Jan
Barciszewski
*
Institute of Bioorganic Chemistry of the Polish Academy of Sciences, Noskowskiego 12, 61-704 Poznań, Poland. E-mail: Jan.Barciszewski@ibch.poznan.pl
First published on 30th October 2015
Abstract
Transfer RNAs (tRNAs) represent a major class of RNA molecules. Their primary function is to help decode a messenger RNA (mRNA) sequence in order to synthesize protein and thus ensures the precise translation of genetic information that is imprinted in DNA. The discovery of tRNA in the late 1950's provided critical insight into a genetic machinery when little was known about the central dogma of molecular biology. In 1965, Robert Holley determined the first nucleotide sequence of alanine transfer RNA (tRNAAla) which earned him the 1968 Nobel Prize in Physiology or Medicine. Today, tRNA is one of the best described and characterized biological molecules. Here we review some of the key historical events in tRNA research which led to breakthrough discoveries and new developments in molecular biology.
1. Introduction
The discovery of the DNA double helix raised a very interesting and important question. How does the structure of DNA relate to the structure of protein? At that time in 1953, there was a general belief that RNA copies of a single strand of DNA were made inside the cell. These would act as templates to encode the amino acid sequence in proteins. However, this raised another question related to protein biosynthesis. How does the translation mechanism work to generate a protein from RNA? There was no chemical similarity or complementarity between amino acids and nucleotides and therefore no means by which they could directly interact with each other. Francis Crick addressed the basic question of how the sequence of amino acids in proteins could be assembled on the basis of an RNA sequence. In his 1955 letter, “On Degenerate Templates and the Adaptor Hypothesis”, which Francis Crick had sent as, “A Note to the RNA Tie Club” (partially published in 1958), he proposed that amino acids might be first attached to short single strands of RNA molecules thereby making the amino acids recognizable to a complementary sequence of nucleotides on the template.1–5 Twenty specific enzymes would catalyze the attachment of twenty different amino acids separately to twenty various adaptor RNA molecules. The ribosome would then use the complementary base pairing between the single-stranded RNA template and amino acid-carrying RNA molecules to precisely polymerize amino acids into protein.1 Later on, these adaptors turned out to be much larger than originally predicted by Crick. The discoverer of these RNA species, Paul Zamecnik, coined them soluble RNA in 1958.2,3 One should note that there was no knowledge of mRNA during the mid-1950's. Only later mRNA was discovered by Jacob and Monod, i.e. in 1961.4In 1962, almost concurrent with the discovery of mRNA, Alexander Rich proposed that some RNAs may have catalytic activity in addition to being carriers of genetic information.5 As a result there were many suggestions that if RNA participated both genetics and catalysis, then the origin of life would be free of the improbability of multiple biopolymers spontaneously emerging together at one time (e.g. DNA and proteins).6–8
Experimental studies on transfer RNA had started as early as 1955 when many people like Zamecnik, DeMoss, Genuth and Novelli, as well as Berg and Newton, showed that amino acids are activated enzymatically to give enzyme-bound aminoacyladenylates. Furthermore, Hoagland, Ogata, Nohara and others succeeded in the binding of radioactive amino acids to a small, soluble molecular weight RNA in a rat-liver enzyme preparation, which was called enzyme pH 5.9–15 The small, soluble molecular weight RNA was in fact tRNA.2,3 Almost at the same time, Robert Holley demonstrated that the pH 5 enzyme catalyzed alanine-dependent ATP-AMP exchange and was sensitive to ribonuclease.16 Soon after, it was demonstrated that there were many amino acid-activating enzymes within the pH 5 fraction capable of transferring activated amino acids to tRNAs. These became to be known as the aminoacyl-tRNA synthetase family of enzymes (E.C.6.1.1.1-). That name was coined by Paul Berg's group in 1958. Each enzyme is specific and attaches to a given amino acid onto a common CCA sequence at the 3′ end of a specific (cognate) tRNA.2,3,8,17 The amino acid binding to the 3′ terminal adenosine residue takes place via an amino acid ester bond to the 2′- or 3′ −OH group of the ribose moiety. This was first shown by Hans G. Zachau.18 Since different amino acids did not compete for the same tRNA attachment site, it seemed most likely that different tRNAs were serving as acceptors for specific amino acids. Therefore, it was interesting to see if different amino acid-bound RNAs would be small enough to permit detailed structural studies, so that the specific functions could be understood. The discovery of tRNAs and assigning the nucleotide sequences of the tRNAs to each of the different 20 amino acids was the first real hope of solving the genetic code.2,3,8
2. The primary structure of tRNAs
Robert Holley started his tRNA structural studies at Caltech in 1956. He later went on to Cornell University to purify tRNAAla from yeast by counter current distribution. A total of 1 g of highly purified tRNAAla was isolated from approximately 260 g of crude yeast tRNAs, which in turn had been obtained by phenol (200 kg) extraction of approximately 140 kg of commercially available baker's yeast.19 At this point it may be of interest to recall a minor, but important observation made by Holley during his early work on tRNA. He found that during the isolation of the specific tRNAAla there sometimes would be a loss of tRNA activity. He attributed this loss of tRNA activity to nuclease contamination that came from his skin. At that time it had not been generally anticipated that such serious contamination by nucleases could result from personal contact of the glassware, dialysis tubing, and so forth.20The structural analysis of tRNA required the identification of all nucleotide residues and their position in sequence. The experimental approach that was used involved cleavage of the polynucleotide chain into small fragments with specific ribonucleases, identification of the small fragments, and then reconstruction of the original nucleotide sequence by determining the order in which the small fragments existed in the tRNA molecule. Pancreatic and Taka-Diastase ribonucleases were used to cleave the RNA chain into fragments ending with pyrimidine nucleotides and guanylic acid, respectively.21 In the mid-sixties of the last century determination of a primary structure of a tRNA fragment required years of time and usually involved the work of several scientists.
The first tRNA sequence had been established by Holley in 1965.21 Partial digestion of the larger oligonucleotides with snake venom phosphodiesterase was necessary to identify the 77 nucleotide sequence in tRNAAla. Three possible secondary structures were suggested for this tRNA, one of which was proposed by Elisabeth Keller is the well-known cloverleaf model.21
The discovery of the tRNAAla and its total sequence determination took Holley and his co-workers a total of nine years. This work was a great achievement in molecular biology and a very important milestone towards solving the genetic code. For that great accomplishment Holly was awarded the Nobel Prize, which he shared with Har G. Khorana and Marshall Nirenberg in 1968.22,23
Ten years later in 1975 Robert Penswick, the co-author of the first paper on tRNA sequencing, together with Guy Dirheimer's group in Strasbourg re-sequenced the same tRNA and found a mistake in the nucleotide sequence. There was an additional GC dinucleotide in the dihydrouridine stem of the yeast tRNAAla.24
In 1966, Zachau succeeded in the determination of the two tRNASer1 and tRNASer2 isoacceptor nucleotide sequences.25 Using similar strategies, the nucleotide sequence of the next three specific tRNAs were determined, including tRNATyr by Madison,26 tRNAPhe by RajBhandary27 and tRNAVal by Baev.28 The sequencing of tRNAs was one of the most interesting topics of that time between 1965 and 1968. Twelve different tRNAs had been sequenced by 1968 as Holley mentioned in his acceptance speech of the Nobel Prize. The availability of the first several tRNA nucleotide sequences and the finding that they could be folded into the same secondary structure proved the cloverleaf model for the secondary structure of all tRNAs (Fig. 1). The strongest evidence for that structure came from the observation that all tRNAs fit the same type of internal complementarity, based on Watson–Crick base pairing. All tRNAs have four hairpin helices and three major loop structures. In every case, the variable loop is inserted between two hairpin structural elements. The 3′ end of all tRNA molecules consist of the strongly conserved CCA sequence.
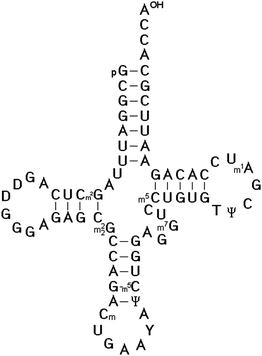 | ||
| Fig. 1 The cloverleaf model for the secondary structure of Saccharomyces cerevisiae tRNAPhe determined by Tom RajBhandary et al.27 It is the most studied and well-known tRNA, for which crystal structures were solved.36–40 | ||
The DNA that encodes tRNAAla in yeast cells was soon chemically synthesized by Khorana and his co-workers.29 In 1983, this tRNAAla gene was used as a prototype template for the polymerase chain reaction developed by Kary Mullis.30
A large amount of data on tRNA sequences has accumulated in the past 50 years. There are now more than 600 different tRNA sequences known and the number of modified nucleosides within these sequences is nearly 120.31–33 We also know the primary structures of these tRNAs. This can be primarily attributed to the availability of high-throughput methods for genome sequencing and the advancement of bioinformatic tools within the last decade. The sequencing of tRNA genes has accelerated in recent years in such a way that currently more than 4000 different nucleotide sequences are known from more than 50 genomes.34
Currently, not very many native tRNAs are isolated using classical approaches in order to use them in various functional and structural studies. The so-called modern or current methods to obtain tRNA are faster and easier to do. Many of these methods such as in vitro transcription, overexpression of specific tRNAs in vivo, isolation through hybridization and others suffer from drawbacks like synthesis of unmodified or partially modified nucleosides and low efficiency of hybridization approaches. It is interesting to underline that only one enzyme, the RNA polymerase III (PolIII), is needed to synthesize all tRNAs, but there are more than 100 enzymes required to modify the tRNAs at the post-transcriptional level.35 Although modifications of tRNA are ubiquitous, their roles have often remained elusive. However, fully modified tRNAs are invariably more stable than unmodified transcripts, certain specific modified bases have functional significance for decoding of the mRNAs and others for the aminoacylation of tRNAs. Thermodynamic folding of unmodified sequences only rarely predicts the cloverleaf as a ground state, although the latter are often kinetically preferred.33,34 Therefore, new experiments may require large quantities of pure tRNA with natively modified nucleosides from classical methods of isolation.
3. Three-dimensional structure
With the complete nucleotide sequence of many tRNAs established, various groups became interested with questions concerning the three-dimensional structure of tRNA.In 1973, Alexander Rich and co-workers showed at a resolution of 4 Å, the L-shaped folding of the yeast tRNAPhe polynucleotide chain.36 It was a breakthrough in tRNA structural determination. Later on, in 1974 the folding of the tRNAPhe molecule was solved at 3 Å resolution in more detail by Aaron Klug37 and Alexander Rich.38 Both groups confirmed the L-shaped folding, even though the lattices were different. The tertiary structure of tRNA is composed of two helices. One is formed by the acceptor stem that is stacked on top of the TψC stem and the second is of the anticodon stem stacked on top of the D-stem. The variable loop is squeezed in between these two major domains. To build the L-shape, the D and TψC loops have to be joined together. There are also non-Watson–Crick base pairs and base triplets which help to stabilize the three-dimensional tRNA overall structure. The significance of this is that the 3′ terminus is separated from the anticodon triplet by more than 70 Å, forming a kind of general binding domain for aminoacyl tRNA synthetases. The sites of the tRNA interaction with the mRNA and the ribosomal peptidyl transferase, where the peptide bond synthesis takes place simultaneously, are located on different corners of the molecule. The tertiary structure of yeast tRNAPhe has been confirmed at 2 Å39 and 1.93 Å40 resolution. Interestingly, the 2 Å resolution structure was determined using 15 year old crystals (Fig. 2).
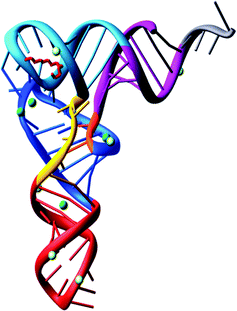 | ||
| Fig. 2 The tertiary structure of yeast tRNAPhe solved from 15 year old crystals.39 | ||
The L-shaped tRNA molecule is now a gold standard of molecular biology, having been found in virtually all tRNA species, also when they are complexed with cognate aminoacyl-tRNA synthetases and other proteins. For understanding the interactions of tRNA molecules the L-shaped folding provides the central information regarding movements within the ribosome during protein biosynthesis. The overall L-shape of the molecule is well conserved, although some variations in the angle between the two domains of the L can be observed.41–43
Currently the crystallographic structures are known for tRNAs of 19 amino acids. This includes the free form or within a supramolecular protein complex.34
4. Functional properties of tRNA
The genes of eukaryotic tRNA are transcribed by PolIII. These transcripts have to be maturated to fully active, native tRNA. This process includes a removal of an extra sequence at the 5′ end with RNase P, trimming of the 3′ trailer sequence with endo- and exonucleases, addition of CCA at the 3′ end, intron splicing and ligation, and post-transcriptional nucleoside modification. It has been shown in various studies that a single mature tRNA folds into a biologically active unique conformation when it interacts with its natural cellular ligand. tRNAs are the most abundant nucleic acid species, constituting up to 10% of all cellular RNAs. They are key components of the transportation system of amino acids to the ribosome to read and translate information encoded in the mRNA sequence. The basic function of tRNA as an adaptor molecule in protein biosynthesis is one of the cornerstone discoveries in the field of molecular biology.44 In addition to their essential role in protein synthesis, tRNAs participate in a plethora of non-translational activities in eukaryotes, bacteria and archaea.41 It is a participant in the first step of heme and chlorophyll biosynthesis. Uncharged tRNAs regulate global gene expression in response to amino acid deficiency. This is controlled by the interaction of general control nonderepressing kinase (Gcn2) protein and non-aminoacylated tRNA in both prokaryotes (stringent response) and eukaryotes.45 A mechanism for recognizing amino acid-deficient foods suggests that tRNA induces the phosphorylation of the eukaryotic initiation factor through its interaction with Gcn2.45 This pathway of a nutritional stress management is active in the brain of mammals and plays a critical role in dietary preference and survival of the organism. tRNAArg mediates amino-terminal arginylation as a tag for protein degradation. This modification may also play a functional role for some proteins in eukaryotes. In prokaryotes leucine is used instead of arginine for tRNA-dependent N-terminal modifications.46 Mitochondrial encoded-tRNAs interfere with a cytochrome c-mediated apoptotic pathway and promote cell survival. tRNA genes may also be acting as insulators in the human genome where they help to separate actively transcribed chromatin domains from the silenced ones; and also specific cellular tRNAs or tRNA fragments can be primers for the reverse transcription of viral genomes.41,42 Furthermore, tRNAs participate in energy metabolism and amino acid biosynthesis, functions that are conserved across all domains of life. In addition, many diseases are caused by mutations in both tRNAs and the proteins interacting with tRNA.42,44 Some of the non-canonical protein synthesis functions of tRNAs are considered to be random due to the high amount of tRNA species within the cell. Sequencing identified that yeast has more than 280 and humans over 500 tRNA genes. On the other hand, for example, a yeast cell contains approximately 3 million tRNA molecules, but in E. coli, there are only 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tRNA molecules.34 The tRNAs are universal macromolecules that encode a great repertoire of various biological and chemical signals which can be attributed to their overall structural properties being similar as well as being diverse.47,48 The presence of a significant amount of modified bases within the structure makes up a panel of signals and impacts the stability of tRNA.49 Interestingly, between 15 and 25% of all nucleosides in eukaryotic tRNAs are modified. These modifications are dynamic and respond to the environmental change.50 Some of these tRNA modifications influence RNA function. They can also act as sensors and cofactors.51
000 tRNA molecules.34 The tRNAs are universal macromolecules that encode a great repertoire of various biological and chemical signals which can be attributed to their overall structural properties being similar as well as being diverse.47,48 The presence of a significant amount of modified bases within the structure makes up a panel of signals and impacts the stability of tRNA.49 Interestingly, between 15 and 25% of all nucleosides in eukaryotic tRNAs are modified. These modifications are dynamic and respond to the environmental change.50 Some of these tRNA modifications influence RNA function. They can also act as sensors and cofactors.51
Detailed knowledge of the tRNA biology in different tissues will improve our understanding of the cellular aspects that affect pathology and diseases.52
5. Summary and perspectives
All these findings have revealed that tRNA has a surprising range of functions beyond being just a simple adaptor molecule. Currently celebrating a jubilee of:– 60 years of the Adaptor Hypothesis,
– 60 years of the tRNA prediction and its discovery,
– 50 years of RNA World Hypothesis,
– 50 years of the first nucleotide sequence determination, and
– 40 years of solution elucidation for crystals and tertiary structure of tRNA.
one may ask two very simple and basic questions:
1. What are the important points, in which we should remember the tRNA?
2. What does knowledge of the tRNA molecule mean today for molecular biology?
tRNA is the best described and characterized biological molecule. It can simply be referred to as the golden standard in molecular biology. Furthermore, tRNA is:
1. The first described native nucleic acid with known functions,
2. The first native nucleic acid with the known primary, secondary and tertiary structures,
3. The first nucleic acid species for which the solution and crystal structure was determined,
4. The first native RNA molecule to confirm the DNA double helix,
5. The first native species to be chemically synthesized – this was the first example of synthetic biology,
6. The first to be chemically synthesized as tDNA.
In addition, one should keep also in mind several structural and functional tRNA peculiarities such as:
1. The synthetic tDNAPhe (rA76) and tDNALys (dA76) can be charged with specific amino acids,
2. The best model for evaluation of new methods to determine nucleotide sequences,
3. The best model for proving methods of chemical synthesis of nucleic acids,
4. The best model for analysis of the structural potential of various biophysical approaches,
5. The model template used for the first polymerase chain reaction (PCR),
6. The source of RNA catalysis (leadzyme and RNase P),
7. RNAs with the structure similar to tRNAs (tRNA-like structure) are recognized with aminoacyl-tRNA synthetases,
8. The origin of macromolecular mimicry,
9. The structural model of the mechanism of protein – nucleic acid recognition studies,
10. The best source of Watson–Crick and non Watson–Crick base pairs,
11. The best reference for anti-parallel and parallel nucleic acid helix structural elements,
12. A marker used in phylogenetic analysis.
Looking at the lists above, it is not difficult to predict that there will be more very interesting things to be discovered about tRNAs.
References
- R. Giege, The early history of tRNA recognition by aminoacyl-tRNA synthetases, J. Biosci., 2006, 31, 477–488 CrossRef CAS PubMed.
- M. B. Hoagland, M. L. Stephenson, J. F. Scott, L. I. Hecht and P. C. Zamecnik, A soluble ribonucleic acid intermediate in protein synthesis, J. Biol. Chem., 1958, 231, 241–257 CAS.
- M. B. Hoagland, Biochemistry or molecular biology? The discovery of ‘soluble RNA’, Trends Biochem. Sci., 1996, 21, 77–80 CAS.
- F. Jacob and J. Monod, Genetic regulatory mechanisms in the synthesis of proteins, J. Mol. Biol., 1961, 3, 318–356 CrossRef CAS PubMed.
- A. Rich, in Horizons In Biochemistry, ed. M. Kasha and B. Pullman, Academic Press, New York, 1962, pp. 103–126 Search PubMed.
- C. R. Woese, The Genetic Code: The Molecular Basis of genetic Expression, Harper & Row, New York, 1967, pp. 179–195 Search PubMed.
- F. H. C. Crick, The origin of the genetic code, J. Mol. Biol., 1968, 38, 367–379 CrossRef CAS PubMed.
- W. Gilbert, Origin of life: The RNA world, Nature, 1986, 319, 618 CrossRef.
- N. Kresge, R. D. Simoni and R. L. Hill, The Discovery of tRNA by Paul C. Zamecnik, J. Biol. Chem., 2005, 280, e37 CAS.
- M. B. Hoagland, An enzymic mechanism for amino acid activation in animal tissues, Biochim. Biophys. Acta, 1955, 16, 288–289 CrossRef CAS.
- M. B. Hoagland, E. B. Keller and P. C. Zamecnik, Enzymatic carboxyl activation of amino acids, J. Biol. Chem., 1956, 218, 345–358 CAS.
- J. A. DeMoss, S. M. Genuth and G. D. Novelli, The enzymatic activation of amino acids via their acyl-adenylate derivatives, Proc. Natl. Acad. Sci. U. S. A., 1956, 42, 325–332 CrossRef CAS.
- P. Berg, Acyl adenylates: the interaction of adenosine triphosphate and L-methionine, J. Biol. Chem., 1956, 222, 1025–1034 CAS.
- K. Ogata and H. Nohara, The possible role of the ribonucleic acid (RNA) of the pH 5 enzyme in amino acid activation, Biochim. Biophys. Acta, 1957, 25, 659–660 CrossRef CAS.
- M. A. Machnicka, A. Olchowik, H. Grosjean and J. M. Bujnicki, Distribution and frequencies of post-transcriptional modifications in tRNAs, RNA Biol., 2014, 11, 1619–1629 CrossRef PubMed.
- R. W. Holley, An alanine-dependent, ribonuclease-inhibited conversion of AMP to ATP, and its possible relationship to protein synthesis, J. Am. Chem. Soc., 1957, 79, 658–662 CrossRef CAS.
- M. B. Hoagland, P. C. Zamecnik and M. L. Stephenson, Intermediate reactions in protein biosynthesis, Biochim. Biophys. Acta, 1957, 24, 215–216 CrossRef CAS.
- H. G. Zachau, G. Acs and F. Lipmann, Isolation of adenosine amino acid esters from a ribonuclease digest of soluble, live rribonucleic acid, Proc. Natl. Acad. Sci. U. S. A., 1958, 44, 885–889 CrossRef CAS.
- R. W. Holley, Large-scale preparation of yeast “soluble” ribonucleic acid, Biochem. Biophys. Res. Commun., 1963, 10, 186–188 CrossRef CAS PubMed.
- R. W. Holley, Evidence for the liberation of a nuclease from human fingers, J. Biol. Chem., 1961, 236, PC42–PC43 CAS.
- R. W. Holley, J. Apgar, G. A. Everett, J. T. Madison, M. Marquisee, S. H. Merrill, J. R. Penswick and A. Zamir, Structure of a ribonucleic acid, Science, 1965, 147, 1462–1465 CAS.
- H. G. Khorana, Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast, Pure Appl. Chem., 1971, 25, 91–118 CrossRef CAS.
- M. Nirenberg, Historical review: Deciphering the genetic code-a personal account, Trends Biochem. Sci., 2004, 29, 46–54 CrossRef CAS PubMed.
- J. R. Penswick, R. Martin and G. Dirheimer, Evidence supporting a revised sequence for yeast alanine tRNA, FEBS Lett., 1975, 50, 28–31 CrossRef CAS PubMed.
- H. G. Zachau, D. Dütting and H. Feldman, The structures of two serine transfer ribonucleic acids, Hoppe-Seyler's Z. Physiol. Chem., 1966, 347, 212–235 CrossRef CAS PubMed.
- J. T. Madison, G. A. Everett and H. Kung, Nucleotide sequence of a yeast tyrosine transfer RNA, Science, 1966, 153, 531–534 CAS.
- U. L. RajBhandary, S. H. Chang, A. Stuart, R. D. Faulkner, R. M. Hoskinson and H. G. Khorana, Studies on polynucleotides, LXVIII the primary structure of yeast phenylalanine transfer RNA, Proc. Natl. Acad. Sci. U. S. A., 1967, 57, 751–758 CrossRef CAS.
- A. A. Baev, A. D. Mirzabekov, V. D. Aksel'rod, T. V. Venkstern, L. Li, A. I. Krutilina, I. Fodor and L. Kazarinova, Primary structure of valine transport RNA 1 in baker's yeast. Partial reconstruction of the molecule, Dokl. Akad. Nauk SSSR, 1967, 173, 204–207 CAS.
- K. Kleppe, E. Ohtsuka, R. Kleppe, I. Molineux and H. G. Khorana, Studies on polynucleotides. XCVI. Repair replications of short synthetic DNA's as catalyzed by DNA polymerases, J. Mol. Biol., 1971, 56, 341–361 CrossRef CAS PubMed.
- K. Mullis and F. A. Faloona, Specific synthesis of DNA in vitrovia a polymerase-catalyzed chain reaction, Methods Enzymol., 1987, 155, 335–350 CAS.
- F. Jühling, M. Mörl, R. K. Hartmann, M. Sprinzl, P. F. Stadler and J. Pütz, tRNAdb 2009: compilation of tRNA sequences and tRNA genes, Nucleic Acids Res., 2009, 37, D159–D162 CrossRef PubMed.
- P. P. Chan and T. M. Lowe, GtRNAdb: a database of transfer RNA genes detected in genomic sequence, Nucleic Acids Res., 2009, 37, D93–D97 CrossRef CAS PubMed.
- S. Li and C. E. Mason, The pivotal regulatory landscape of RNA modifications, Annu. Rev. Genomics Hum. Genet., 2014, 15, 127–150 CrossRef CAS PubMed.
- R. Giegé, F. Jühling, J. Pütz, P. Stadler, C. Sauter and C. Florentz, Structure of transfer RNAs: similarity and variability, Wiley Interdiscip. Rev.: RNA, 2012, 3, 37–61 CrossRef PubMed.
- E. Borek, Modification of nucleic acids in relation to differentiation, Trends Biochem. Sci., 1977, 2, 3–6 CrossRef CAS.
- S. H. Kim, G. J. Quigley, F. L. Suddath, A. McPherson, D. Sneden, J. J. Kim, J. Weinzierl and A. Rich, Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain, Science, 1973, 179, 285–288 CAS.
- J. D. Robertus, J. E. Ladner, J. T. Finch, D. Rhodes, R. S. Brown, B. F. Clark and A. Klug, Structure of yeast phenylalanine tRNA at 3 Å resolution, Nature, 1974, 250, 546–551 CrossRef CAS PubMed.
- S. H. Kim, F. L. Suddath, G. J. Quigley, A. McPherson, J. L. Sussman, A. H. Wang, N. C. Seeman and A. Rich, Three-Dimensional Tertiary Structure of Yeast Phenylalanine Transfer RNA, Science, 1974, 185, 435 CAS.
- L. Jovine, S. Djordjevic and D. Rhodes, The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: cleavage by Mg(2+) in 15-year old crystals, J. Mol. Biol., 2000, 301, 401–414 CrossRef CAS PubMed.
- H. J. Shi and P. B. Moore, The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structurerevisited, RNA, 2000, 6, 1091–1105 CrossRef CAS PubMed.
- M. Raina and M. Ibba, tRNAs as regulators of biological processes, Front. Genet., 2014, 5, 171 Search PubMed.
- C. Megel, G. Morelle, S. Lalande, A. M. Duchêne, I. Small and L. Maréchal-Drouard, Surveillance and cleavage of eukaryotic tRNAs, Int. J. Mol. Sci., 2015, 16, 1873–1893 CrossRef CAS PubMed.
- U. L. RajBhandary and C. Köhrer, Early days of tRNA research: discovery, function, purification and sequence analysis, J. Biosci., 2006, 31, 439–451 CrossRef CAS PubMed.
- S. Kirchner and Z. Ignatova, Emerging roles of tRNA in adaptive translation, signalling dynamics and disease, Nat. Rev. Genet., 2015, 16, 98–112 CrossRef CAS PubMed.
- S. Hao, J. W. Sharp, C. M. Ross-Inta, B. J. McDaniel, T. G. Anthony, R. C. Wek, D. R. Cavener, B. C. McGrath, J. B. Rudell, T. J. Koehnle and D. W. Gietzen, Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex, Science, 2005, 307, 1776–1778 CrossRef CAS PubMed.
- R. Rai and A. Kashina, Identification of mammalian arginyltransferases that modify a specific subset of protein substrates, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10123–10128 CrossRef CAS PubMed.
- T. Weinert and A. K. Hopper, tRNA traffic meets a cell-cycle checkpoint, Cell, 2007, 131, 838–840 CrossRef CAS PubMed.
- D. M. Thompson and R. Parker, Stressing out over tRNA cleavage, Cell, 2009, 138, 215–219 CrossRef CAS PubMed.
- A. G. Torres, E. Batlle and L. Ribas de Pouplana, Role of tRNA modifications in human diseases, Trends Mol. Med., 2014, 20, 306–314 CrossRef CAS PubMed.
- X. Wang and C He, Dynamic RNA modifications in posttranscriptional regulation, Mol. Cell, 2014, 56, 5–12 CrossRef CAS PubMed.
- M. Helm and J. D. Alfonzo, Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland, Chem. Biol., 2014, 21, 174–185 CrossRef CAS PubMed.
- J. A. Abbott, C. S. Francklyn and S. M. Robey-Bond, Transfer RNA and human disease, Front. Genet., 2014, 5, 158 Search PubMed.
Footnote |
| † This paper is dedicated to the pioneers in tRNA studies who passed away recently Brian F. C. Clark (October 6, 2014), Alexander Rich (April 27, 2015) and Volker A. Erdmann (September 11, 2015). |
| This journal is © The Royal Society of Chemistry 2016 |
