Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Recent advances in X-ray compatible microfluidics for applications in soft materials and life sciences
Aghiad
Ghazal†
 a,
Josiane P.
Lafleur†
b,
Kell
Mortensen
a,
Jörg P.
Kutter
b,
Lise
Arleth
*a and
Grethe V.
Jensen‡
*a
a,
Josiane P.
Lafleur†
b,
Kell
Mortensen
a,
Jörg P.
Kutter
b,
Lise
Arleth
*a and
Grethe V.
Jensen‡
*a
aNiels Bohr Institute, University of Copenhagen, Universitetsparken 5, DK-2100 Copenhagen, Denmark. E-mail: gvjensen@udel.edu; arleth@nbi.ku.dk
bDept. of Pharmacy, University of Copenhagen, Universitetsparken 2, DK-2100 Copenhagen, Denmark
First published on 12th October 2016
Abstract
The increasingly narrow and brilliant beams at X-ray facilities reduce the requirements for both sample volume and data acquisition time. This creates new possibilities for the types and number of sample conditions that can be examined but simultaneously increases the demands in terms of sample preparation. Microfluidic-based sample preparation techniques have emerged as elegant alternatives that can be integrated directly into the experimental X-ray setup remedying several shortcomings of more traditional methods. We review the use of microfluidic devices in conjunction with X-ray measurements at synchrotron facilities in the context of 1) mapping large parameter spaces, 2) performing time resolved studies of mixing-induced kinetics, and 3) manipulating/processing samples in ways which are more demanding or not accessible on the macroscale. The review covers the past 15 years and focuses on applications where synchrotron data collection is performed in situ, i.e. directly on the microfluidic platform or on a sample jet from the microfluidic device. Considerations such as the choice of materials and microfluidic designs are addressed. The combination of microfluidic devices and measurements at large scale X-ray facilities is still emerging and far from mature, but it definitely offers an exciting array of new possibilities.
1 Introduction
With the development of several X-ray synchrotron sources with increasingly narrow and brilliant beams, the opportunities for investigating very large amounts of samples and sample conditions have virtually exploded in the past decade. The modern facilities allow for very efficiently answering a broad range of questions within, e.g., materials science, colloid science, pharmaceutical formulation, and structural biology. Large experimental parameter spaces, in terms of buffer composition or external parameters such as temperature, pressure or magnetic field, may be investigated. Moreover, increasingly strong X-ray beam intensities, available at both synchrotron and X-ray free electron laser (XFEL) facilities, have enabled novel types of data on the fast dynamics of the investigated systems to be obtained and thereby direct insight into the underlying self-assembly and functioning principles.1–3However, with this opportunity, a general demand for approaches to efficiently produce the very large numbers of samples required for the scanning of such a large parameter space, to position them in the synchrotron beam, and – for time-resolved experiments – to time the triggering of time-resolved processes with the data acquisition exists; all this with minimal sample consumption, which is a general requirement for the highly interesting but often only sparsely available samples. Overcoming beam damage to retain the integrity of samples and sample devices is another crucial requirement resulting from ever more brilliant beams.
Microfluidic-based approaches designed for systematic mixing and manipulating of small amounts of samples are often mentioned as an answer to these demands and have been pioneered by several groups in the context of synchrotron radiation during the past decade. The present review provides a status of the field. The development of microfluidics-based sample manipulation approaches has, to a large extent, been driven by the broader desire to develop the so-called “lab-on-a-chip”.4 The name “lab-on-a-chip” is a catchy moniker for devices that are small and incorporate all necessary functions to perform a chemical analysis, an assay or a synthesis on a small footprint, preferably in a hand-held device, to be used at the location where it is necessary, required or convenient.5 In diagnostics, this is known as point of care, and lab-on-a-chip devices for this application are considered to constitute a prospective future market.6 However, the name “lab-on-a-chip” has – to a certain degree – also limited the general recognition of alternative ways to exploit the underlying technology of microfluidics and of the fact that the ultimate goal is not necessarily only to shrink the entire lab and carry everything in the palm of your hand.
In the present review, it is shown that microfluidics has strong potential as a sophisticated and powerful sample preparation, manipulation, and delivery method in connection with large, and certainly not hand-held, X-ray probes and detection infrastructures. The versatility of microfluidics for production of a large number of low volume samples for specific investigations has been illustrated in combination with both mass spectrometry7 and NMR8 and also with X-ray9 and neutron scattering techniques,10 mainly small-angle X-ray scattering (SAXS) and crystallography. Add in the possibility for automation and intelligent feedback,11 and it is clear that microfluidic devices indeed hold great potential for making the best use of the expensive X-ray infrastructures, where access and measurement time are at a premium, by providing a highly efficient sample preparation and manipulation system.
The general advantages of microfluidics are manifold and have been praised several times before. Some are obvious, while others require more thinking, experiments, and engineering skills to exploit properly. Microfluidic devices are inherently useful for working with small sample volumes, which is an advantage for both rare samples and/or performing many measurements.5 Microfluidic devices are used extensively in the context of measurements at synchrotron beamlines in sample preparation and mixing steps. Making use of laminar flow and diffusion and carefully tuning longitudinal (axial) and transverse (radial) transport, microfluidics can be used to precisely mix small amounts of reagents.12,13 Scanning along the microfluidic channel makes time slices available for the investigation of the sample under certain conditions or of ongoing reactions. Likewise, droplet-based microfluidics can “freeze” certain conditions (and limit dispersion) and present solutions at various (but clearly defined and reproducible) stages of mixing and reacting.14 The ease of mixing, possibility for automation and ability to process multiple samples in parallel on microfluidic platforms can help investigate a large parameter space more effectively to identify conditions and pathways for, e.g., crystallization15 or protein unfolding.11 The flow profile in the microfluidic channels may also be used actively, e.g. to shear the sample, leading to structural reordering or alignment, which can then also be investigated using the X-ray probe.16,17
Combining a microfluidic sample preparation system with an X-ray beam setup is indeed a marriage of the small and the large, and naturally, this requires some considerations and precautions. The interface is, of course, the main important piece here, assuming that the microfluidic part has been carefully constructed, and the detection part is understood and controlled as well. In the case of connecting a microfluidic device with an X-ray beam line, it is important to look at aspects such as compatible materials, designs of the detection window, path lengths, and the interplay of flow rate (flow velocity) and exposure time to the beam. However, equally important, beam sizes, fluxes and the potential of damage to the sample caused by the beam have to be taken into consideration for successful setups.18,19
The intent of this review is to shed some light on most of the abovementioned aspects that are important when using microfluidic devices in connection with X-ray beamlines. The review is split into four main sections. Section 2 focuses on materials and fabrication methods. Section 3 focuses on how microfluidics is applied to map large parameter spaces. Here, some of the first applications were in the field of protein crystallography, while later, applications in protein formulation were proposed by combining microfluidics and solution small-angle X-ray scattering. Section 4 focuses on how microfluidics may be used to perform time resolved studies, typically in the microsecond to second range, and provides an overview of the physical limitations associated with this approach. Section 5 focuses on microfluidics used for sample manipulation and gives examples of how the extreme flow fields in the narrow channels can be used to introduce alignment or other types of sample modulation, which can then be probed via the narrow synchrotron X-ray beams.
Technical (materials, design, interface, etc.) as well as application aspects and examples are summarized in large tables, and selected papers are briefly discussed in the main text to highlight unique solutions or specific applications. The tables feature exclusively references where synchrotron data collection is performed in situ, i.e. the microfluidic device containing the sample or a free sample jet from the microfluidic device is placed directly in the X-ray beam path of a synchrotron source for data collection. The list of cited references is comprehensive to the best of our knowledge and covers the past 15 years.
2 Materials, fabrication methods and beam considerations
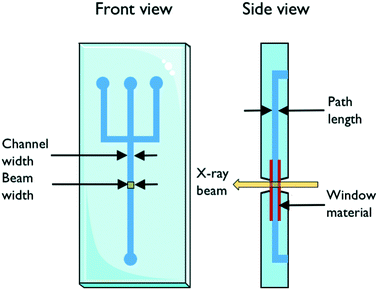
The design of the microfluidic chip, choice of material and fabrication method are important factors to consider in the fabrication of X-ray compatible microfluidic devices. A schematic with important parameters is shown in Fig. 1. The width of the microfluidic channel should be wide (the dimension which is perpendicular to the X-ray beam) enough to accommodate the size of the beam in order to minimize reflection and scattering on the channel edges. This requirement is slowly alleviated by the advent of powerful beams with small dimensions down to 1 μm;20,21 however, it can still be a limiting factor. The X-ray path length through the sample is determined by the depth (the dimension of the channel which is parallel to the beam direction) of the channel. The transmission of X-rays through the sample depends on this path length together with the sample properties and should be considered when designing a microfluidic chip for X-ray exposure. The transmission T through a sample of path length d follows T ∼ exp(−Ad), where A is the linear attenuation coefficient for the given X-ray wavelength and sample material. The scattering S, on the other hand, is proportional to the scattering volume, which for a given beam size means proportional to the sample path length: S ∼ d. Therefore, the detected scattering will be proportional to I/I0 ∼ d × exp(−Ad), resulting in a maximum intensity for d = 1/A, which is then the ideal path length for optimal signal. For an aqueous solution and an X-ray wavelength of 1.5 Å (8 keV), the optimal path length is ca. 1 mm, and for 1.2 Å (10 keV), it is ca. 2 mm.22 For an optimized signal using an ∼10 micrometer beam, the microfluidic channel can therefore be very deep, relative to its width.An ideal chip material should allow for easy, affordable and reproducible production of high quality chips. Ideally, it is also optically transparent and compatible with X-rays. However, as this is often not the case, a common strategy is using the so-called “windows” made of a different, thinner, X-ray compatible material. This material should have high transparency (low attenuation), optimal scattering properties in terms of low level of scattering and resistance to X-ray radiation, while also featuring material properties allowing easy integration/bonding to the material forming the body of the microfluidic chip. X-ray compatible microfluidic chips have been produced using different materials. Silicon is the traditional material for microfluidics, stemming from the field of microelectronics. In the field of X-ray compatible microfluidics, it has mainly been used for microfluidic mixers.23,24 The fabrication techniques are based on photolithography in combination with etching and layer deposition. These techniques are very well established but also quite demanding as many processes are involved, including several steps of chemical cleaning. In addition, the aspect ratio of the channels that can be produced in one etching process is limited.25 Highly transparent silicon nitride windows have been integrated successfully into microfluidic devices.26,27
Polymeric materials provide a cheaper alternative to silicon. Poly(dimethylsiloxane) (PDMS) is a popular material as it is X-ray compatible and easy to cure on a pre-fabricated mold. It has been used for protein crystallization screening.24,28,29 UV-curable adhesives used either alone,30,31 in combination with silicon nitride windows,26 or with polystyrene windows32 have also been used for the fabrication of X-ray chips for X-ray studies. Both PDMS and UV-curable adhesives can be patterned via soft lithography, ensuring an easy replication of the chips. In the case of off-stoichiometry thiol-ene (OSTE),32 the flexibility of the final cured material can be tuned by varying the composition of the reactants.
Toft et al. investigated several potential polymer-based microfluidic materials for use in combination with SAXS, from both the microfluidic (machinability, bonding and resistance to solvents) and the X-ray (transparency, scattering and X-ray endurance) viewpoints.11 They found that polymethylmethacrylate (PMMA), polycarbonate (PC), polyvinylchloride (PVC) and polystyrene (PS) all exhibited low X-ray scattering, however with very poor transmission for PVC. Of these four, PS exhibited the best combined properties with respect to machinability, bonding and solvent resistance and was therefore chosen for SAXS experiments.11,33 The polyimide material with the trademark name Kapton®, which is a material traditionally used for X-ray windows, featured a higher degree of X-ray scattering than any of these four. Kapton® is, however, very durable towards X-rays. Barrett et al. found that Kapton® microdevices could resist prolonged exposure (1 h) to an intense microfocused X-ray beam with no observable degradation (no leaks, no change in refractive index, no burn stains).34 In X-ray compatible microfluidics, it has been used as a window for chips made of stainless steel,35,36 and entire chips have been fabricated using this material.37
Apart from the interaction between the chip material and the X-ray beam, the chip material can also affect the chemical reactions or biological processes ongoing in the chip. For example, crystallization conditions can be affected as water can evaporate through permeable polymers such as polydimethylsiloxane (PDMS). For this reason, COC and cyclic olefin polymer (COP) have been widely used to serve as a barrier against water evaporation.38 COP/COC absorb X-rays, but only over a narrow range of the wavelength spectrum from 5.4 to 5.1 Å,15,39 while PDMS absorbs at 7.5 Å,15 both far from the 1–2 Å X-ray wavelengths typically used in SAXS and crystallography instruments. Guha et al. investigated the variation in the linear attenuation coefficient as a function of X-ray energy as well as the transmission factor as a function of thickness for COC and PDMS (Fig. 2).15 At 12.4 keV (1 Å), COC attenuates X-rays seven times less than PDMS and SiO2. Maeki et al. investigated two different wall thicknesses and material capillaries for in situ X-ray diffraction (80 μm-thick poly(tetrafluoroethylene) (PTFE, commonly known as Teflon) and a 10 μm-thick glass capillary) and observed no difference in the results obtained from these two kinds of capillaries.40
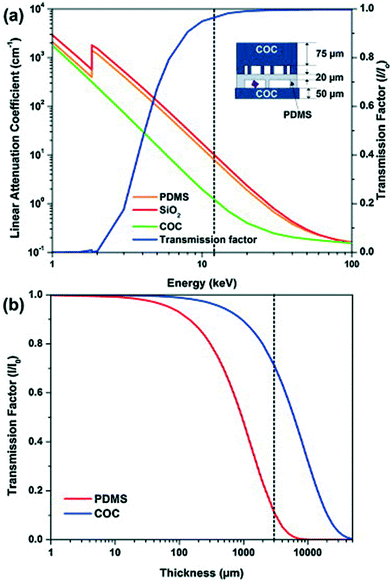 | ||
| Fig. 2 (a) Linear attenuation coefficient for PDMS, COC, and SiO2 (quartz) as a function of photon energy. The transmission factor I/I0 as a function of photon energy for a typical device architecture as shown in the schematic. (b) Transmission factor I/I0 as a function of film thickness for PDMS and COC. The transmission factor was calculated at a photon energy of 12.4 keV corresponding to a wavelength of 1 Å (reproduced with permission from the International Union of Crystallography; Guha et al., 2012).15 | ||
Protein adsorption to the channel walls can also be an issue. It can be addressed by confining the protein solution in droplets41 or in a sheet surrounded by pure buffer,42 thereby avoiding contact with the channel walls and X-ray window.
Not only can the microfluidic chip material be damaged upon intense X-ray exposure, but the sample can also be damaged. Beam damage, which is the damage inflicted on samples due to ionisation by the intense X-ray beam, is a major challenge. This is especially relevant when using extremely high intensity X-ray beams such as those available at XFEL facilities. Weierstall2 thoroughly discusses this aspect and emphasizes that the replenishment rate of the sample should match the XFEL pulses, the background scattering should be minimized by placing the sample in a vacuum, and general care should be taken to avoid that fragile samples (such as crystals) are damaged during sample handling by shear forces, charging, etc. In SAXS, radiation damage limits the data acquisition time for many sample types, including protein solutions.19 This damage may also be prevented by continuously refreshing the sample by flowing it through the beam,18,43–45 which is an intrinsic feature for most microfluidic systems. Cryo-SAXS has also been shown to be an alternative solution to this problem.46 Throughout the review, summary tables are provided for each section. The summary tables include detailed information about the chip and window material, as well as the chip fabrication methods, used for each paper reviewed.
3 Combining microfluidics and X-ray scattering to map large parameter spaces in crystallography and small-angle scattering
Some experiments require that a large amount of different samples can be prepared/mixed in a controlled fashion and with minimal sample consumption. In crystallography applications, for example, samples need to be screened against an array of precipitants to identify appropriate crystallization conditions. The study of unfolding or other conformational changes of proteins in response to variations in their chemical environment by SAXS also requires the investigation of a large parameter space, not least, because the conditions which promote structural changes are often unknown a priori. In a conventional crystallography or SAXS experiment, only part of the experimental space of the sample may be investigated due to constraints of a very practical nature. Not only are many samples too sparsely available to allow a thorough parameter space investigation, but the amount of beam time that can be obtained at synchrotron facilities is also limited due to its high cost (and high demand). Until a few years ago, it was common practice for both X-ray crystallography and SAXS experiments that a multitude of samples needed to be prepared manually or with the help of a pipetting robot prior to the experimental beam time and then mounted manually during the experiment. As a result, there was a significant down-time between the individual measurements and only a fraction of the parameter space of a sample could realistically be investigated during the allocated beamline time due to time-consuming and cumbersome manual sampling. While this sample loading problem, to a large extent, has been solved with advanced liquid sample handling robots at most modern synchrotron facilities,47,48 the existing methods still rely on pre-mixed samples. It is therefore still highly relevant to evaluate how microfluidics can help investigate a large parameter space more effectively to identify conditions for, e.g., crystallization or protein unfolding.a. Applications in crystallography
Since crystallization conditions are not known a priori, it is often necessary to screen samples against a wide range of precipitants in order to identify suitable crystallization conditions. Table 1 summarizes advances in crystallization using microfluidic devices featuring in situ X-ray data collection. Microbatch, vapour diffusion and free interface diffusion (FID)/counter-diffusion traditional crystallization methods have been adapted to microfluidics. In the microbatch method, the protein and precipitant are mixed together in a small volume, while in vapour diffusion and FID/counter-diffusion, the small volume containing the protein is allowed to equilibrate with a larger, concentrated precipitant reservoir, or a concentration gradient causes both protein and precipitant to gradually diffuse under the influence of a concentration gradient. In all these cases, as nuclei form and crystals start to grow, the concentration of the protein gradually decreased, preventing further nuclei from forming. Microfluidic devices allow precise control over diffusion and mixing conditions, bringing a clear advantage to the table for the in situ study of single crystals by X-ray crystallography. Miniaturized platforms can offer integrated fluid handling properties to rapidly generate complex mixtures with minimal sample consumption. Several reviews report recent advances in miniaturization for crystallography.49–52 Although miniaturized platforms have been used extensively in the past decade to screen crystallization conditions and/or produce crystals under reproducible conditions, examples where data are acquired in situ are more scarce. In conventional approaches, crystals need to be manually harvested, cryocooled and mounted on a goniometer for X-ray studies. Here, we focus on examples where X-ray data are acquired in situ at synchrotron sources. This approach eliminates crystal handling, minimizing damage during harvesting and mounting of the crystals. In combination with in situ X-ray data acquisition, microbatch methods have been miniaturized by either using pneumatic valves (discussed in more detail below) to mix precipitants and proteins or inside microdroplets where the concentration of protein solution and precipitant can be varied by adjusting their flow rate ratios in order to create an array of droplets containing each a different crystallization trial. Vapor diffusion has successfully been implemented with in situ X-ray data collection in microdroplets by allowing diffusion of water between adjacent droplets.53 FID methods, which are normally difficult to implement and rarely performed,49 have also been successfully implemented in conjunction with in situ X-ray determination (for several references, see Table 1), where a precise control over diffusion times is achievable simply by varying microchannel dimensions.| Chip features | Chip material/fabrication | Window material and thickness | Sample volume/consumption | Crystallization method | Temperature | Beamline, source | Ref. |
|---|---|---|---|---|---|---|---|
| a Abbreviations: RT, room temperature; COC, cyclic olefin copolymer; COP, cyclic olefin homopolymer; PDMS, polydimethylsiloxane; PMMA, polymethylmethacrylate; PTFE, polytetrafluoroethylene; FID, free interface diffusion; APS, Advanced Photon Source; ESRF, European Synchrotron Radiation Facility; EMBL, European Molecular Biology Laboratory; SSRL, Stanford Synchrotron Radiation Lightsource; SAGA-LS, SAGA Light Source; CHESS, Cornell High Energy Synchrotron Source. | |||||||
| Droplet-based; evaporation is eliminated and diffusion of water between droplets is controlled to screen crystallization conditions | Composite PDMS/glass capillary microfluidic device | 20 μm thick X-ray glass capillary | nL–μL droplets | Microbatch and vapour-diffusion method | RT | Beamlines 14-BM-C and 14-ID-B, APS | Zheng et al. (2004)53 |
| PDMS plug generator | PDMS chip (rapid prototyping) with a thin glass capillary and a PTFE (Teflon) capillary | Glass or PTFE (Teflon) | The thin glass capillaries are filled with approximately 100 (replicates or unique) 20 nl microbatch trials | Microbatch | 277 K | Beamline GM/CA-CAT, APS | Yadav et al. (2005)67 |
| Valved microfluidic system; the device has channel lengths ranging from 300 to 2400 μm | PDMS | PDMS membrane | 10 nL reactors | FID | Cryocooled at 100 K | Beamline 12.3.1, APS | Hansen et al. (2006)28 |
| Simple crystallization/mixing channel | PDMS/replica casting, PMMA/laser ablation and COC/hot embossing | 250 μm for rigid polymers, 1 mm for PDMS | 150 nL of biomolecule solution | Counter-diffusion | RT | Beamline FIP-BM30A, ESRF | Sauter, Dhouib, and Lorber (2007)72 and Dhouib et al. (2009)54 |
| Microfluidic formulator device with valves | PDMS | PDMS membrane | High-resolution structure: >1 mg of protein; solubility screening and phase diagram measurements: ∼200 μg of protein; crystal screening and optimization: ∼250 μg; crystal scale-up: ∼450 μg | FID | Cryocooled | Beamline 11-3, SSRL | Anderson et al. (2007)64 |
| Centrifugal microfluidic disc enabling up to 100 different crystallization experiments in parallel | COC/hot embossing | COC | 600 nL of total protein solution for 100 experiments using 100 arbitrary precipitants | FID | RT | High throughput crystallization facility, EMBL Hamburg | Steinert et al. (2007)73 |
| Each prototype plate contains nine microchannels for crystallization by diffusion (0.10 to 0.3 mm wide, 20–60 mm long and 0.1 mm high). | COC/micro-injection molding | COC | 200 nl to 1.96 μl protein solution for each crystallization experiment | Counter-diffusion equilibration | RT (283 K) | Beamline 1-5, SSRL | Ng et al. (2008)74 |
| Liquid handling in this system is performed in 2 mm thin transparent cards and actuated via centrifugal microfluidics | COP/laser cut | COP | 500–320 nL chambers per card | Microbatch, vapour-diffusion and FID protocols | RT | Beamline FIP-BM30A, ESRF | Emamzadah et al. (2009)39 |
| Nanodroplet-based crystallization method | PDMS droplet generator connected to a PTFE (Teflon) or a glass capillary | 80 μm-thick PTFE (Teflon) capillary and 10 μm-thick glass capillary | 200 μm-diameter (2 nL) spherical microdroplets | Mixing with precipitant solution in droplets | RT | Beamline BL 07, SAGA-LS | Maeki et al. (2012)40 |
| Array chips consisting of a series of separate half-wells for protein and precipitant solutions arranged in columns; each of these individual wells is a separate crystallization trial and is isolated from the rest of the wells using a series of normally closed valves | Thin hybrid microfluidic chip composed of layers of COC (control layer) and PDMS (fluidic layer)/photolithography, soft lithography and hot embossing | A typical device has an X-ray path length of 125 μm of COC and 20 μm of PDMS (50 μm COC substrate, 20 μm PDMS membrane, 75 μm COC control layer) | Each half-well contains 50 nL of solution and the entire chip uses just 1.4 μL of protein solution for a 24-well design; the platform allows screening of up to 100 crystallization conditions | FID | Room T (add small data wedges) & cryogenic conditions | Beamline 21-ID, APS | Guha et al. (2012)15 |
| Integrated, two-layer microfluidic chip allowing metering of protein and reagents, mixing, and incubation of crystallization trials | Thin hybrid microfluidic chip composed of layers of COC, PDMS and Dura-lar®/photolithography, soft lithography and hot embossing (fabrication based on Guha et al., 2012 (ref. 15)) | 50 μm COC or 12 μm Dura-lar® substrate, thin PDMS membrane, 100 μm COC control layer | 6 μL of protein per 96-well microfluidic chip (96 wells that are each less than 20 nL in volume) | FID | Room T (add small data wedges) | Beamline 21-ID, APS | Perry et al. (2013)57 |
| 12-well chip; each well relies on diffusion for mixing | Fabrication based on Guha et al., 2012 (ref. 15) | COC and thin PDMS membrane | 60 nL of the protein solution per crystallization trial | FID | RT (adds wedges) | Beamline 21-ID-F, APS | Khvostichenko et al. (2014)38 |
| The microfluidic chips consist of separate half-wells for protein and precipitant solutions; dedicated valve lines for each set of half-wells enable independent filling of protein and precipitate | Fabrication based on Guha et al., 2012 (ref. 15) and Perry et al. (2013)57 | COC (50 μm), PDMS (45 μm) and Dura-Lar® (13 μm) | Crystallization is performed in both 24- and 96-well microfluidic chips | FID | RT; merge small data slices (in situ serial Laue diffraction) | Beamline 14-ID-B, APS | Perry et al. (2014)59 |
| Single crystals formed in droplets | Fabrication based on Guha, 2012 | COC (75 μm) or COC and Kapton® foil (8 μm) and a thin PDMS membrane | pL to nL droplets | Mixing in droplets | Serial crystallography with non-cryocooled crystals | Beamline F1, CHESS | Heymann et al. (2014)58 |
| 96-well chip for mixing | Fabrication based on Guha et al., 2012 (ref. 15) | COC and thin PDMS membrane | One complete chip required 6 μl of protein/precipitant solution and 6 μl of microseed solution | FID | RT; serial crystallography (time-resolved serial Laue diffraction) | Beamline 14-ID-B, APS | Pawate et al. (2015)60 |
| Microfluidic chip featuring 6–24 crystallization chambers allowing mixing via normally closed valves | COC–PDMS microfluidic chip (240 μm thick)/photolithography/soft lithography (fabrication based on Guha et al., 2012 (ref. 15) and Perry et al. (2013)57 | 100 μm COC + thin PDMS membrane | The size of the crystallization chamber is 0.67 mm (width) × 0.75 mm (height) × 50 μm (depth) | Counter-diffusion mixing via mixing valve | 100 and 277 K | Beamline BL 07, SAGA-LS | Maeki et al. (2015)56 |
PDMS is ubiquitous in the field of microfluidics, and it is therefore without surprise that many of the devices used in crystallography applications rely on this material even though its properties are far from ideal for this type of applications. Dhouib et al. reported that crystallization chips made from PDMS were too flexible for handling, insufficiently watertight to prevent dehydration during crystallization, and not transparent enough to X-rays.54 They obtained better performance using thin sheets of cyclic olefin copolymer (COC) and poly(methylmethacrylate) (PMMA) for chip material. However, other groups have used this water permeability of PDMS to their advantage by using it to change the solute concentration of fluids stored in a microfluidic network to map phase diagrams.29,55 Guha et al. reported that crystallization trials could easily be performed for a week and sometimes even two weeks despite loss of water by adsorption into the thin layer of PDMS present in their device (Fig. 2).15 Zheng et al.,53 on their part, opted for PDMS/glass capillary hybrid microfluidic devices to eliminate evaporation of solutions during crystallization trials. They found that in contrast to the case in which droplets were incubated in PDMS channels, droplets and crystals incubated in sealed glass capillaries were stable even in the absence of humidity control and did not show signs of evaporation over six months.53
In crystallography, crystals which are not cryocooled are also highly susceptible to radiation damage. Maeki et al. worked on the development of an on-chip contactless cryoprotection procedure to eliminate potential damage to samples.56 Another clever approach to avoid radiation damage upon X-ray exposure, while also avoiding tedious on-chip cryocooling, has been used by the group of Kenis et al.15,38,57–60 Serial crystallography and other similar methods in which small data wedges from a large number of crystals can be collected at room temperature and later merged to form a complete data set are also used to avoid radiation damage. This method is easily implemented in microfluidic devices since a large number of crystals can be created easily and reproducibly.
Some very complex microfluidic devices have been developed to screen a multitude of crystallization conditions with minimal sample consumption and maximum throughput. The group of Quake developed valved microfluidic systems61–63 for parallel control of large fluidic arrays, providing a platform for rapid screening of protein crystallization conditions. In an early iteration, a chip featuring 480 active valves could perform 144 parallel reactions with only 10 nL of protein samples.61 However, the device is complex and still requires manual handling as crystals had to be extracted from the device for X-ray analysis. Later versions of the device28,64 allowed cutting out small sections of the PDMS membranes containing the protein crystals for X-ray analysis.
Water/oil two-phase flows can be used advantageously in microfluidic devices to form a stream of immiscible aqueous plugs or droplets in a hydrophobic oil carrier or simply to prevent migration of reactants between samples. For example, the addition of an oil phase can be used to form arrays of chemically independent droplets for long term storage of crystals.65 The group of Fraden developed a two-phase flow chip,55,66 which allowed precise control over the nucleation and crystal growth process. The system was subsequently refined to allow in situ X-ray analysis of single crystals generated inside the droplets.58Fig. 4 shows the formation of single crystals in emulsion droplets. The group of Ismagilov also took advantage of two-phase flows in microfluidics to generate nanoliter-scale water-in-oil droplets containing mixtures of protein and crystallization solutions, adjusting the mixing ratios by changing the corresponding flow rates. The device allowed screening hundreds of protein crystallization conditions using less than 4 nL of protein solution for each crystallization trial.41 The device was made compatible with in situ X-ray measurement by the addition of an X-ray glass53,67 or polytetrafluoroethylene (PTFE, commercially known as Teflon) capillary.67 The same group made further efforts in improving and controlling the stages of nucleation and growth in protein crystallization using a microfluidic device68,69 and in simplifying chip operation with the SlipChip70,71 but without using these devices for in situ X-ray analysis. The group of Kenis developed an array chip consisting of a series of separate wells for protein and precipitant solutions arranged in columns (Fig. 3). Each of these individual wells is a separate crystallization trial and is isolated from the rest of the wells using a series of normally closed valves. The device is designed to have minimal X-ray cross section and was used for in situ X-ray data acquisition at both room temperature and under cryogenic conditions.15,57 In order to reduce the diffusional path length, Khvostichenko et al. layered the protein solution on top of the lipid, significantly reducing the diffusional path and, consequently, the mixing time compared to the traditional side-by-side placement of microfluidic compartments.38 The same group also demonstrated the use of a microfluidic crystallization platform for the serial time-resolved Laue diffraction analysis of macroscopic crystals.60Table 1 summarizes advances in crystallization using microfluidic devices featuring in situ X-ray data collection.
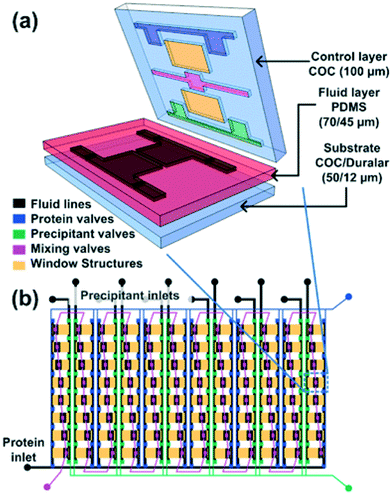 | ||
Fig. 3 (a) 3D exploded view showing the different materials used for various layers of the hybrid device. (b) Schematic design of a 96-well array chip showing the various valve lines for filling in different components as well as the protein and precipitant chambers. The ratio of the protein to precipitant chambers varies from 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1 1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 to allow for screening of a larger number of conditions. Reproduced from Perry et al., 2013 (ref. 57) with permission from the Centre National de la Recherche Scientifique (CNRS) and The Royal Society of Chemistry. 4 to allow for screening of a larger number of conditions. Reproduced from Perry et al., 2013 (ref. 57) with permission from the Centre National de la Recherche Scientifique (CNRS) and The Royal Society of Chemistry. | ||
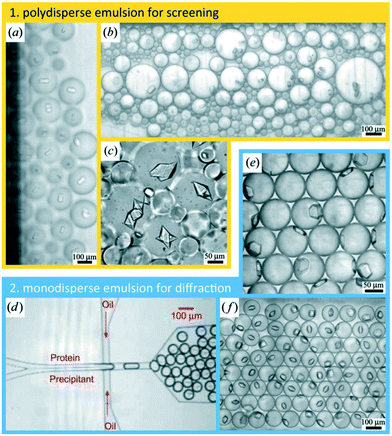 | ||
| Fig. 4 Protein crystallization in emulsion droplets stabilized by a surfactant. (a–c) Polydisperse emulsions. (d) Protein and precipitant solutions were introduced in a co-flow geometry under laminar flow conditions that prevents mixing upstream of the nozzle where both solutions became encapsulated into emulsion droplets. (e and f) Monodisperse emulsions (reproduced with permission from the International Union of Crystallography; Heymann et al., 2014 (ref. 58)). | ||
b. Applications using SAXS
Small-angle scattering measures the structure of samples at length-scales from 1 nm to 100–500 nm.75 The technique is often used to investigate problems in structural biology, where it has a large advantage that it is compatible with investigating samples under native-like solution conditions.76 For synchrotron SAXS experiments, where typical measurement times are on the order of seconds, efficient sample changing is absolutely essential in order to maximize sample throughput. As mentioned above, SAXS can provide low-resolution structural information on proteins directly in solution, and it is therefore a preferred tool for analyzing structural changes of proteins and complex systems in response to variations in their chemical environment. However, in a traditional SAXS experiment, only a limited part of the experimental space of a sample can be investigated as samples have to be prepared prior to the experimental campaign and structural information becomes apparent only upon measurement at the synchrotron beamline.During the past decade, there have been significant efforts in implementing sample changing robots on SAXS beamlines in order to increase throughput and minimize sample consumption.47,48 However, this approach does not have the significant advantages offered by microfluidic devices, where sample conditions can easily be changed in the course of an experiment in response to changes in the observed SAXS signal. As summarized in Table 2, several groups have taken advantage of microfluidic devices to map large parameter spaces in SAXS experiments, mostly for protein structure determination.
| Chip features | Chip material/fabrication | Window material and thickness | Sample volume/consumption | Sample investigated | Beamline, source, beam characteristics | Ref. |
|---|---|---|---|---|---|---|
| a Abbreviations: SLS, Swiss Light Source; APS, Advanced Photon Source; CHESS, Cornell High Energy Synchrotron Source. | ||||||
| Microfluidic chip featuring an integrated X-ray transparent 200 nL sample chamber and diffusion-based mixing of protein and buffer solutions; fully automated fluidic control, data acquisition, and data analysis | Polystyrene (PS)/micromilling | Two 175 μm thick PS films | 200–500 nL sample chamber; 170 μL sample consumption to fill tubing and equilibrate pressure + 36 μL per sample measurement (for 5 minute exposure time) | Protein dilution series | Beamline I711, MAXlab, wavelength 1.05 Å; beam size 0.4 × 0.10 μm2 or 0.5 × 0.15 μm2; path length 1 mm; | Toft et al. (2008)11 |
| Microfluidic platform including hardware and software for fluidic control, sample mixing by diffusion, automated X-ray exposure control, UV absorbance measurements and automated data analysis | Polystyrene (PS)/micromilling | Two 125 μm thick PS films | As little as 15 μl of sample is required to perform a complete analysis cycle, including sample mixing, SAXS measurement, continuous UV absorbance measurements, and cleaning of the channels and X-ray cell with buffer | Protein dilution series | Beamline cSAXS, SLS, beam size: 200 × 400 mm (vertical × horizontal); path length 1.7 mm | Lafleur et al. (2011)33 |
| Microfluidic device, which combines metering and active mixing using pneumatic valves | Fabrication based on Guha et al., 2012 (ref. 15) | 150 μm COC + 30 μm thin PDMS membrane | ∼80 nL per sample/the chip required a total volume of 200 nL of each material (lipid, detergent solution, diluent) for filling | Phase behaviour of lipidic mesophases | Beamline 21-ID-D, APS | Khvostichenko et al. (2013)79 |
| Microfluidic device based on the pervaporation of water through a PDMS membrane allowing formulation of continuous and steady concentration gradients on the nanolitre scale | PDMS/photolithography, soft lithography | Two 50 μm PDMS membranes | The volume of the microfluidic evaporator lies in the 1 nL–1 μL range | Phase behavior of a binary aqueous molecular mixture and of a model complex fluid (triblock copolymer solution) | Beamline SWING, SOLEIL (French national synchrotron), energy 12 keV; beam size 200 × 50 μm2; path length: 60 μm | Daubersies et al. (2013)29 |
| Remote-controlled microfluidic device allowing simultaneous SAXS and ultraviolet absorption measurements during protein dialysis; the dialysis and detection are performed on two separate chip modules | PDMS/micromilling and replica moulding | Two 25 μm thick PS films | ∼67 μL needed to perform a concentration series | Protein concentration series | Beamlines F2 and G1, CHESS, energy 10 keV; beam size: 250 × 250 μm2; path length: 1.0 mm | Skou et al. (2014)80 |
| Centrifugal microfluidic disk | COC/three-layer thermoforming process | 70 μm + 115 μm COC | 2.5 μL of protein solution to generate 120 different measurement conditions | Protein structure analysis under varying protein and salt concentrations | Beamline P12, PETRA III, energy 10 keV; beam size 50 × 50 μm2; path length 0.86 mm | Schwemmer et al. (2016)78 |
Toft et al. were among the pioneers in the field and reported a high-throughput device for protein structural analysis on a relatively simple microfluidic device named the bioXTAS chip,11 which enabled automated mixing of samples by diffusion in a long meandering channel with fluid flow actuated using syringe pumps. Fluidic control was coupled with control of the beamline shutter system, such that X-ray exposure and data collection happened in sequence with sample preparations. Lafleur et al. further improved the bioXTAS platform by the addition of an on-chip sample reservoir to minimize protein consumption, motor-controlled rotary valves to reduce the impact of fluidic capacitances and allow accurate on-chip sample preparation, as well as integrated optical fibers to allow simultaneous sample monitoring via UV-vis spectrophotometry (Fig. 5).33 An open-source software program developed in-house, ECON, allowed synchronized control of the microfluidic device and the X-ray beamline shutter systems, while the open-source software bioXTAS RAW77 took care of in situ processing and first analysis of the isotropic SAXS data in terms of background subtraction, plotting and indirect Fourier transform, providing users with invaluable real time feedback. BioXTAS RAW has afterwards gained broader applicability in more general synchrotron SAXS. Very recently, Schwemmer et al. introduced a centrifugal microfluidic (LabDisk) device.78 The platform (illustrated in Fig. 6) can be used to provide SAXS data for basic analysis, such as measurement of the radius of gyration, and for more advanced methods, such as the ab initio calculation of 3D structures. The absence of external actuation mechanisms, such as syringe pumps and tubing, eliminates dead volumes and allows for a reduction in sample consumption. The platform requires only 2.5 μL of protein stock solution to prepare 120 different dilution/screening conditions. With this type of centrifugal platform, however, the composition of the dilutions have to be pre-decided as the channels and reservoirs need to be engraved in the discs in a specific way to provide a certain dilution ratio. This system therefore does not allow for on-the-fly adjustments during the course of an experiment as would be possible with computer controlled syringe pumps. After sample preparation, the disc containing the samples is transferred to a SAXS positioner platform for in situ detection.
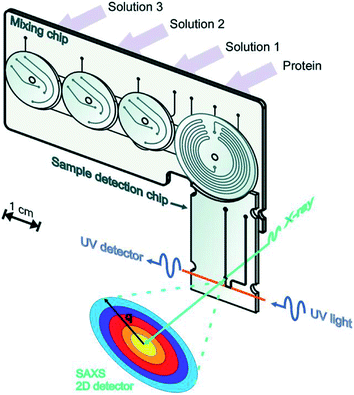 | ||
| Fig. 5 Illustration of the bioXTAS lab-on-a-chip system. The bioXTAS platform allows the continuous variation and monitoring (UV and SAXS) of experimental solution conditions to promote and identify different structural states in proteins in a high-throughput fashion. Reproduced with permission from the International Union of Crystallography; Lafleur et al. (2011).33 | ||
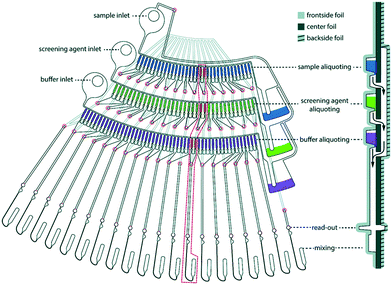 | ||
| Fig. 6 One dilution matrix of the LabDisk for SAXS. The left side shows the top view of the dilution matrix, and the right side shows the cross-section. The red circles indicate through holes in the center foil. After metering the three input liquids, the rotational frequency is increased. Aliquots from the sample and screening agent are transferred through holes in the center foil (red circles), transported over the backside fluidic layer and combined with the buffer aliquots in the read-out and mixing section. Aliquots are combined in 20 sets of six aliquots each, comprising different combinations of sample, screening and buffer aliquots. The arrows indicate liquid flow. Schwemmer et al. (2016),78 published with permission from the Royal Society of Chemistry. | ||
There are several more subtle ways to create different experimental conditions and concentration gradients in microfluidic devices. As an example, the water permeability of PDMS can be used advantageously in microfluidics to create concentration gradients and map phase diagrams (as already mentioned previously). Selimović et al. described a PDMS device in which the solute concentration within nanolitre sized aqueous droplets could be altered by migration of water through a PDMS membrane to map phase diagrams of aqueous samples.55 Daubersies et al. took advantage of the same characteristic water permeability of PDMS for the in situ formulation of viscous fluids by controlled solvent evaporation.29
4 Combining microfluidics-based fast mixing and X-ray scattering for time resolved structural studies
The study of structural kinetics of proteins and soft matter is an interesting combined application of microfluidics and X-ray scattering techniques.81,82 A structural response is initiated by fast mixing on the chip of two solutes under continuous flow. The structures at times Δt after mixing can then be probed at different distances from the mixing point Δx, where the time is given via the flow velocity v as Δt = Δx/v. The time resolution that can be obtained depends on the mixing time but also on the flow rate and the X-ray beam size.As mentioned before, small-angle X-ray scattering (SAXS) probes the nanometre structure of particles in solution, e.g., proteins, polymers, and micelles. In combination with microfluidic mixing devices, SAXS has mainly been used to study folding of proteins9,83–91 and RNA24,92–95 but also compaction36,96 and complex formation97 of DNA, as well as self-assembly of filaments,31 fibrils,98 and lyotropic liquid crystals.37
Compared to the microfluidic chips used for spectroscopic techniques, longer path lengths are typically necessary in order to maximize the X-ray scattering intensity. For most of the reported microfluidic platforms, path lengths on the order of 0.2–0.4 mm have been chosen (see Table 3). As described in section 2 on general considerations when combining microfluidics with X-rays, the path length giving the optimal signal would be 1–2 mm (depending on X-ray wavelength), however, at the cost of increased sample consumption. The channels are kept narrow, ranging from 0.05 to 0.4 mm, to minimize sample consumption, which is particularly relevant for protein samples that are often only sparsely available. This is especially important considering that protein concentrations on the order of one to several milligrams per millilitre are typically required to obtain a sufficiently good scattering signal. For diffusional mixing (as described below), narrow channels are also crucial to obtain laminar flow as well as short diffusion distances, leading to the fastest possible mixing.99 Generally, the channel widths are limited on the low side by the size of the X-ray beam: if the beam is larger than the exposure channel, it will hit the channel edges, leading to unwanted scattering ‘streaks’.
| Microfluidic chip | Sample | X-ray beamline | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mixer type | Chip features | Materials and fabrication | Window material | Channel dimensions (width × path length) (mm) | Accessible kinetic times, time resolution, time of mixing (ms) | System | Flow rate (mm s−1), typical sample consumption | Beamline, source, X-ray energy | Beam dimensions (mm) | q range investigated (Å−1) | Exposure time (s) | Ref. |
| a Values derived from reported numbers. Abbreviations: CHESS, Cornell High Energy Synchrotron Source; APS, Advanced Photon Source; ESRF, European Syxnchrotron Radiation Facility; SLS, Swiss Light Source; SSRL, Stanford Synchrotron Radiation Laboratory; BESSY, Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung. | ||||||||||||
| Laminar | Simple cross mixer | Etched silicon | Silicon | Folding of cyt-C (pH jump) | 300 mm s−1 | Beamline D, CHESS | 0.04 × 0.12 | Pollack et al. (1999)9 | ||||
| 17.5 μL min−1 protein solutiona | ||||||||||||
| Laminar | Simple cross mixer | Etched silicon | Silicon | 0.05 × 0.39 | 1–8, 0.24, 2 | Folding of beta lactoglobulin (diluting the solution with urea) | 166 mm s−1 | Beamline IMM-CAT, APS, 7.65 keV, pink beam | 0.01 × 0.04 | 0.04–0.5 | 40 × 2 minimum | Pollack et al. (2001)83 |
| 19.4 μL min−1 protein solutiona | ||||||||||||
| Laminar | 3D hydrodynamic focusing | Etched silicon | PDMS | 5–45, 0.4, 0.4 | Folding of RNA (mixing with Mg2+) | 86 mm s−1 | Beamline 8-ID, APS, pink beam | 0.01 × 0.04 | 60 | Russell et al. (2002)24 | ||
| Contact lithography and deep reactive ion etching | ||||||||||||
| Laminar | 3D hydrodynamic focusing | Etched silicon | PDMS | 2–150, 0.46, N/A | Folding of RNA (mixing with Mg2+) | Beamline 8-ID, APS, pink beam | 0.01 × 0.04 | 60 | Das et al. (2003)92 | |||
| Laminar | 3D hydrodynamic focusing | Etched silicon | PDMS | Folding of RNA (mixing with Mg2+) | Beamline 8-ID, APS, pink beam | 0.01 × 0.04 | Kwok et al. (2006)93 | |||||
| Laminar | 3D hydrodynamic focusing | Etched silicon | PDMS | 1.3–168, N/A, N/A | Folding of RNA (mixing with Mg2+) | Beamline 8-ID G1, APS, pink beam | 0.01 × 0.04 | 0.025–0.25 | 25 × 30 | Lamb et al. (2008)94 | ||
| Laminar | 3D hydrodynamic focusing | Etched silicon | PDMS | 1.3–160, N/A, N/A | Folding of RNA (mixing with Mg2+) | 15–200 mm s−1 | Beamline 8-ID, APS, pink beam | 0.05 × 0.013 | Schlatterer et al. (2008)95 | |||
| Laminar | Simple cross mixer | Spark eroded stainless steel | Kapton, 20 μm | 0.1–0.15 × 0.2–0.3, 3 mm long | 40–6000, 40, N/A | DNA compaction by mixing with dendrimers | 0.5 mm s−1 | Beamline ID10B, ESRF, 8 keV | 0.02 × 0.02 | 30 | Dootz et al. (2006)36 | |
| Laminar | Simple cross mixer | Spark eroded stainless steel | Adhesive Kapton | 0.1–0.15 × 0.2–0.3, 3 mm long | 200–3 × 104, 200, N/A | Formation of lipoplexes after mixing of DNA with lipids | 0.01–0.1 mm s−1 | Beamline ID10B, ESRF, 8 keV | 0.02 × 0.02 | 30 | Evans et al. (2007)97 | |
| Laminar | Simple cross mixer | Spark eroded stainless steel | Kapton | 0.05–0.15 × 0.3, 3 mm long | 40–6000, 40, N/A | DNA compaction by mixing with dendrimers | 0.5 mm s−1 | Beamline ID10B, ESRF, 8 keV | 0.02 × 0.02 | 30 | Pfohl et al. (2007)96 | |
| Laminar | Simple cross mixer | Spark eroded stainless steel | Adhesive Kapton | 0.15 × 0.3 | N/A, 2.85–0.123, N/A | Self-assembly of collagen fibrils (pH jump) | 2.1–48.6 mm s−1 | Beamline ID10B with compound refractive lenses, ESRF, 8 keV | 0.02 × 0.02 | 0.02–0.25 | 30 | Köster et al. (2008)98 |
| Laminar | 3D hydrodynamic focusing | UV-cured polymeric adhesive | UV-cured adhesive (integrated) | 0.3 × 0.24 | N/A, 26, N/A | Vimentin filament assembly | 0.77 mm s−1, 0.67 μL min−1 protein solution | Beamline cSAXS, SLS | 0.005 × 0.02 | 5 × 1 | Brennich et al. (2011)31 | |
| Laminar | Simple cross mixer | Kapton, 125 μm, laser cut, laminated | Kapton, 25 μm | 0.11 × 0.115 | 25 (1.6 is the limit)–1150, 1.6, 380 | Lyotropic phase transitions in block copolymer micelle liquid crystals | 19.8 mm s−1, 1.67 μL min−1 protein solution | Beamline MiNaXs P03, Petra III, wavelength 1.088 Å | 0.031 × 0.022 | 1 | With et al. (2014)37 | |
| Laminar | 3D hydrodynamic focusing, two sets of inlet side channels | UV-cured polymeric adhesive | UV-cured adhesive (integrate) (130 μm) | 0.3 × 0.24 | Vimentin filament assembly | 17–55 mm s−1, 20–80 μL min−1 protein solution | Beamline ID13, ESRF, 12.4 keV | 0.005 × 0.005 | 0.024–0.921 | 5 | Saldanha et al. (2016)30 | |
| Laminar | Simple cross mixer | Thiol-ene moulding using PMMA and PDMS masters | Polystyrene | 0.32 × 0.22 | Structure of lipidic cubosomes upon exposure to Ca2+ | 3.7 mm s−1, 16 μL min−1 | Beamline I911-SAXS, MAX-lab, 1.5 GeV | 0.25 × 200 | 60 | Ghazal et al. (2016)32 | ||
| Turbulent | 300 μm nozzle with a capillary | Quartz capillary, 550 μm | Quartz capillary, 550 μm | 14–100, N/A, 3 mm = 14 ms dead time | Folding of lysozyme after pH jump or dilution of sample with GdnHCl denaturant | 5000–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mm s−1 (according to Chan et al., 1997 (ref. 124)) 000 mm s−1 (according to Chan et al., 1997 (ref. 124)) |
Beamline 4-2, SSRL, 8.98 keV | 600–900 | Segel et al. (1999)84 | |||
| Turbulent | T mixer with a 33 μm nozzle | Stainless steel | Mylar polyester, 22 μm | 0.2 × 0.4 | 0.16–6, 0.096, 0.07 | Folding of cytochrome c after pH jump | 4170 mm s−1, 20 mL min−1 protein solution | Beamline 45XU, SPring-8, 8.3 keV | 0.4 × 0.4 | 30 × 0.022 | Akiyama et al. (2002)85 | |
| Turbulent | T mixer with a 33 μm nozzle | Stainless steel | Kapton 25 and 50 μm | 1 (path length) | 0.3–22, 0.08, 0.13 | Folding of apomyoglobin after pH jump | 840–2600 mm s−1 | Beamline 45XU, SPring-8, 8.3 keV | 0.4 × 0.4 | 8–12 | Uzawa et al. (2004)86 | |
| Turbulent | T mixer with a 33 μm nozzle | Stainless steel | Kapton | 0.6–N/A, N/A, N/A | Folding of heme oxygenase after pH jump | SPring-8, beamline 45XU, 8.3 keV | Uzawa et al. (2006)87 | |||||
| Turbulent | T mixer | Stainless steel | Polyester | 0.3–10, 0.3, N/A | Folding of dihydrofolate reductase | 10–20 mL min−1 total, 1 mL min−1 protein solution minimum | Beamline 18-ID Bio-CAT, APS, 3.5–13 keV | 1 | Arai et al. (2007)88 | |||
| Turbulent | T mixer | Stainless steel | Polyester | 0.15–5.0, 0.15, — | Folding of TIM barrel protein | 10–20 mL min−1 total | Beamline 18-ID Bio-CAT, APS, 3.5–13 keV | 1 | Wu et al. (2008)89 | |||
| Turbulent | Arrow mixer | Stainless steel, 400 um | Kapton, 8 μm | 0.1 × 0.4 | 0.1–2.4, 0.02 (0.0024 is the limit), 0.093 | Folding of cyt-C by diluting solution with GdnHCl | 8333 mm s−1, 20 mL min−1 protein | Beamline 18-ID Bio-CAT with compound refractive lenses, | 0.02 × 0.005 | 0.015–0.35 or 0.025–0.55 | Continuous scanning | Graceffa et al. (2013)91 |
| 0.03 in mixing region | ||||||||||||
| APS, 3.5–13 keV | ||||||||||||
| Droplet | Inlets with aqueous solutions, leading to oil flow | PDMS mixer attached to a glass capillary | Glass capillary for X-ray exposure (10 μm walls) | 0.1 in. PDMS chip, 0.3 in. capillary, 25 mm long | 3200–5000 (no kinetics shown), 4.2 s dwell time | Formation of Au nanoparticles from solution | 4.9 mm s−1, 25 μL min−1 protein | Beamline 7 T-MPW-SAXS, BESSY II, 7.5 and 8.0 keV | 0.3 × 0.05 | 0.01–0.25 | 900 | Stehle et al. (2013)116 |
| Free jet | PMMA sandwich with a PEMA adhesive layer, deep X-ray lithography | Impact modified PMMA, 500 μm | 0.5 outlet, narrow to 0.008 at exit nozzle; 0.01 × 0.06 in chip, 0.025 diameter of jet | 0.075–0.77 (5 is the maximum due to jet break-up), 0.075, <0.04 | Performance test only | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mm s−1, 22.5 μL min−1 protein 000 mm s−1, 22.5 μL min−1 protein |
Austrian SAXS beamline, ELETTRA, 8 keV | 1 × 0.8 | 80 | Marmiroli et al. (2009)128 | ||
The investigation of structural kinetics using microfluidic chips under continuous flow should be discussed in relation to the classical stopped-flow technique, where two solutions are pumped into a turbulent mixer and then further into an observation capillary, where the flow is stopped, and the structural evolution over time is probed. The method of observation is often spectroscopic, but the setup is also routinely applied at SAXS beamlines.91,100–102 The time of mixing is a couple of milliseconds, which is rather long compared to many kinetic processes of both proteins and soft matter. In addition, the required sample volume is relatively high, at typically 100 μL per mixing event, often leading to a total sample consumption of several millilitres. The time resolution is limited by the detector readout time between consecutive measurements or, alternatively, by beam damage of the sample, which might require further waiting time between consecutive measurements. The resulting time gaps can be covered by performing a set of measurements with a different delay time between mixing and the collection of the first frame, which, however, requires consumption of more sample. For slower kinetic processes, taking place on the order of seconds to minutes, these issues are less of a problem.
It should be mentioned that X-ray techniques for studying much faster kinetics, down to the nanosecond time scale, also exist, following a photoinitiated structural transition of proteins in solution.103–112 The high time resolution of this so-called pump-probe technique is obtained by precise control of the time from the photoactivation by a laser flash (the pump) to the arrival of the X-ray pulse at the sample (the probe). The fast structural rearrangements of the protein sub-domains are probed in the wide-angle X-ray scattering (WAXS) domain. Measurements are repeated at up to several Hz and up to hundreds of times for each delay time104,110 to obtain a sufficient scattering signal. Therefore, this approach is limited to proteins with a photo-activated and reversible structural transition. Ligand-induced transitions might also be addressed by mixing the protein with caged ligands, which are then photo-released. Compared to the microfluidic-based studies reviewed here, the time resolution is much higher; however, highly concentrated protein samples are required, which are relatively robust towards hours of exposure to room temperature while being exposed to laser- and X-ray flashes.
Depending on the sample of interest, microfluidic mixing under continuous flow may be more suitable than the stopped-flow technique for probing kinetics on the millisecond time scale.12 The two main categories of microfluidic mixers operate under laminar and turbulent flows, respectively, as given by the Reynolds number Re = ρvl/η, where ρ is the liquid density, v is the liquid flow velocity, l is the channel width, and η is the liquid viscosity.99,113 The transition between laminar and turbulent flows takes place in the range of Re from 102 to 104.114 The two scenarios are illustrated in Fig. 7. Turbulent mixing is relatively fast, on the order of microseconds, but requires a high Reynolds number, and therefore a high flow velocity or wide(r) channels, leading to consumption of potentially very large sample volumes (tens of millilitres per minute).81 A laminar flow requires a low Reynolds number and hence a low flow velocity or narrow channels. This results in a low sample consumption,82 typically at a few microliters per minute. However, the mixing occurs by passive diffusion, which is relatively slow, on the order of milliseconds for typical ligands diffusing over micrometer distances. A third approach is mixing in droplets formed on the microfluidic chip and suspended in an oil phase.115,116 This setup can be optimized to give slightly faster mixing than laminar mixers, while the confinement in droplets means that the problem of sample dispersion, which is an issue for laminar mixing as described in section a below, is avoided. However, only a few examples of a combination with X-ray scattering are published to date. We speculate that this could be due to issues with parasitic scattering from the droplet surfaces.
The applications of the different mixer types are reviewed in the following sections. Examples and perspectives are also given for an approach where the mixing is performed on the chip, while the mixed sample jet is outside the chip, and the scattering data are collected on this uncovered, ‘free’ jet.
a. Laminar mixers
Laminar mixers are based on hydrodynamic focusing. Many advanced chip designs have been suggested for optimal mixing,13 but the simplest possible design, suggested by the Austin group114,117 and later also introduced at an X-ray beamline by the same group,9 has channels forming a cross, with three inlets and one outlet (Fig. 7B). Pollack continued this work,24,83,92–95 and a similar design formed the basis of the work by the Pfohl group.36,96–98,118 A solution of the target species (e.g. proteins) is pumped into the chip through the centre inlet, and a solution of the molecules that will initiate the structural transition is pumped into the two side channels. The two solutions meet at an intersection and flow side by side in the mixing channel towards the outlet. Relatively low flow rates are applied (see Table 3), from a few to a couple of hundreds of microliters per minute, resulting in low Reynolds numbers and a laminar flow with the solutions flowing in parallel sheets.The protein solution is hydrodynamically focused to a thin sheet in the centre, and the mixing happens by passive diffusion of the initiating molecules into the centre sheet and depends on the protein having a much larger diffusion constant than the initiating molecule if dilution of the protein should be avoided. The speed of the mixing process is limited by the relatively slow process of diffusion (e.g. the diffusion constant of a calcium ion is 7.92 × 10−10 m2 s−1, leading to an average time of 15 ms for diffusing 5 μm).119 Sub-millisecond mixing times have been achieved,24,98 but mixing times of several milliseconds are more common (see Table 3). While this greatly reduces the types of kinetics that can be probed, the advantage is that the low flow rates result in a correspondingly low sample consumption.
As mentioned, the flow velocities, v (see Table 3), give the translation factor between the distance from the mixing point, Δx, and the associated kinetic time Δt according to Δt = Δx/v. However, the flow velocity v is not constant across the channel cross-section. For a circular cross-section, it will be parabolic, and similar profiles are found for rectangular channel cross-sections, with the maximum velocity at the channel center.114 The protein molecules will therefore travel with different velocities in different streamlines and will also, depending on their diffusion constant, diffuse across the channel and visit other streamlines, leading to dispersion of the kinetic times. This phenomenon, known as Taylor dispersion, leads to smearing of the kinetic times.120 Rather than corresponding to one well-defined time, a position in the channel will represent an average time corresponding to the average velocity of the probed sample molecules. For sheet widths comparable to the channel width, the dispersion will be significant and thereby smearing of the time resolution across the channel.121 Brennich and Köster illustrated how the effect could be addressed by simulation techniques to determine the average kinetic time associated with each position in the chip.122
Kinetics on time scales comparable to the mixing times will be convoluted with the mixing process, which must also be taken into account when relating each position in the microfluidic chip with kinetic time.122 This issue was also addressed by Brennich and Köster122 using detailed simulations of the flow in the chip channels. They showed how reduction of the total flow rate in the channel slowed down the mixing, and how this could be used to decouple the mixing kinetics from the reaction kinetics.
A more narrow sample sheet leads to faster mixing, reducing the issues of overlap between mixing and structural kinetics. However, the narrow sheet requires a comparatively narrow X-ray beam to probe mainly the sample sheet rather than the surrounding sheets, thereby avoiding relative weakening of the sample scattering signal. Therefore, microfocus X-ray setups such as compound refractive lenses are often used for these measurements. This is the case in studies by both the Pollack group24,83,92–95 and the Pfohl group.36,96–98,118
The first reported microfluidic laminar mixers were designed by the Austin group and were characterized using fluorescence microscopy methods.114,117 Pollack et al. continued their pioneering work within the field of microfluidic laminar mixers optimized for X-ray scattering investigations. This work is based on a chip made in etched silicon. The first reported results were obtained using a chip with 100 μm wide channels and an X-ray path length of 390 μm, concerning folding of equine cytochrome c after a jump in pH.9 It was followed through the evolution of the protein radius of gyration and through Kratky plots, showing the initial chain-like structure and the final more compact structure but also capturing one frame showing an intermediate state. A very narrow protein sheet was obtained, leading to a sub-millisecond pH transition, as confirmed by confocal microscopy measurements using a pH sensitive fluorescent dye.9 The time resolution was also sub-millisecond, owing to the relatively high flow rates. However, only two kinetic times were probed, one in the interval of 0.15–0.5 ms and one at 10 ms, corresponding to the final, folded state. The same chip design, but now with 50 μm wide channels, was used to investigate the folding of beta lactoglobulin.83 The protein solution contained urea in concentrations that ensured an unfolded protein, and the transition was initiated by diluting with buffer without urea. The time of onset for the transition was defined as the time when the urea concentration had decreased below a target value of 3 M, giving a faster onset than if complete mixing had been required.
The fast (millisecond) compaction of RNA or RNA fragments during folding, induced by mixing with magnesium, was the subject of a series of studies by the Pollack group.24,82,92–95 A modification of the chip was required to avoid beam damage: a laminating layer of buffer along the X-ray windows was created, as illustrated in Fig. 8 (reproduced from Russell et al.24). Using this way of avoiding direct contact between protein solution and X-ray window, deposition of radiation-damaged protein was minimized. The window material was also changed from being an integrated part of the silicon chip9 to a softer PDMS material, which has been shown to better prevent the adhesion of protein.123 The RNA compaction was probed at kinetic times in the interval of 1–160 ms. The data could be interpreted using singular value decomposition,24 as a combination of the initial and the collapsed states. However, a combination with stopped-flow measurements, giving the kinetics on longer time scales, revealed an additional kinetic phase, showing that the initially formed globular state is an intermediate.24 The initial collapse was independent of specific protein mutations and addition of various cations, which led to the conclusion that it is of electrostatic nature,92 whereas the following phase is related to formation of specific tertiary contacts, as concluded from complementary investigations.
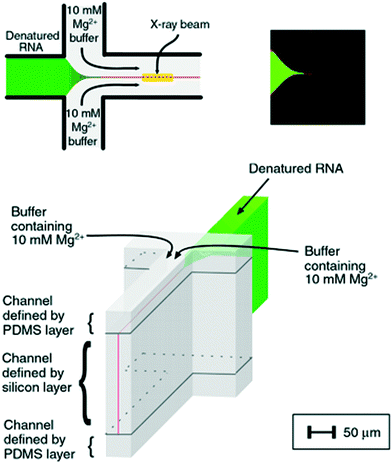 | ||
| Fig. 8 Illustration of a laminar flow mixer used following the denaturing of RNA by Mg2. Upper left: Schematic of the inlets and mixing channel, showing one possible position of the X-ray beam. Upper right: Multiphoton microscopy image showing the signal when magnesium from the side channels has mixed with a calcium sensitive dye in the centre channel. Bottom: 3D schematic of the channels, highlighting the 3D focusing, resulting in a buffer covering the protein sheets towards the X-ray windows (demarcation between the buffer and the sample indicated by black lines) (reproduced from Russell, R. et al.,24 copyright (2002), National Academy of Sciences, U.S.A.). | ||
Another chip development was presented by Park et al.,42 also from the Pollack group, introducing a five-inlet channel, with an additional set of channels, placed between the central protein channel and the side channels containing the magnesium ligand. These diagonal channels contained pure buffer, pumped at a very low flow rate, resulting in a thin buffer sheet between the protein sheet and the ligand. By simulation, confirmed by fluorescence imaging, it was shown that this buffer sheet prevented premature mixing of the protein and the ligand. A buffer flow rate of just 0.2% of the total flow rate was enough to move the position where the first well-determined kinetic time could be probed without significant influence of the mixing event, from 130 μm to 66 μm. The channel width and path length were 50 and 100 μm, respectively.
Pfohl and Köster et al. also made significant contributions to the field, fabricating a chip in spark eroded steel with Kapton windows. It was applied to follow the structural response of DNA to mixing with dendrimers36,96 or lipids.97 The chip had wider channels as compared to the chip by the Pollack group, and considerably lower flow rates were applied. This resulted in lower sample consumption while covering longer kinetic times. At the shortest distances from the intersection point, the collected data reflect the mixing process and thereby a gradient in the concentration of the ligand. Rather than aiming for avoiding this gradient by faster mixing, the X-ray scattering results were discussed in the light of the different local mixing ratios. Formation of compact and oriented liquid crystal-like DNA–dendrimer complexes was reported as well as a study on formation of multilamellar lipid bilayers with intercalated DNA.97 The same setup was also applied to follow the self-assembly of collagen fibrils, following a mixing-induced jump in pH.98 A diffraction peak is observed at certain pH values, corresponding to a repeat distance which is consistent with pentameric fibrils of collagen trimers.
A chip with 3D hydrodynamic focusing, along the lines of Russell et al.24 as described above, was presented by Brennich et al.,31 fabricated in a UV-cured adhesive, based on thiol-ene cross-linking, resulting in a polymeric material as compared to the silicon and steel seen so far. The window was integrated into the chip and hence also composed of UV-cured adhesive, which showed very good characteristics with respect to X-ray transmission and minimal X-ray scattering. The chip had a channel width of 300 μm and was working at low flow velocities (below 1 mm s−1), covering relatively long time scales. It was applied to follow the assembly of vimentin filaments and the growth of their cross-section. A more sophisticated version of this chip, from the same research group, featured two sets of inlet side channels, allowing for investigation of the vimentin fiber structure upon subsequent addition of potassium and magnesium, respectively.30 Times up to 20 seconds were covered, as determined from simulations of the flow conditions in the chip.
Ghazal et al.32 also used chips of UV-cured thiol-ene material, however with polystyrene as window material. The channels were coated on the inside with photocurable polymers to avoid adsorption of the lipids. They studied the effect of exposing negatively charged lipids to Ca2+ ions, which is relevant for certain biological processes such as protein regulation. It was possible to follow the internal structural transitions of the lipid cubosome nanoparticles from cubic Pn3m to Im3m phases upon mixing with calcium on a time scale ranging from 0.5 to 5.5 seconds.
The application of microfluidic mixers for colloids and soft matter besides proteins is limited, likely because of the more readily available samples, making sample consumption less of an issue. A recent example, though, is presented by With et al.37 using a chip fabricated solely from a Kapton film, used for investigating the formation of a lyotropic liquid crystal of block copolymer micelles.
b. Turbulent mixers
In turbulent mixers, two solutions are pumped into the chip to meet at the mixing point, where turbulent mixing takes place, resulting in a homogeneously mixed solution, flowing towards the outlet (Fig. 7A). The formation of chaotic eddies requires a high Reynolds number, which in micrometre sized channels must be obtained by high flow velocities. Typically, velocities on the order of several metres per second are applied, which, of course, requires the availability of large sample volumes (tens of millilitres per minute). The advantage of the turbulent approach is that mixing is about an order of magnitude faster than the diffusional mixing in the laminar mixers, typically at hundreds of microseconds. In addition, because the resulting mixed solution homogeneously fills the X-ray exposure channel, the concentration of the target species will not decrease over time by diffusion out of the X-ray beam.While the concept of turbulent mixing of liquids is obviously not new,99 the combination with microfluidics and in particular with X-ray scattering has evolved within the past decades. A group of scientists around Hofrichter and Kiefhaber initiated the process with their turbulent mixers, which were not based on microfluidic (planar) chips but on a mixing nozzle.84,124 The solutions are led to the nozzle through two concentric needles, with one solution in the inner needle and the other in the outer needle, surrounding the inner one. The original mixer was not applied for X-ray studies but for fluorescence quenching, which was used to provide information on the folding of cytochrome c. The folding was initiated by dilution of a sample containing a denaturant at concentrations ensuring an unfolded initial state.124 The same system was also studied with Raman spectroscopy and fluorescence quenching, using a T-shaped turbulent mixer.125
SAXS measurements were first performed on a modified version of the needle-based mixer. The nozzle size was increased to 300 μm, and it was connected to a quartz capillary for the SAXS measurements. The folding of lysozyme was initiated by either a jump in pH, or by dilution of a sample containing the denaturant guanidinium hydrochloride, GdnHCl. The folding process was followed from 14 to 100 ms. By singular value decomposition, an intermediate structural state was identified and characterized as alpha-helical based on the results from circular dichroism (CD). By using a combination with stopped-flow SAXS measurements on longer time scales, the entire folding process was described, and rate constants were determined based on the temporal evolution of the radius of gyration.
The inclusion on a microfluidic chip of both a turbulent mixer and an X-ray observation channel was made by the Fujisawa group.85 Their chip was produced in stainless steel, with a T-shaped hole cut to serve as inlet channels and a mixing channel and with Kapton or Mylar windows86 (see Fig. 9). A narrowing of the channels from 200 μm to 33 μm was introduced just after the mixing point to increase the turbulence (whereas l decreases, v increases with 1/l2, leading to an increased Reynolds number, Re = ρvl/η). Mixing times as short as 70 μs were achieved. Folding of cytochrome c was followed after a pH jump, and intermediates were identified from the temporal evolution of the protein radius of gyration.85 The same chip was used to study the folding after the pH jump of apomyoglobin86 and heme oxygenase,87 respectively. For apomyoglobin, three kinetic phases are observed by time-resolved CD spectroscopy (using the mixing chip modified for CD by using quartz windows), whereas only two (one within the probed time range) are observed by SAXS. The combination of the two techniques led to the conclusion that an initial fast collapse is followed by a conformational search, including the formation of alpha helices. An equivalent approach was taken for the results on heme oxygenase, also leading to conclusions on a fast initial collapse.
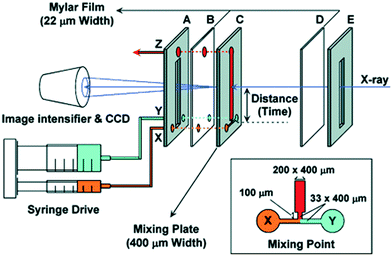 | ||
| Fig. 9 Schematic illustration of the setup by Akiyama et al. for turbulent mixing and X-ray scattering of protein solutions (reproduced from Akiyama et al.,85 copyright (2002), National Academy of Sciences, U.S.A.). | ||
The Bilsel group performed similar experiments, concerning the folding of the enzyme dihydrofolate reductase, which also features an initial collapse of the chain. They used a similar mixing chip, which is, however, not specified in detail.88 In a following paper,89 they studied the folding of a TIM barrel (named after triosephosphate isomerase and composed of both alpha helices and beta strands), initiated by dilution of a sample containing the denaturant urea. The folding was monitored with SAXS, FRET and CD.
As an interesting further development, an arrow shaped channel design (see Fig. 10) was introduced by the Bilsel group126 to obtain mixing times below 50 μs and thereby possibly allow access to the fast initial collapse processes during protein folding that were observed for several proteins as described above. The arrow formation of the channels increases the mixing efficiency by the introduced change in momentum. The first published study126 did not include X-ray scattering but was tested by mixing-induced fluorescence quenching. Indeed, the target mixing time of 50 ms was reached for a flow rate of 10 mL min−1, and even faster mixing could be achieved for higher flow rates, where other turbulent mixer designs might require 30–60 mL min−1. A version of the arrow mixer chip, produced in stainless steel with Kapton windows, was used for a SAXS study of the folding of cytochrome c by dilution of a sample containing GdnHCl.91 A time of mixing below 100 μs was obtained for a 20 mL min−1 flow rate, which is comparable to the results obtained by the Fujisawa group.85 The time resolution was very high at 20 μs, owing to the small X-ray beam applied.
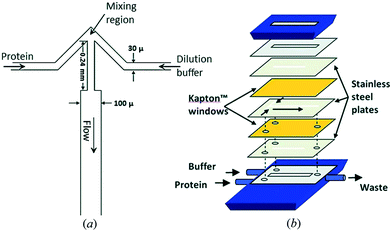 | ||
| Fig. 10 a) Illustration of the design of the arrow turbulent mixer. b) The assembly of mixer from layers of different materials. Reproduced with permission of the International Union of Crystallography, Graceffa, R. et al. (2013).91 | ||
As reviewed elsewhere,81 several other approaches to reducing the mixing time in turbulent mixers are also given in the literature, however, so far without having been combined with X-ray techniques.
c. Mixing inside droplets
In this approach, a stream of liquid (dispersed phase) is created in a microchannel, and its periodic break-up is induced by flow focusing with a second, immiscible fluid (continuous phase), creating monodisperse, micrometre-sized droplets. Different components that are injected into these vessels will rapidly mix by diffusion after the droplet pinch-off.14 In a linear flow channel, the shear by the channel walls leads to formation of streamlines forming circulating loops within each half of the droplet. When the droplet travels through a serpentine section of the channel, asymmetric circulation takes place within the droplet, and a volume from one side of the droplet will be folded into the other side. By introducing a series of turns to form a serpentine channel, the mixing time is greatly reduced and can be calculated for a given droplet size.127The confinement of the sample in droplets ensures that there is no dispersion of the sample, which is a great advantage compared to laminar mixers (Fig. 11), as described in section 4a.
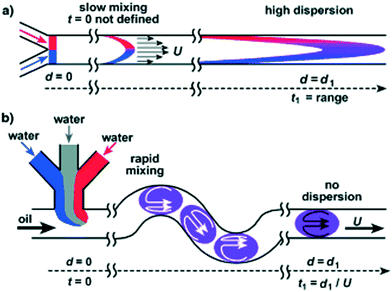 | ||
| Fig. 11 Illustration of (a) a laminar mixer and (b) mixing in a droplet-based mixer, resulting in faster mixing and a lower degree of dispersion of the sample in the flow direction compared to the laminar mixer, therefore avoiding smearing of the probed kinetic times. Reprinted (adapted or reprinted in part) with permission from Song, H. et al.121 Copyright [2003/John Wiley and Sons] [Biopolymers/John Wiley and Sons]. | ||
Only one example, by Stehle et al., was found for microfluidic droplet mixing in combination with X-ray scattering for kinetic studies.116 It concerns formation of gold nanoparticles from solution. A train of aqueous droplets is transferred by a continuous oil phase from the chip to a quartz capillary where the SAXS data are collected. This introduces a dead time, and the accessible time scale for the setup is 3.2 to 5 seconds, all covered in one data point. A background scattering is obtained due to the oil–water surfaces of the droplets passing the beam, which is, however, not an issue for the reported results since it is not highly significant compared to the intense scattering from gold. However, for other sample types, such as the protein samples discussed in the previous sections, it would complicate the measurements. Continuous collection of short data frames and the subsequent selections of the ones where no surface is in the beam would then be required.
The advantages of droplet-based mixing, as outlined by Song and Ismagilov et al., are, however, compelling.115,121 Firstly, it does not give rise to dispersion of the sample along the flow direction, as opposed to the laminar mixers, where the velocity profile is parabolic, leading to smearing of the kinetic times. Regarding the mixing time, as illustrated in Fig. 11, it is limited by diffusion as for the laminar mixer. The flow lines of each of the mixing constituents of the dispersed phase will stay in separate loops also after the droplet has formed. However, by designing the chip with zig-zag channels, the merging of the two volumes is improved. Therefore, mixing inside droplets might hold potential for future applications for kinetics and X-ray scattering.
d. Free-jet mixers
The microfluidic mixers described so far all featured an X-ray exposure channel for collection of scattering data or a connection from the chip to a measurement capillary. In recent papers, the possibility for microfluidic formation of liquid jets in free air or under vacuum is also explored. With this approach, fouling of the surfaces of the exposure channel is avoided and background scattering is minimized. Marmiroli et al.128 showed how this could be used to study fast chemical reactions, in this case, the formation of calcium carbonate nanoparticles. The chip was designed as a laminar mixer, featuring channels in a cross layout (Fig. 7A), with one reagent solution entering the central inlet channel, hydrodynamically focused by the other reagent solution entering from the side channels. After a short (500 μm) output channel, a free jet is formed, with a diameter of 25 μm. Owing to the small dimensions of the output channel (10 μm × 60 μm), very fast mixing of less than 40 μs was obtained. In an X-ray exposure channel, these small dimensions might have resulted in significant parasitic scattering from the channel walls, whereas this is no issue for the free jet.Another interesting aspect of a free jet is its compatibility with X-ray free electron laser (XFEL) beams that are extremely bright and may destroy virtually any kind of chip material. Microfluidic mixers can therefore not be inserted in the beam but can provide a free jet with the continuous supply of fresh sample that is required for XFEL applications. Wang et al.129 designed a device for mixing and subsequent formation of a free jet, which was based on three concentric tubes. The solution of the particles or crystals of interest is pumped through the centre tube and is hydrodynamically focused by the triggering ligand in the second tube, which is slightly longer. Both these tubes are contained in the third tube, with a gas stream, focusing the mixing solution to a free jet of only 3 μm width. Mixing times down to 150 μs were obtained. As always for continuous flow mixers, different kinetic times are probed by varying the distance from the point of mixing to the point of measurement, in this case by sliding the inner tube. With the applied flow rates, kinetic times from 10 to 1000 ms could be obtained. The same interval would, of course, be reached using a synchrotron source instead of an XFEL. Trebbin et al.130 presented a PDMS microfluidic chip with the same function, mixing of solutions and formation a free jet using a gas flow. Compared to the concentric tubes of Wang et al., this provides better fabrication reproducibility of channel dimensions and design. It was shown that the chip could operate under vacuum, making it compatible with an evacuated chamber at an X-ray beamline. The mixing ability of the chip was mainly intended for advanced sample delivery, as opposed to the kinetic applications mentioned by Wang et al.129
5 Sample processing
a. Sample handling and delivery
Although a clear advantage of using microfluidic devices is the ability to precisely control fluid flows as well as the possibility of generating a multitude of conditions for scanning large parameter spaces, microfluidic devices can also be used as sample delivery or capture devices for minute amounts of samples prepared off-chip. Most of the reported applications within this category are within delivery of microcrystals for collection of diffraction data. Table 4 summarizes recent advances in microfluidic devices for sample delivery with in situ X-ray data collection.| Chip features | Chip material/fabrication | Window material and thickness | Sample volume/consumption | Sample investigated | X-Ray technique | Beamline, source, X-ray energy | Ref. |
|---|---|---|---|---|---|---|---|
| a Abbreviations: LCLS, Linac Coherent Light Source; BESSY, Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung; SSRL, Stanford Synchrotron Radiation Lightsource; APS, Advanced Photon Source. | |||||||
| Nozzles (purchased from Microliquids GmbH) | Convergent quartz glass nozzle pulled out over oxygen–hydrogen flame; 10–15 μm diameter jet | Vacuum (no window, free jet) | Liquid phase molecular dynamics and chemistry | X-ray spectroscopy | LCLS, Stanford, 0.54 keV | Kunnus et al. (2012)140–143 | |
| Silica capillary | Electrospun microjet, 50 μm diameter internal capillary, 50 μm jet | Vacuum (no window, free jet) | Photosystem II | X-ray diffraction and spectroscopy | LCLS, Stanford, 9.7 keV | Kern et al. (2013)144 and Sierra et al. (2012)145 | |
| PDMS droplet generator connected to a thin glass tube capillary via a piece of polyethylene tubing for in situ data collection | PDMS/photolithography and soft-lithography | 10 μm thick glass capillary | ∼100 μm size droplets | Gold nanoparticles | SAXS | Beamline 7T-MPW-SAXS, BESSY II, 7.5 and 8.0 keV | Stehle et al. (2013)116 |
| Microfluidic device allowing cell-culture and in situ X-ray imaging | UV-curable adhesive, Kapton® and silicon nitride/photolithography and soft lithography | Silicon nitride | Hydrated cells | Scanning X-ray diffraction | Beamline P10, PETRA III, 7.9 keV, beam size: 360 × 280 nm2 | Weinhausen and Köster (2013)26 | |
| Hollow-core fused silica optical fibre as the inner capillary, borosilicate glass forms the outer housing | Two capillaries inserted into one another to form droplets, 360 μm outer diameter and 20–50 μm inner diameter, 1.2 mm outer diameter by 0.9 mm inner diameter | Vacuum (no window, free jet) | NA | Performance test only | X-ray diffraction | LCLS, Stanford, 9.4 keV | Abdallah et al. (2015),146 DePonte et al. (2008),139 Weierstall et al. (2012)147 and Redecke et al. (2013)148 |
| Microfluidic chip, which can capture microcrystals at fixed, addressable points in a trap array from a small volume (<10 μl) of a pre-existing slurry grown off-chip | PDMS/photolithography and replica molding | 250 μm PMMA + 50 μm PDMS | <5 μl sample per chip | Protein microcrystals | X-ray crystallography, Room temperature without flash cooling | X-ray Pump Probe endstation, LCLS and beamline 12-2, SSRL | Lyubimov et al. (2015)131 |
| Features hydrophobic and hydrophilic surface patterns to help localize microcrystals | Three layer chip composed of silicon nitride, a photoresist and Kapton®/photolithography | 150 nm silicon nitride and 8 μm Kapton® | <2 μL | Protein microcrystals | X-ray crystallography, Room temperature | Beamline 12-2, SSRL and X-ray Pump Probe endstation, LCLS and beamline 23-ID, APS | Murray et al. (2015)27 |
Lyubimov et al. were able to efficiently capture microcrystals from a small volume of slurry (<10 μL) into a hydrodynamic trap array for X-ray data collection (Fig. 12).131 Murray et al. used hydrophobic and hydrophilic surface patterns to help localize microcrystals.27 Oberti et al. used acoustic forces to move crystals in micromachined channels towards an orifice where they could be removed using a nylon loop.132 Acoustophoresis could be used advantageously for sample manipulation and delivery in future crystallography applications as the process is gentle and does not damage the crystals.
 | ||
| Fig. 12 A microfluidic trap array for protein microcrystals. (a) Schematic representation of the crystal-capturing device design. (b) Close-up of the general scheme for trap-and-bypass hydrodynamic crystal capture. (c) Schematic of a single hydrodynamic trap [labelled in (b); WT is the width of the trap channel and LT is the length of the trap channel]. (d) Light micrograph of a representative section of a fabricated crystal-capture chip. (e) Light-micrograph series showing single and multiple high-purity egg-white lysozyme (HEWL) microcrystals immobilized in hydrodynamic traps (reproduced with permission from the International Union of Crystallography; Lyubimov et al., 2015 (ref. 131)). | ||
Weinhausen et al. developed an X-ray compatible microfluidic device for X-ray measurement on hydrated cells based on a commercially available UV-curable adhesive (NOA 81, Norland Optical Adhesives, Cranbury, NJ).26 The device is sealed with Kapton® foil and features X-ray compatible silicon nitride windows as a growth substrate for cells. That is, the sample is not delivered by the chip but grown on the chip, readily available for X-ray inspection.
Another use of microfluidics as a sample delivery method is the use of microjet for X-ray free electron lasers (XFEL). XFELs, with their extremely intense beam, are gaining importance in serial crystallography and as a method for addressing the structure of proteins that fail to produce crystals large enough to be examined using traditional synchrotron X-ray radiation. To outrun radiation damage, an approach by Neutze et al.133 commonly referred to as ‘diffraction before destruction’ is used. Neutze et al. proposed this approach already in year 2000 based on simulations and theoretical considerations. Through femtosecond laser pulses, the protein structure is acquired before the crystal is obliterated by the intense XFEL radiation.134,135 In a pioneering work, Chapman et al.136 used the diffraction-before-destruction technique to solve the structure of membrane protein photosystem I with a sub-nanometre resolution.136 A typical setup of microfluidics coupled with XFEL is presented in Fig. 13. Nanocrystals are injected and form a 4 μm diameter jet that flows at 10 m s−1 perpendicular to the XFEL beam that is focused on the jet.
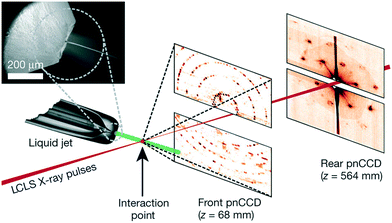 | ||
| Fig. 13 Typical setup of a microjet coupled with XFEL. Inset: SEM image of the nozzle. Two pairs of high-frame-rate pnCCD detectors137 record low- and high-angle diffraction from single X-ray FEL pulses, at the FEL repetition rate of 30 Hz. Reprinted by permission from Macmillan Publishers Ltd. [Nature] (Chapman et al.136), copyright (2011). | ||
In another example, Abdallah et al. used a free-jet microfluidic system138,139 similar to the one used by Chapman et al. to study photosystem I nanocrystal fractions that range between 200 and 600 nm in size. Other examples of the combination of liquid jet with XFEL are presented in Table 4; however, it should be noted that the examples mentioned in the table do not cover all papers utilizing the combination of microfluidic free-jet systems with XFEL but rather are a representation of the most common techniques.
b. Confinement and alignment
In microfluidic systems, confinement of the flowing sample can either take place near a solid surface (the microchannel walls and pillars etc.)16,149–152 or near a liquid–liquid interface, created by hydrodynamic focusing.16,17,96,152–154 SAXS measurements were then collected at different positions along the length of the microchannel. Systems that were studied under alignment and confinement by microfluidics include liquid crystals,153,155 cellulose nanofibrils,17,154 filamentous proteins,152 DNA61 and surfactant micelles.13,112,149,150Flow alignment can be exploited to investigate material properties that are usually not accessible from the non-oriented sample. Furthermore, new and unexpected phenomena can emerge once confinement and alignment under non-equilibrium conditions in microfluidic systems are achieved. However, still only a limited number of studies exploited the advantages associated with microfluidics coupled with X-ray techniques for confinement and alignment studies (see Table 5).
| Chip features | Chip material/fabrication | Window material and thickness | Sample volume/consumption/flow rates | Sample and sample orientation | Beam dimensions | Source/beamline | Ref. |
|---|---|---|---|---|---|---|---|
| The channel sizes are 300 μm in width and 40 μm in depth | Laser ablated polyimide (Kapton) | Kapton KJ used as windows and was thermally bonded to the standard Kapton film | 1–60 μL h−1, all the micelles are aligned with the flow | Wormlike micelles under shear | 10 × 40 μm2 | Microfocusing endstation ID18F, ESRF | Barrett et al. (2006)34 |
| A network of channels meeting at a cross; uniform channel height of 150 μm while the width changes from 50 to 150 μm | PDMS/molding using photolithographic techniques (SU-8) | Adhesive polyimide (Kapton) windows (53 μm thick) | 10–500 μm s−1 | Smectic liquid crystal n-octyl-4-cyanobiphenyl (8CB) | 20 μm in diameter | ID10B at the European Synchrotron Radiation Facility (ESRF) | Dootz et al. (2007)153 |
| A network of channels meeting at a cross; uniform channel height of 300 μm and width of 150 μm | Spark eroded stainless steel covered with Kapton | Adhesive polyimide (Kapton) windows | 100 μm s−1 | DNA compacted using cationic dendrimers, strong orientation preferentially along the flow direction | 20 μm diameter | ID10B at the European Synchrotron Radiation Facility (ESRF) | Pfohl et al. (2007)96 |
| The channel sizes were approximately 70 μm in height, 16 cm in length and with widths varying from 100 to 2000 μm | PDMS and glass, with SU-8 as master | 150 μm thick glass + varying PDMS thicknesses ranging from approximately 0.9 mm to 1.8 mm | 1 mL h−1 corresponding to 7 mm s−1 | Surfactant mixtures containing SDS and CTAC | 150 × 100 μm2 | I22 beamline at Diamond Light Source, Oxfordshire, UK | Martin et al. (2010)150 |
| Microfluidic devices with channels of 100 μm height, with width varying from 150 to 50 μm and of 16 cm overall length | PDMS microfluidic chip covered with Kapton tape | Kapton tape | 32.4 μL h−1 corresponding to 360 μm s−1 | Cylindrical block copolymer micelles, anisotropic colloidal particles align perpendicular to the flow direction | 20 × 30 μm2 | Beamlines BW4 and P03 at HASYLAB/DESY | Trebbin et al. (2013)151 |
| A network of channels meeting at a cross; the channel cross section is a square with width and depth of 1 mm | Stainless steel sandwiched between two Kapton sheets | Polyimide (Kapton) windows | 10–14 μL s−1 | Cellulose nanofibrils | 24 × 11 μm2 | P03 beamline, at the PETRA III, DESY, Hamburg, Germany | Håkansson et al. (2014, 2015)17,154 |
| A network of channels consists of 4 inlets meeting at a cross; the channel cross section is square with a width of 100 um | Cyclic olefin copolymer (COC) (commercially available) | 0.1–3 μL min−1 | Nematic liquid crystals, aligned in the direction of the flow with a small tilt angle of 11° | 6 × 40 μm2 | cSAXS, Swiss Light Source, Paul Scherrer Institute | Silva et al. (2015)16 | |
| 30 × 300 μm2 | |||||||
| SAXS/D beamline Stanford Synchrotron Radiation Light Source | |||||||
| 2 mm wide channel with a single constriction of 400 μm width and a constant depth of 540 μm | Thiol-ene copolymer (NOA 81) within thin borosilicate plates | 300 μm borosilicate glass | 1 mL h−1 | Lamellar and multilamellar vesicle phases of the linear sodium alkylbenzenesulfonate surfactant | Approx. 300 × 500 μm2 | I22 beamline, Diamond Light Source (UK); photon energy of 12.4 and 17 keV | Poulos, S. et al. (2016)160 |
| Microfluidic devices with channels of 70 μm height, with width varying from 100 to 2000 μm and of 16 cm overall length | PDMS microfluidic chip made by soft lithography, SU-8 master is used | 1 mm thick PDMS window | 0.1 to 5 mL h−1, corresponding to fluid residence times of approximately 2–400 s | Single-chain cationic surfactant, a short-chain alcohol and water; orientation varies between parallel and perpendicular depending on the position in the channel | 150 × 150 μm2 | Beamline I22, Diamond Light Source (Oxfordshire, UK) | Martin et al. (2016)149 |
Small-angle X-ray scattering (SAXS) was used to probe the real-time dynamic evolution of the supramolecular interactions between DNA and cationic dendrimers (Fig. 14), which assemble with an orientation transverse to the flow direction. DNA was injected in the center channel (depicted in green in Fig. 14) using a custom-built syringe setup and was then focused by two side streams containing dendrimers.
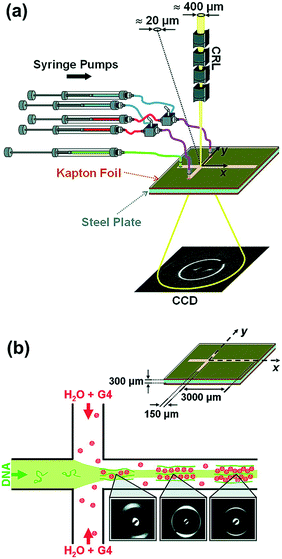 | ||
| Fig. 14 Experimental setup of the DNA compaction experiment. (a) The microdevice was placed in a microfocused X-ray beam, and a diffraction pattern was acquired; (b) close-up schematics of the device showing relevant length scales. Reprinted (adapted) with permission from Pfohl et al.96 Copyright (2007), American Chemical Society. | ||
Other biological applications include the alignment of filamentous proteins through confinement in microchannels.152 Similar devices to the one used in Fig. 14 are commonly used in mixing applications as mentioned in section 4 on mixing-induced kinetics62,66,86,98 but can also be used for aligning the DNA molecules. The microchannels were cut using spark erosion into stainless steel plates then covered with thin adhesive Kapton foils. All SAXS images showed an orientation of the dendrimer–DNA complexes aligned transverse to the flow direction at a point in the microdevice where the strain rate is highest.
Håkansson et al. used a hydrodynamic-focusing microfluidic system to produce homogeneous and smooth filaments from a dispersion of cellulose nanofibrils.17,154 Cellulose pulp fibers with fully aligned fibrils can be comparable to glass fibers in strength156 and comparable to poly(p-phenylene terephthalamide) (PPTA) (commercially known as Kevlar or Twaron) in specific stiffness.157 The microfluidic system was combined with SAXS to understand and control the formation process of such filaments. Different flow rate ratios, salt concentrations, and sample concentrations were used for this study, with a sample consumption of ca. 5 μL s−1. The microfluidic system was prepared by cutting out channels in 1 mm thick stainless steel which was then sandwiched between two polyimide (Kapton) windows. The system uses hydrodynamic focusing for alignment, in a similar way to Pfohl96 and Silva;16 however, somewhat surprisingly, the device must be bigger if good alignment of fibrils is to be achieved. Therefore, dimensions of 1 mm were applied for both width and height of the channel, which is comparable to length scales in some of the traditional macroscopic rheological techniques. However, we still included the work in this review because traditional microfabrication techniques were applied and because microchannels of smaller dimensions were also investigated in order to determine the optimal channel dimensions. The filaments are prepared by utilizing a surface-charge-controlled gel transition.158,159Fig. 15 shows an idealized description of the assembly phase of the filament-forming process, as derived from the scattering data. Above and below the illustration of flowing nanofibrils, the mechanical (above) and electrochemical (below) processes affecting the fibrils are illustrated. In the liquid dispersion, fibrils are fairly free to rotate (a), due to strong electrostatic repulsion (e). An accelerating flow causes the fibrils to align in the flow direction65,112 (b). Before the alignment is lost due to Brownian diffusion (c), the electrostatic repulsion between the particles is reduced by an electrolyte diffusing into the suspension (f–h). Finally, the aligned structure is frozen as a gel (d).
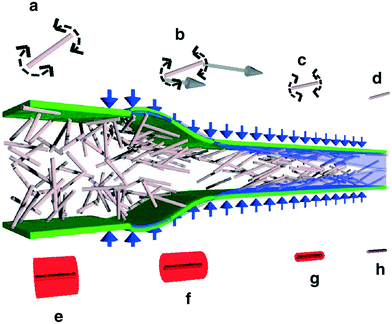 | ||
| Fig. 15 Illustration for the use of hydrodynamic-focusing microfluidics to produce filaments from a dispersion of cellulose nanofibrils; (reproduced from) reprinted with permission from Macmillan Publishers Ltd. [Nature] (Håkansson et al.17), copyright (2014). | ||
The orientation of flowing liquid crystals using microfluidics and SAXS was studied by Silva et al.16 The microfluidic system used was made of cyclic olefin copolymer (COC or Topas) and commercially available. The system features a network of four channels meeting at a cross. The same system was used to study shear effects of a nematic liquid crystal under confinement by either a solid wall or a liquid wall. SAXS data were collected using the setup presented in Fig. 15. For solid-wall confinement, the sample is flowed at a flow rate of 3 μL min−1 at the inlet of the long channel (depicted in Fig. 16a) and collected at the other three outlets. For liquid-wall confinement, hydrodynamic focusing of the nematic liquid crystal solution between sheets of Triton X solutions was obtained by using three channels as inlets (Ch. 1–3, see Fig. 16d). By collecting scattering data at various positions on the chip, the effect on the nematic ordering by flow rates, wall-confinement and surfactant could be probed.
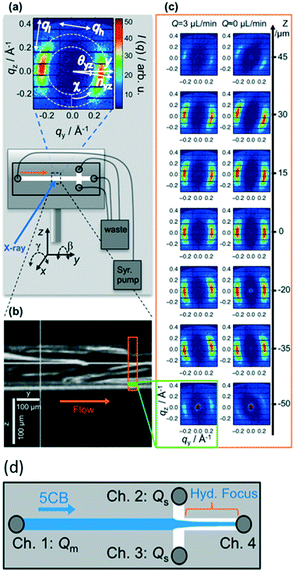 | ||
| Fig. 16 Summary of the microfluidic SAXS experiments by Silva et al. (a) Typical SAXS pattern (top) and schematic of the microfluidic device. A typical SAXS pattern at small q is dominated by the nematic phase correlation peak. (b) Snapshot of 5CB liquid crystals flowing slowly before data acquisition. (c) Representative scattering patterns of a flowing nematic phase recorded along the width (z direction) of the microchannel at a flow rate of 3 μL min−1 (left) and at rest (right). (d) Liquid-wall confinement. Reprinted with permission from Silva et al.16 Copyright (2015), ACS Publications. | ||
In another work, Martin et al.149 combined microfluidics with SAXS to study concentrated surfactant mixtures. They studied the influence of model flow fields (contraction–expansion and rotational flows) on the microstructure and relaxation of a surfactant mixture in microdevices. Four different microfluidic designs were used for this study (Fig. 17). The channel of each design is patterned differently with constrictions such as the case in MC2, array of circular posts in MC3 and bends in MC4. MC1 consists of a 3 cm long and 1 mm wide unpatterned channel, followed by a 13 cm long and 2 mm wide channel. MC2, on the other hand, consists of four constrictions of 100 μm width and 820 μm length each. MC3 comprises an array (5 × 4) of circular posts with a diameter of 150 μm and an inter-post spacing of 50 μm. MC4 comprises a meandering channel with 22 bends with an outside diameter of 1750 μm, forming a total average flow path length of 13.8 mm.
 | ||
| Fig. 17 Microfluidic channel designs with different patterns such as constrictions, array of circular posts and bends (reproduced from Martin et al.149 with permission from the Centre National de la Recherche Scientifique (CNRS) and The Royal Society of Chemistry). | ||
The alignment in this work is based on the effect that geometric objects (such as the circular posts) have on the fluid rather than shear forces induced by the focusing fluids from the sides. The SAXS data shown in Fig. 18 reveal a degree of alignment of the lamellar phase. The lamellar structures align with the channel walls and remain mostly isotropic along the channel centreline. The geometric constriction forces the lamellar stacks to align parallel to the flow; however, upon expansion at the exit of the constriction, the lamellae flip to a perpendicular orientation to the flow and return to a less ordered, ‘polycrystalline’ arrangement compared to the single crystalline order present in the constriction. A similar study was carried out by Poulos et al.160 on lamellar and multilamellar vesicle phases.
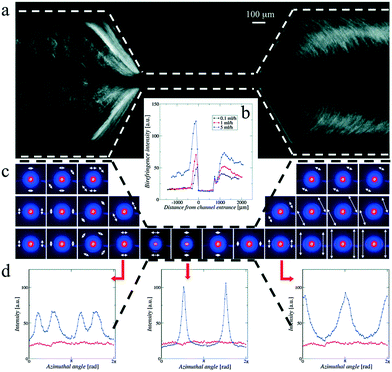 | ||
| Fig. 18 CTAC/pentanol/water system under flow through the first constriction of MC2 from a width of 1000 μm to 100 μm at a flow rate of 5 mL h−1; (a) a cross-polarized optical micrograph; (b) birefringence intensity along the channel from the constriction entrance; (c) 2-D SAXS map; (d) azimuthal average of the intensity for 2-D SAXS for the surfactant system (blue symbols) and for the background (red symbols) at constriction entrance (left), within the constriction (middle), and upon exit (right) (reproduced from Martin et al., 2016 (ref. 149) with permission from the Centre National de la Recherche Scientifique (CNRS) and The Royal Society of Chemistry). | ||
Priebe et al.161 studied the shear induced alignment of multilamellar lipid assemblies in a microfluidic jet, formed by a nozzle under different shear conditions, which can be controlled by varying the flow rate and nozzle diameter. The orientation of the lipid was investigated across the jet, and showed strong alignment parallel to the direction of the flow, as presented in Fig. 19. The degree of alignment was highest at the centre of the jet. This was related to the fact that the elongational strain was highest at the centre of the nozzle, whereas the shear rate was highest near the nozzle walls. For the applied volumetric flow rate of 1 ml min-1 and two different jet widths of 40 μm and 13 μm, only a small degree of relaxation of the alignment was observed along the direction of the jet flow, before the jet breaks up into droplets ca. 25 μs after exiting the nozzle.
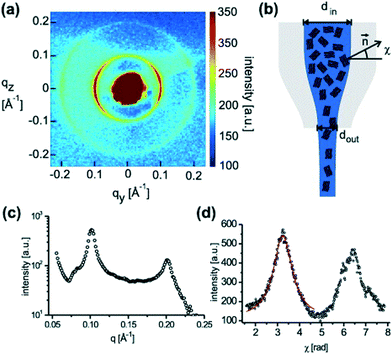 | ||
| Fig. 19 (a) Typical diffraction pattern (raw data) of a multilamellar sample with the circular segments of n = 1 and n = 2 lamellar orders, recorded by the CCD. (b) Schematic of the shear-induced orientation of lamellar domains. (c) The radial intensity profile derived from (a), showing two lamellar reflections. (d) The azimuthal distribution, reflecting the sample alignment, fitted to a Lorentzian with a linear background (solid line). Reprinted with permission from Priebe et al.161 Copyright (2010), New J. Phys., IOP Science. | ||
Another interesting possibility of utilizing shear forces present inside a microchannel was demonstrated by Barrett et al., who used shear forces to promote structural transitions of pre-formed micelles.34
Microfluidics for alignment and shear force studies is a field where not only X-ray scattering but also neutron scattering shows great potential. Neutron scattering provides complementary scattering contrasts compared to X-ray scattering, including the possibility of contrast variation by selective isotope labeling. The microfluidic chips might be prepared using a wide variety of materials, including metals, owing to the high transmission of neutrons through these. Unfortunately, the large neutron beams, typical beam diameters are in the range of 8–20 mm, are not immediately compatible with micrometer to sub-millimeter sized microfluidic channels. However, the high neutron intensity at the upcoming European Spallation Source Facility gives promise also in this area. This has sparked a few pioneering experiments in the field and the first results, investigating model surfactant systems under shear flow, using a 500 μm neutron beam, have been reported by Lopez et al.10 Judging from the current initiatives in the field, there are more to follow.162–164
6 Discussion and outlook
We have provided a comprehensive review of the past 15 years of development within the field of microfluidics for X-ray analysis at synchrotron beamlines, including a few applications of XFEL sources as well. An overall picture of a field that is still in its youth and highly dominated by a few pioneering groups collaborating closely with highly dedicated synchrotron beamlines emerges.The review shows that microfluidics has obvious applications when it comes to efficiently producing many different samples allowing the investigation of the interaction between a biological molecule and its surrounding buffer. As shown in section 3, this may be relevant when identifying crystallisation conditions for a protein and in a protein formulation context when optimising the buffer for a given solution structure state of the protein. Additionally, microfluidics may also be used as an integrated sample preparation and delivery platform at a synchrotron X-ray beamline.
Some of the first devices were designed for investigating protein or DNA folding in a time-resolved fashion. Here, various microfluidic devices were used to trigger a structural change in the bio-macromolecule through pH changes or diffusion of ligands. As discussed in section 4, the time-scales that can be studied using this approach are limited by the diffusion time of the involved molecules. This sets a lower limit at about 0.1 millisecond for the fastest accessible processes which makes the approach suitable to investigate larger structural rearrangements for a broad range of biomolecules. However, it also suggests that additional development is required to cover some of the faster structural transitions.
During the past ten years, there has been some focus on using the rather extreme flow fields within microfluidic channels to shear-align highly anisotropic samples. As shown in section 5, this is a very interesting development that provides unique data and also a lot of opportunities for further fundamental studies.
Based on our review of the field, it is relevant to ask whether the combination of microfluidics and X-ray techniques is really as promising as proposed by some of the pioneers within the field. Our review shows that it is not possible to provide a single answer to this question; instead, the answer depends largely on which application this combination is going to be used for and which scientific challenge it is addressing. Our review also shows that it is maybe still too early to fully assess the potential of the combined approach.
We believe that there are four major lines of applications when using this combination: 1) high-throughput sample mixing and delivery such as in the case of protein crystallography where microfluidics is used as a platform that in principle allows for investigating a large parameter space very effectively to identify different crystallization conditions.28 2) Obtaining a dynamic sample preparation environment, which allows for tuning the buffer conditions during an experiment and based on feedback from the experimental results.11 3) Obtaining insight into the dynamics of a sample through controlled microfluidics enabled mixing81,82 and 4) as a means for investigating sample behavior as a function of a strong shear field.160
Regarding 1), the early development of microfluidics based crystallisation platforms was initiated about 20 years ago when there was a very large demand for efficient tools for this purpose. However, in parallel with the microfluidics development, there has also been a very strong development within liquid handling robots for the same purpose,47,165 standardised and highly effective screening kits for different types of proteins,166 along with sample loading robotic devices for crystallography data acquisition.167,168 Most of the modern synchrotron based high throughput facilities for protein crystallography (e.g. at Diamond and ESRF) are based on this combination of setups. Thus, while the underlying problem cannot yet be regarded as fully solved, and while there is still room for further development, most of the present activity is focused on the very fast and robust liquid handling robots and sample loading devices. However, as shown from examples in this review, there is an interesting recent development in the use of combined microfluidics and serial crystallography where microfluidic platforms can be used to both prepare and load the crystals without the necessity for cryocooling. This opens up the possibility of using gentle microfluidic methods, such as acoustophoresis, to manipulate fragile crystals132 and place them in the beam path.
Regarding 2), microfluidics enables a dynamic sample preparation environment. This means that the sample composition and hence the experiment design may, in principle, be optimised during the experiment and as a response to the incoming experimental results. This idea was first proposed in a couple of articles by some of the authors of this review11,33 and suggestions for microfluidic platforms for this purpose were developed. However, the challenge of systematically integrating these with fully automated data analysis in order to provide automated feedback to the sample preparation units still remains. More recent solutions have focused instead on the preparation of a large number of samples with pre-defined conditions,78 sacrificing flexibility for more simplicity and minimal sample consumption.
Regarding 3), microfluidics enables time resolved studies of mixing induced structural kinetics. For mixing on the millisecond time scale, the use of laminar flow conditions drastically reduces the required sample volume as compared to classical stopped-flow techniques, which is essential especially for many biological samples. Sub-millisecond time scales can also be reached, however, requiring much higher sample volumes to enable fast turbulent mixing. Since the functioning of proteins, as well as the properties of soft materials in general, are closely related to their structure and dynamics, we believe that this application of microfluidic chips will continue to grow.
Regarding 4), the use of synchrotron X-ray on conventional macroscopic rheological setups has been extensively explored in the past few decades. However, coupling microscopic rheological setups (microchannels) with synchrotron X-ray techniques is still in its infancy, and compared to the other microfluidic applications discussed in this review, it is the least explored avenue of research. However, the studies published so far show how the approach has great potential for exploring non-Newtonian phenomena that are not easily studied with other techniques.
As mentioned above, the general combination of microfluidics and X-rays is still in its early years, and therefore, most synchrotron X-ray beamtime sessions, where microfluidics are applied, are still highly experimental and require immense efforts from both users and local beamline staff, while yielding less data compared to standard experiments. According to our experience, set-up times of the microfluidic experiments ranging from 6 to 24 hours are not unusual before any results can be acquired. In addition, since the experimental setup is configured manually with the help of a local beam scientist, even minor configuration changes can cost considerable amounts of time. An additional challenge associated with the combination of microfluidics and X-rays is the need for rather different fields of expertise on the experimental team. Due to the limited availability of standardized microfluidic solutions, a microfluidic expert is required on the team to design and manufacture the customized microfluidic system that can address the group's specific needs. However, the fact that a multidisciplinary team with expertise in microfluidics, X-ray experiments, and biological (or other) systems need to collaborate to complete a successful experiment may also help bring together researchers and scientists with different backgrounds to develop radically novel solutions for specific challenges.
It is also noteworthy that the field of combining microfluidics and synchrotron X-ray so far is highly dominated by universities and academic research groups but lacks the serious involvement of industrial partners. This is probably due to the advanced, edge-of-science nature of the experiments, which may not be compatible with the expertise and resources available to companies working in the field. Another important factor is certainly the fact that application cases that are sufficiently convincing to industry still need to be developed.
To summarize, the combination of microfluidics and synchrotron X-ray techniques should be considered an emerging method still under development. Just like any new technology, it has to go through a considerable amount of optimization and maturation in order to reach a larger audience. The good news is that many synchrotron facilities around the world have started to build beamlines with very narrow beams that are optimally suited for microfluidic applications. Also, the recent advancements in microfluidic large scale integration (mLSI),169 where thousands of integrated micromechanical components such as valves can be integrated together, are promising for application in biology and chemistry to perform fast, reliable analysis and replace mechanical automation solutions such as fluid-handling robots. Moreover, with more and more microfluidic companies, standardized, affordable and reliable microfluidic solutions should become available for a broader community in the coming years.170
Acknowledgements
The authors would like to acknowledge funding to beamtime travels from the DANSCATT programme under the Danish Research Infrastructure Programme, CoNeXT funding from the UCPH-2016 Programme of Excellence. Furthermore, Lise Arleth wishes to acknowledge the Danish Council for Independent Research for a Sapere Aude Starting Grant.References
- R. Neutze and K. Moffat, Curr. Opin. Struct. Biol., 2012, 22, 651–659 CrossRef CAS PubMed.
- U. Weierstall, Philos. Trans. R. Soc., B, 2014, 369, 20130337 CrossRef PubMed.
- B. W. J. McNeil and N. R. Thompson, Nat. Photonics, 2010, 4, 814–821 CrossRef CAS.
- R. Daw and J. Finkelstein, Nature, 2006, 442, 367 CrossRef CAS.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS PubMed.
- W. Jung, J. Han, J.-W. Choi and C. H. Ahn, Microelectron. Eng., 2015, 132, 46–57 CrossRef CAS.
- D. Gao, H. Liu, Y. Jiang and J.-M. Lin, Lab Chip, 2013, 13, 3309 RSC.
- E. Hughes, A. A. Maan, S. Acquistapace, A. Burbidge, M. L. Johns, D. Z. Gunes, P. Clausen, A. Syrbe, J. Hugo, K. Schroen, V. Miralles, T. Atkins, R. Gray, P. Homewood and K. Zick, J. Colloid Interface Sci., 2013, 389, 147–156 CrossRef CAS PubMed.
- L. Pollack, M. W. Tate, N. C. Darnton, J. B. Knight, S. M. Gruner, W. A. Eaton and R. H. Austin, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 10115–10117 CrossRef CAS.
- C. G. Lopez, T. Watanabe, A. Martel, L. Porcar and J. T. Cabral, Sci. Rep., 2015, 5, 7727 CrossRef CAS PubMed.
- K. N. Toft, B. Vestergaard, S. S. Nielsen, D. Snakenborg, M. G. Jeppesen, J. K. Jacobsen, L. Arleth and J. P. Kutter, Anal. Chem., 2008, 80, 3648–3654 CrossRef CAS PubMed.
- E. A. Mansur, M. Ye, Y. Wang and Y. Dai, Chin. J. Chem. Eng., 2008, 16, 503–516 CrossRef CAS.
- Y. K. Suh and S. Kang, Micromachines, 2010, 1, 82–111 CrossRef.
- S.-Y. Teh, R. Lin, L.-H. Hung and A. P. Lee, Lab Chip, 2008, 8, 198–220 RSC.
- S. Guha, S. L. Perry, A. S. Pawate and P. J. A. Kenis, Sens. Actuators, B, 2012, 174, 1–9 CrossRef CAS PubMed.
- B. F. B. Silva, M. Zepeda-Rosales, N. Venkateswaran, B. J. Fletcher, L. G. Carter, T. Matsui, T. M. Weiss, J. Han, Y. Li, U. Olsson and C. R. Safinya, Langmuir, 2015, 31, 4361–4371 CrossRef CAS PubMed.
- K. M. O. Håkansson, A. B. Fall, F. Lundell, S. Yu, C. Krywka, S. V. Roth, G. Santoro, M. Kvick, L. Prahl Wittberg, L. Wågberg and L. D. Söderberg, Nat. Commun., 2014, 5, 4018 Search PubMed.
- C. M. Jeffries, M. A. Graewert, D. I. Svergun and C. E. Blanchet, J. Synchrotron Radiat., 2015, 22, 273–279 CAS.
- S. Kuwamoto, S. Akiyama and T. Fujisawa, J. Synchrotron Radiat., 2004, 11, 462–468 CrossRef CAS PubMed.
- T. R. Schneider, HFSP J., 2008, 2, 302–306 CrossRef CAS PubMed.
- R. Moukhametzianov, M. Burghammer, P. C. Edwards, S. Petitdemange, D. Popov, M. Fransen, G. McMullan, G. F. X. Schertler and C. Riekel, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2008, 64, 158–166 CrossRef CAS PubMed.
- J. H. Hubbell, Int. J. Appl. Radiat. Isot., 1982, 33, 1269–1290 CrossRef CAS.
- L. Pollack, M. W. Tate, N. C. Darnton, J. B. Knight, S. M. Gruner, W. A. Eaton and R. H. Austin, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 10115–10117 CrossRef CAS.
- R. Russell, I. S. Millett, M. W. Tate, L. W. Kwok, B. Nakatani, S. M. Gruner, S. G. J. Mochrie, V. Pande, S. Doniach, D. Herschlag and L. Pollack, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4266–4271 CrossRef CAS PubMed.
- K. F. Lei, in RSC Detection Science, ed. F. H. Labeed and H. O. Fatoyinbo, Royal Society of Chemistry, Cambridge, 2014, pp. 1–28 Search PubMed.
- B. Weinhausen and S. Köster, Lab Chip, 2013, 13, 212–215 RSC.
- T. D. Murray, A. Y. Lyubimov, C. M. Ogata, H. Vo, M. Uervirojnangkoorn, A. T. Brunger and J. M. Berger, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2015, 71, 1987–1997 CAS.
- C. L. Hansen, S. Classen, J. M. Berger and S. R. Quake, J. Am. Chem. Soc., 2006, 128, 3142–3143 CrossRef CAS PubMed.
- L. Daubersies, J. Leng and J.-B. Salmon, Lab Chip, 2013, 13, 910 RSC.
- O. Saldanha, M. E. Brennich, M. Burghammer, H. Herrmann and S. Köster, Biomicrofluidics, 2016, 10, 24108 CrossRef PubMed.
- M. E. Brennich, J.-F. Nolting, C. Dammann, B. Noding, S. Bauch, H. Herrmann, T. Pfohl and S. Koster, Lab Chip, 2011, 11, 708–716 RSC.
- A. Ghazal, M. Gontsarik, J. Kutter, J. P. Lafleur, A. Labrador, K. Mortensen and A. Yaghmur, J. Appl. Crystallogr., 2016, 49 DOI:10.1107/S1600576716014199.
- J. P. Lafleur, D. Snakenborg, S. S. Nielsen, M. Møller, K. N. Toft, A. Menzel, J. K. Jacobsen, B. Vestergaard, L. Arleth and J. P. Kutter, J. Appl. Crystallogr., 2011, 44, 1090–1099 CAS.
- R. Barrett, M. Faucon, J. Lopez, G. Cristobal, F. Destremaut, A. Dodge, P. Guillot, P. Laval, C. Masselon and J.-B. Salmon, Lab Chip, 2006, 6, 494–499 RSC.
- T. Uzawa, S. Akiyama, T. Kimura, S. Takahashi, K. Ishimori, I. Morishima and T. Fujisawa, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1171–1176 CrossRef CAS PubMed.
- R. Dootz, A. Otten, S. Köster, B. Struth and T. Pfohl, J. Phys.: Condens. Matter, 2006, 18, S639–S652 CrossRef CAS.
- S. With, M. Trebbin, C. B. A. Bartz, C. Neuber, M. Dulle, S. Yu, S. V. Roth, H.-W. Schmidt and S. Förster, Langmuir, 2014, 30, 12494–12502 CrossRef CAS PubMed.
- D. S. Khvostichenko, J. M. Schieferstein, A. S. Pawate, P. D. Laible and P. J. A. Kenis, Cryst. Growth Des., 2014, 14, 4886–4890 CAS.
- S. Emamzadah, T. J. Petty, V. De Almeida, T. Nishimura, J. Joly, J.-L. Ferrer and T. D. Halazonetis, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 913–920 CrossRef CAS PubMed.
- M. Maeki, S. Yoshizuka, H. Yamaguchi, M. Kawamoto, K. Yamashita, H. Nakamura, M. Miyazaki and H. Maeda, Anal. Sci., 2012, 28, 65 CrossRef CAS PubMed.
- B. Zheng, L. S. Roach and R. F. Ismagilov, J. Am. Chem. Soc., 2003, 125, 11170–11171 CrossRef CAS PubMed.
- H. Y. Park, X. Qiu, E. Rhoades, J. Korlach, L. W. Kwok, W. R. Zipfel, W. W. Webb and L. Pollack, Anal. Chem., 2006, 78, 4465–4473 CrossRef CAS PubMed.
- P. Pernot, P. Theveneau, T. Giraud, R. N. Fernandes, D. Nurizzo, D. Spruce, J. Surr, S. McSweeney, A. Round, F. Felisaz, L. Foedinger, A. Gobbo, J. Huet, C. Villard and F. Cipriani, J. Phys.: Conf. Ser., 2010, 247, 12009 CrossRef.
- A. Martel, P. Liu, T. M. Weiss, M. Niebuhr and H. Tsuruta, J. Synchrotron Radiat., 2012, 19, 431–434 CrossRef CAS PubMed.
- S. S. Nielsen, M. Møller and R. E. Gillilan, J. Appl. Crystallogr., 2012, 45, 213–223 CrossRef CAS PubMed.
- S. P. Meisburger, M. Warkentin, H. Chen, J. B. Hopkins, R. E. Gillilan, L. Pollack and R. E. Thorne, Biophys. J., 2013, 104, 227–236 CrossRef CAS PubMed.
- A. R. Round, D. Franke, S. Moritz, R. Huchler, M. Fritsche, D. Malthan, R. Klaering, D. I. Svergun and M. Roessle, J. Appl. Crystallogr., 2008, 41, 913–917 CrossRef CAS PubMed.
- G. L. Hura, A. L. Menon, M. Hammel, R. P. Rambo, F. L. Poole II, S. E. Tsutakawa, F. E. Jenney Jr, S. Classen, K. A. Frankel, R. C. Hopkins, S. Yang, J. W. Scott, B. D. Dillard, M. W. W. Adams and J. A. Tainer, Nat. Methods, 2009, 6, 606–612 CrossRef CAS PubMed.
- L. Li and R. F. Ismagilov, Annu. Rev. Biophys., 2010, 39, 139–158 CrossRef CAS PubMed.
- C. Hansen, Curr. Opin. Struct. Biol., 2003, 13, 538–544 CrossRef CAS PubMed.
- M. Maeki, H. Yamaguchi, M. Tokeshi and M. Miyazaki, Anal. Sci., 2016, 32, 3–9 CrossRef CAS PubMed.
- B. Zheng, C. J. Gerdts and R. F. Ismagilov, Curr. Opin. Struct. Biol., 2005, 15, 548–555 CrossRef CAS PubMed.
- B. Zheng, J. D. Tice, L. S. Roach and R. F. Ismagilov, Angew. Chem., Int. Ed., 2004, 43, 2508–2511 CrossRef CAS PubMed.
- K. Dhouib, C. K. Malek, W. Pfleging, B. Gauthier-Manuel, R. Duffait, G. Thuillier, R. Ferrigno, L. Jacquamet, J. Ohana, J.-L. Ferrer, A. Théobald-Dietrich, R. Giegé, B. Lorber and C. Sauter, Lab Chip, 2009, 9, 1412–1421 RSC.
- Š. Selimović, F. Gobeaux and S. Fraden, Lab Chip, 2010, 10, 1696–1699 RSC.
- M. Maeki, A. S. Pawate, K. Yamashita, M. Kawamoto, M. Tokeshi, P. J. A. Kenis and M. Miyazaki, Anal. Chem., 2015, 87, 4194–4200 CrossRef CAS PubMed.
- S. L. Perry, S. Guha, A. S. Pawate, A. Bhaskarla, V. Agarwal, S. K. Nair and P. J. A. Kenis, Lab Chip, 2013, 13, 3183 RSC.
- M. Heymann, A. Opthalage, J. L. Wierman, S. Akella, D. M. E. Szebenyi, S. M. Gruner and S. Fraden, IUCrJ, 2014, 1, 349–360 CrossRef CAS PubMed.
- S. L. Perry, S. Guha, A. S. Pawate, R. Henning, I. Kosheleva, V. Srajer, P. J. A. Kenis and Z. Ren, J. Appl. Crystallogr., 2014, 47, 1975–1982 CrossRef CAS PubMed.
- A. S. Pawate, V. Šrajer, J. Schieferstein, S. Guha, R. Henning, I. Kosheleva, M. Schmidt, Z. Ren, P. J. A. Kenis and S. L. Perry, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2015, 71, 823–830 CrossRef CAS PubMed.
- C. L. Hansen, E. Skordalakes, J. M. Berger and S. R. Quake, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16531–16536 CrossRef CAS PubMed.
- C. L. Hansen, M. O. A. Sommer and S. R. Quake, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14431–14436 CrossRef CAS PubMed.
- M. J. Anderson, C. L. Hansen and S. R. Quake, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 16746–16751 CrossRef CAS PubMed.
- M. J. Anderson, B. DeLaBarre, A. Raghunathan, B. O. Palsson, A. T. Brunger and S. R. Quake, Biochemistry, 2007, 46, 5722–5731 CrossRef CAS PubMed.
- B. T. C. Lau, C. A. Baitz, X. P. Dong and C. L. Hansen, J. Am. Chem. Soc., 2007, 129, 454–455 CrossRef CAS PubMed.
- J. Shim, G. Cristobal, D. R. Link, T. Thorsen and S. Fraden, Cryst. Growth Des., 2007, 7, 2192–2194 CAS.
- M. K. Yadav, C. J. Gerdts, R. Sanishvili, W. W. Smith, L. S. Roach, R. F. Ismagilov, P. Kuhn and R. C. Stevens, J. Appl. Crystallogr., 2005, 38, 900–905 CrossRef CAS PubMed.
- C. J. Gerdts, V. Tereshko, M. K. Yadav, I. Dementieva, F. Collart, A. Joachimiak, R. C. Stevens, P. Kuhn, A. Kossiakoff and R. F. Ismagilov, Angew. Chem., Int. Ed., 2006, 45, 8156–8160 CrossRef CAS PubMed.
- L. Li, D. Mustafi, Q. Fu, V. Tereshko, D. L. Chen, J. D. Tice and R. F. Ismagilov, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19243–19248 CrossRef CAS PubMed.
- W. Du, L. Li, K. P. Nichols and R. F. Ismagilov, Lab Chip, 2009, 9, 2286–2292 RSC.
- L. Li, W. Du and R. F. Ismagilov, J. Am. Chem. Soc., 2010, 132, 112–119 CrossRef CAS PubMed.
- C. Sauter, K. Dhouib and B. Lorber, Cryst. Growth Des., 2007, 7, 2247–2250 CAS.
- C. P. Steinert, J. Mueller-Diekmann, M. Weiss, M. Roessle, R. Zengerle and P. Koltay, in IEEE 20th International Conference on Micro Electro Mechanical Systems, MEMS, 2007, pp. 561–564 Search PubMed.
- J. D. Ng, P. J. Clark, R. C. Stevens and P. Kuhn, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2008, 64, 189–197 CrossRef CAS PubMed.
- O. Glatter and O. Kratky, Small Angle X-ray Scattering, Academic Press, 1982 Search PubMed.
- D. I. Svergun and M. H. J. Koch, Rep. Prog. Phys., 2003, 66, 1735 CrossRef CAS.
- S. S. Nielsen, K. N. Toft, D. Snakenborg, M. G. Jeppesen, J. K. Jacobsen, B. Vestergaard, J. P. Kutter and L. Arleth, J. Appl. Crystallogr., 2009, 42, 959–964 CrossRef CAS.
- F. Schwemmer, C. E. Blanchet, A. Spilotros, D. Kosse, S. Zehnle, H. D. T. Mertens, M. A. Graewert, M. Rössle, N. Paust, D. I. Svergun, F. von Stetten, R. Zengerle and D. Mark, Lab Chip, 2016, 16, 1161–1170 RSC.
- D. S. Khvostichenko, E. Kondrashkina, S. L. Perry, A. S. Pawate, K. Brister and P. J. A. Kenis, Analyst, 2013, 138, 5384–5395 RSC.
- M. Skou, S. Skou, T. G. Jensen, B. Vestergaard and R. E. Gillilan, J. Appl. Crystallogr., 2014, 47, 1355–1366 CrossRef CAS PubMed.
- S. V. Kathuria, A. Chan, R. Graceffa, R. Paul Nobrega, C. Robert Matthews, T. C. Irving, B. Perot and O. Bilsel, Biopolymers, 2013, 99, 888–896 CrossRef CAS PubMed.
- L. Pollack and S. Doniach, in Methods in Enzymology, Academic Press, 2009, vol. 469, pp. 253–268 Search PubMed.
- L. Pollack, M. W. Tate, A. C. Finnefrock, C. Kalidas, S. Trotter, N. C. Darnton, L. Lurio, R. H. Austin, C. A. Batt, S. M. Gruner and S. G. J. Mochrie, Phys. Rev. Lett., 2001, 86, 4962–4965 CrossRef CAS PubMed.
- D. J. Segel, A. Bachmann, J. Hofrichter, K. O. Hodgson, S. Doniach and T. Kiefhaber, J. Mol. Biol., 1999, 288, 489–499 CrossRef CAS PubMed.
- S. Akiyama, S. Akiyama, S. Takahashi, T. Kimura, K. Ishimori, I. Morishima, Y. Nishikawa and T. Fujisawa, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 1329–1334 CrossRef CAS PubMed.
- T. Uzawa, T. Uzawa, S. Akiyama, T. Kimura, S. Takahashi, K. Ishimori, I. Morishima and T. Fujisawa, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1171–1176 CrossRef CAS PubMed.
- T. Uzawa, T. Kimura, K. Ishimori, I. Morishima, T. Matsui, M. Ikeda-Saito, S. Takahashi, S. Akiyama and T. Fujisawa, J. Mol. Biol., 2006, 357, 997–1008 CrossRef CAS PubMed.
- M. Arai, E. Kondrashkina, C. Kayatekin, C. R. Matthews, M. Iwakura and O. Bilsel, J. Mol. Biol., 2007, 368, 219–229 CrossRef CAS PubMed.
- Y. Wu, E. Kondrashkina, C. Kayatekin, C. R. Matthews and O. Bilsel, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 13367–13372 CrossRef CAS PubMed.
- M. Arai, M. Iwakura, C. R. Matthews and O. Bilsel, J. Mol. Biol., 2011, 410, 329–342 CrossRef CAS PubMed.
- R. Graceffa, R. P. Nobrega, R. A. Barrea, S. V. Kathuria, S. Chakravarthy, O. Bilsel and T. C. Irving, J. Synchrotron Radiat., 2013, 20, 820–825 CrossRef CAS PubMed.
- R. Das, L. W. Kwok, I. S. Millett, Y. Bai, T. T. Mills, J. Jacob, G. S. Maskel, S. Seifert, S. G. J. Mochrie, P. Thiyagarajan, S. Doniach, L. Pollack and D. Herschlag, J. Mol. Biol., 2003, 332, 311–319 CrossRef CAS PubMed.
- L. W. Kwok, I. Shcherbakova, J. S. Lamb, H. Y. Park, K. Andresen, H. Smith, M. Brenowitz and L. Pollack, J. Mol. Biol., 2006, 355, 282–293 CrossRef CAS PubMed.
- J. Lamb, L. Kwok, X. Qiu, K. Andresen, H. Y. Park and L. Pollack, J. Appl. Crystallogr., 2008, 41, 1046–1052 CAS.
- J. C. Schlatterer, L. W. Kwok, J. S. Lamb, H. Y. Park, K. Andresen, M. Brenowitz and L. Pollack, J. Mol. Biol., 2008, 379, 859–870 CrossRef CAS PubMed.
- T. Pfohl, A. Otten, S. Köster, R. Dootz, B. Struth and H. M. Evans, Biomacromolecules, 2007, 8, 2167–2172 CrossRef CAS PubMed.
- H. M. Evans, R. Dootz, S. Köster, B. Struth and T. Pfohl, Bull. Pol. Acad. Sci.: Tech. Sci., 2007, 55, 217–227 CAS.
- S. Köster, H. M. Evans, J. Y. Wong and T. Pfohl, Biomacromolecules, 2008, 9, 199–207 CrossRef PubMed.
- P. Regenfuss, R. M. Clegg, M. J. Fulwyler, F. J. Barrantes and T. M. Jovin, Rev. Sci. Instrum., 1985, 56, 283 CrossRef CAS.
- P. Panine, S. Finet, T. M. Weiss and T. Narayanan, Adv. Colloid Interface Sci., 2006, 127, 9–18 CrossRef CAS PubMed.
- T. Fujisawa, K. Inoue, T. Oka, H. Iwamoto, T. Uruga, T. Kumasaka, Y. Inoko, N. Yagi, M. Yamamoto and T. Ueki, J. Appl. Crystallogr., 2000, 33, 797–800 CrossRef CAS.
- H. Amenitsch, M. Rappolt, M. Kriechbaum, H. Mio, P. Laggner and S. Bernstorff, J. Synchrotron Radiat., 1998, 5, 506–508 CrossRef CAS PubMed.
- S. Westenhoff, E. Malmerberg, D. Arnlund, L. Johansson, E. Nazarenko, M. Cammarata, J. Davidsson, V. Chaptal, J. Abramson, G. Katona, A. Menzel and R. Neutze, Nat. Methods, 2010, 7, 775–776 CrossRef CAS PubMed.
- M. Cammarata, M. Levantino, M. Wulff and A. Cupane, J. Mol. Biol., 2010, 400, 951–962 CrossRef CAS PubMed.
- M. Cammarata, M. Levantino, F. Schotte, P. A. Anfinrud, F. Ewald, J. Choi, A. Cupane, M. Wulff and H. Ihee, Nat. Methods, 2008, 5, 881–886 CrossRef CAS PubMed.
- M. Andersson, E. Malmerberg, S. Westenhoff, G. Katona, M. Cammarata, A. B. Wöhri, L. C. Johansson, F. Ewald, M. Eklund, M. Wulff, J. Davidsson and R. Neutze, Structure, 2009, 17, 1265–1275 CrossRef CAS PubMed.
- S. Ahn, K. H. Kim, Y. Kim, J. Kim and H. Ihee, J. Phys. Chem. B, 2009, 113, 13131–13133 CrossRef CAS PubMed.
- H. S. Cho, N. Dashdorj, F. Schotte, T. Graber, R. Henning and P. Anfinrud, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7281–7286 CrossRef CAS PubMed.
- P. L. Ramachandran, J. E. Lovett, P. J. Carl, M. Cammarata, J. H. Lee, Y. O. Jung, H. Ihee, C. R. Timmel and J. J. van Thor, J. Am. Chem. Soc., 2011, 133, 9395–9404 CrossRef CAS PubMed.
- E. Malmerberg, Z. Omran, J. S. Hub, X. Li, G. Katona, S. Westenhoff, L. C. Johansson, M. Andersson, M. Cammarata, M. Wulff, D. van der Spoel, J. Davidsson, A. Specht and R. Neutze, Biophys. J., 2011, 101, 1345–1353 CrossRef CAS PubMed.
- T. W. Kim, J. H. Lee, J. Choi, K. H. Kim, L. J. van Wilderen, L. Guerin, Y. Kim, Y. O. Jung, C. Yang, J. Kim, M. Wulff, J. J. van Thor and H. Ihee, J. Am. Chem. Soc., 2012, 134, 3145–3153 CrossRef CAS PubMed.
- H. Takala, A. Björling, O. Berntsson, H. Lehtivuori, S. Niebling, M. Hoernke, I. Kosheleva, R. Henning, A. Menzel, J. A. Ihalainen and S. Westenhoff, Nature, 2014, 509, 245–248 CrossRef CAS PubMed.
- S. V. Kathuria, L. Guo, R. Graceffa, R. Barrea, R. P. Nobrega, C. R. Matthews, T. C. Irving and O. Bilsel, Biopolymers, 2011, 95, 550–558 CrossRef CAS PubMed.
- J. P. Brody, P. Yager, R. E. Goldstein and R. H. Austin, Biophys. J., 1996, 71, 3430–3441 CrossRef CAS PubMed.
- H. Song and R. F. Ismagilov, J. Am. Chem. Soc., 2003, 125, 14613–14619 CrossRef CAS PubMed.
- R. Stehle, G. Goerigk, D. Wallacher, M. Ballauff and S. Seiffert, Lab Chip, 2013, 13, 1529 RSC.
- J. B. Knight, A. Vishwanath, J. P. Brody and R. H. Austin, Phys. Rev. Lett., 1998, 80, 3863–3866 CrossRef CAS.
- S. Köster, J. B. Leach, B. Struth, T. Pfohl and J. Y. Wong, Langmuir, 2007, 23, 357–359 CrossRef PubMed.
- P. Ander, L. Leung-Louie and F. Silvestri, Macromolecules, 1979, 12, 1204–1207 CrossRef CAS.
- B. Wunderlich, D. Nettels and B. Schuler, Lab Chip, 2013, 14, 219–228 RSC.
- H. Song, J. D. Tice and R. F. Ismagilov, Angew. Chem., Int. Ed., 2003, 42, 768–772 CrossRef CAS PubMed.
- M. E. Brennich and S. Köster, Microfluid. Nanofluid., 2014, 16, 39–45 CrossRef.
- D. C. Duffy, J. C. McDonald, O. J. A. Schueller and G. M. Whitesides, Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS PubMed.
- C. K. Chan, Y. Hu, S. Takahashi, D. L. Rousseau, W. A. Eaton and J. Hofrichter, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 1779–1784 CrossRef CAS.
- S. Takahashi, S.-R. Yeh, T. K. Das, C.-K. Chan, D. S. Gottfried and D. L. Rousseau, Nat. Struct. Biol., 1997, 4, 44–50 CrossRef CAS PubMed.
- O. Bilsel, C. Kayatekin, L. A. Wallace and C. R. Matthews, Rev. Sci. Instrum., 2005, 76, 14302 CrossRef.
- J. Wang, J. Wang, L. Feng and T. Lin, RSC Adv., 2015, 5, 104138–104144 RSC.
- B. Marmiroli, G. Grenci, F. Cacho-Nerin, B. Sartori, E. Ferrari, P. Laggner, L. Businaro and H. Amenitsch, Lab Chip, 2009, 9, 2063 RSC.
- D. Wang, U. Weierstall, L. Pollack and J. Spence, J. Synchrotron Radiat., 2014, 21, 1364–1366 CrossRef CAS PubMed.
- M. Trebbin, K. Krüger, D. DePonte, S. V. Roth, H. N. Chapman and S. Förster, Lab Chip, 2014, 14, 1733 RSC.
- A. Y. Lyubimov, T. D. Murray, A. Koehl, I. E. Araci, M. Uervirojnangkoorn, O. B. Zeldin, A. E. Cohen, S. M. Soltis, E. L. Baxter, A. S. Brewster, N. K. Sauter, A. T. Brunger and J. M. Berger, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2015, 71, 928–940 CAS.
- S. Oberti, D. Möller, S. Gutmann, A. Neild and J. Dual, J. Appl. Crystallogr., 2009, 42, 636–641 CAS.
- R. Neutze, R. Wouts, D. van der Spoel, E. Weckert and J. Hajdu, Nature, 2000, 406, 752–757 CrossRef CAS PubMed.
- M. S. Hunter and P. Fromme, Methods, 2011, 55, 387–404 CrossRef CAS PubMed.
- M. S. Hunter, D. P. DePonte, D. A. Shapiro, R. A. Kirian, X. Wang, D. Starodub, S. Marchesini, U. Weierstall, R. B. Doak, J. C. H. Spence and P. Fromme, Biophys. J., 2011, 100, 198–206 CrossRef CAS PubMed.
- H. N. Chapman, P. Fromme, A. Barty, T. A. White, R. A. Kirian, A. Aquila, M. S. Hunter, J. Schulz, D. P. DePonte, U. Weierstall, R. B. Doak, F. R. N. C. Maia, A. V. Martin, I. Schlichting, L. Lomb, N. Coppola, R. L. Shoeman, S. W. Epp, R. Hartmann, D. Rolles, A. Rudenko, L. Foucar, N. Kimmel, G. Weidenspointner, P. Holl, M. Liang, M. Barthelmess, C. Caleman, S. Boutet, M. J. Bogan, J. Krzywinski, C. Bostedt, S. Bajt, L. Gumprecht, B. Rudek, B. Erk, C. Schmidt, A. Hömke, C. Reich, D. Pietschner, L. Strüder, G. Hauser, H. Gorke, J. Ullrich, S. Herrmann, G. Schaller, F. Schopper, H. Soltau, K.-U. Kühnel, M. Messerschmidt, J. D. Bozek, S. P. Hau-Riege, M. Frank, C. Y. Hampton, R. G. Sierra, D. Starodub, G. J. Williams, J. Hajdu, N. Timneanu, M. M. Seibert, J. Andreasson, A. Rocker, O. Jönsson, M. Svenda, S. Stern, K. Nass, R. Andritschke, C.-D. Schröter, F. Krasniqi, M. Bott, K. E. Schmidt, X. Wang, I. Grotjohann, J. M. Holton, T. R. M. Barends, R. Neutze, S. Marchesini, R. Fromme, S. Schorb, D. Rupp, M. Adolph, T. Gorkhover, I. Andersson, H. Hirsemann, G. Potdevin, H. Graafsma, B. Nilsson and J. C. H. Spence, Nature, 2011, 470, 73–77 CrossRef CAS PubMed.
- L. Strüder, S. Epp, D. Rolles, R. Hartmann, P. Holl, G. Lutz, H. Soltau, R. Eckart, C. Reich, K. Heinzinger, C. Thamm, A. Rudenko, F. Krasniqi, K.-U. Kühnel, C. Bauer, C.-D. Schröter, R. Moshammer, S. Techert, D. Miessner, M. Porro, O. Hälker, N. Meidinger, N. Kimmel, R. Andritschke, F. Schopper, G. Weidenspointner, A. Ziegler, D. Pietschner, S. Herrmann, U. Pietsch, A. Walenta, W. Leitenberger, C. Bostedt, T. Möller, D. Rupp, M. Adolph, H. Graafsma, H. Hirsemann, K. Gärtner, R. Richter, L. Foucar, R. L. Shoeman, I. Schlichting and J. Ullrich, Nucl. Instrum. Methods Phys. Res., Sect. A, 2010, 614, 483–496 CrossRef.
- B. G. Abdallah, N. A. Zatsepin, S. Roy-Chowdhury, J. Coe, C. E. Conrad, K. Dörner, R. G. Sierra, H. P. Stevenson, F. Camacho-Alanis, T. D. Grant, G. Nelson, D. James, G. Calero, R. M. Wachter, J. C. H. Spence, U. Weierstall, P. Fromme and A. Ros, Struct. Dyn., 2015, 2, 41719 CrossRef PubMed.
- D. P. DePonte, U. Weierstall, K. Schmidt, J. Warner, D. Starodub, J. C. H. Spence and R. B. Doak, J. Phys. D: Appl. Phys., 2008, 41, 195505 CrossRef.
- K. Kunnus, I. Rajkovic, S. Schreck, W. Quevedo, S. Eckert, M. Beye, E. Suljoti, C. Weniger, C. Kalus, S. Grübel, M. Scholz, D. Nordlund, W. Zhang, R. W. Hartsock, K. J. Gaffney, W. F. Schlotter, J. J. Turner, B. Kennedy, F. Hennies, S. Techert, P. Wernet and A. Föhlisch, Rev. Sci. Instrum., 2012, 83, 123109 CrossRef PubMed.
- A. Charvat, E. Lugovoj, M. Faubel and B. Abel, Rev. Sci. Instrum., 2004, 75, 1209 CrossRef CAS.
- M. Faubel, B. Steiner and J. P. Toennies, J. Chem. Phys., 1997, 106, 9013 CrossRef CAS.
- M. Faubel, K. R. Siefermann, Y. Liu and B. Abel, Acc. Chem. Res., 2012, 45, 120–130 CrossRef CAS PubMed.
- J. Kern, R. Alonso-Mori, R. Tran, J. Hattne, R. J. Gildea, N. Echols, C. Glockner, J. Hellmich, H. Laksmono, R. G. Sierra, B. Lassalle-Kaiser, S. Koroidov, A. Lampe, G. Han, S. Gul, D. DiFiore, D. Milathianaki, A. R. Fry, A. Miahnahri, D. W. Schafer, M. Messerschmidt, M. M. Seibert, J. E. Koglin, D. Sokaras, T.-C. Weng, J. Sellberg, M. J. Latimer, R. W. Grosse-Kunstleve, P. H. Zwart, W. E. White, P. Glatzel, P. D. Adams, M. J. Bogan, G. J. Williams, S. Boutet, J. Messinger, A. Zouni, N. K. Sauter, V. K. Yachandra, U. Bergmann and J. Yano, Science, 2013, 340, 491–495 CrossRef CAS PubMed.
- R. G. Sierra, H. Laksmono, J. Kern, R. Tran, J. Hattne, R. Alonso-Mori, B. Lassalle-Kaiser, C. Glöckner, J. Hellmich, D. W. Schafer, N. Echols, R. J. Gildea, R. W. Grosse-Kunstleve, J. Sellberg, T. A. McQueen, A. R. Fry, M. M. Messerschmidt, A. Miahnahri, M. M. Seibert, C. Y. Hampton, D. Starodub, N. D. Loh, D. Sokaras, T.-C. Weng, P. H. Zwart, P. Glatzel, D. Milathianaki, W. E. White, P. D. Adams, G. J. Williams, S. Boutet, A. Zouni, J. Messinger, N. K. Sauter, U. Bergmann, J. Yano, V. K. Yachandra and M. J. Bogan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 1584–1587 CAS.
- B. G. Abdallah, N. A. Zatsepin, S. Roy-Chowdhury, J. Coe, C. E. Conrad, K. Dörner, R. G. Sierra, H. P. Stevenson, F. Camacho-Alanis, T. D. Grant, G. Nelson, D. James, G. Calero, R. M. Wachter, J. C. H. Spence, U. Weierstall, P. Fromme and A. Ros, Struct. Dyn., 2015, 2, 41719 CrossRef PubMed.
- U. Weierstall, J. C. H. Spence and R. B. Doak, Rev. Sci. Instrum., 2012, 83, 35108 CrossRef CAS PubMed.
- L. Redecke, K. Nass, D. P. DePonte, T. A. White, D. Rehders, A. Barty, F. Stellato, M. Liang, T. R. M. Barends, S. Boutet, G. J. Williams, M. Messerschmidt, M. M. Seibert, A. Aquila, D. Arnlund, S. Bajt, T. Barth, M. J. Bogan, C. Caleman, T.-C. Chao, R. B. Doak, H. Fleckenstein, M. Frank, R. Fromme, L. Galli, I. Grotjohann, M. S. Hunter, L. C. Johansson, S. Kassemeyer, G. Katona, R. A. Kirian, R. Koopmann, C. Kupitz, L. Lomb, A. V. Martin, S. Mogk, R. Neutze, R. L. Shoeman, J. Steinbrener, N. Timneanu, D. Wang, U. Weierstall, N. A. Zatsepin, J. C. H. Spence, P. Fromme, I. Schlichting, M. Duszenko, C. Betzel and H. N. Chapman, Science, 2013, 339, 227–230 CrossRef CAS PubMed.
- H. P. Martin, N. J. Brooks, J. M. Seddon, P. F. Luckham, N. J. Terrill, A. J. Kowalski and J. T. Cabral, Soft Matter, 2016, 12, 1750–1758 RSC.
- H. P. Martin, N. J. Brooks, J. M. Seddon, N. J. Terrill, P. F. Luckham, A. J. Kowalski and J. T. Cabral, J. Phys.: Conf. Ser., 2010, 247, 12050 CrossRef.
- M. Trebbin, D. Steinhauser, J. Perlich, A. Buffet, S. V. Roth, W. Zimmermann, J. Thiele and S. Forster, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6706–6711 CrossRef CAS PubMed.
- N. F. Bouxsein, L. S. Hirst, Y. Li, C. R. Safinya, Z. Abu Samah, N. C. MacDonald and R. Pynn, Appl. Phys. Lett., 2004, 85, 5775 CrossRef CAS.
- R. Dootz, H. Evans, S. Köster and T. Pfohl, Small, 2007, 3, 96–100 CrossRef CAS PubMed.
- K. M. O. Håkansson, RSC Adv., 2015, 5, 18601–18608 RSC.
- B. F. B. Silva, M. Zepeda-Rosales, N. Venkateswaran, B. J. Fletcher, L. G. Carter, T. Matsui, T. M. Weiss, J. Han, Y. Li, U. Olsson and C. R. Safinya, Langmuir, 2015, 31, 4361–4371 CrossRef CAS PubMed.
- T. Saito, R. Kuramae, J. Wohlert, L. A. Berglund and A. Isogai, Biomacromolecules, 2013, 14, 248–253 CrossRef CAS PubMed.
- I. Sakurada, Y. Nukushina and T. Ito, J. Polym. Sci., 1962, 57, 651–660 CrossRef CAS.
- A. B. Fall, S. B. Lindström, O. Sundman, L. Ödberg and L. Wågberg, Langmuir, 2011, 27, 11332–11338 CrossRef CAS PubMed.
- A. B. Fall, S. B. Lindström, J. Sprakel and L. Wågberg, Soft Matter, 2013, 9, 1852–1863 RSC.
- A. S. Poulos, M. Nania, P. Lapham, R. M. Miller, A. J. Smith, H. Tantawy, J. Caragay, J. Gummel, O. Ces, E. S. J. Robles and J. T. Cabral, Langmuir, 2016, 32, 5852–5861 CrossRef CAS PubMed.
- M. Priebe, S. Kalbfleisch, M. Tolkiehn, S. Köster, B. Abel, R. J. Davies and T. Salditt, New J. Phys., 2010, 12, 43056 CrossRef.
- the_future_of_soft_matter_sans_2012.pdf. Available at: https://europeanspallationsource.se/sites/default/files/the_future_of_soft_matter_sans_2012.pdf. (Accessed: 9th July 2016).
- M. A. Calabrese and N. J. Wagner, New insights from Rheo-SANS, Royal Society of Chemistry, 2016 Search PubMed.
- M. Calabrese and N. Wagner, in Wormlike micelles: systems, characterization and applications, Royal Society of Chemistry, 2016 Search PubMed.
- C. E. Blanchet, A. Spilotros, F. Schwemmer, M. A. Graewert, A. Kikhney, C. M. Jeffries, D. Franke, D. Mark, R. Zengerle, F. Cipriani, S. Fiedler, M. Roessle and D. I. Svergun, J. Appl. Crystallogr., 2015, 48, 431–443 CrossRef CAS PubMed.
- Hampton Research, Available at: https://www.hamptonresearch.com/menus.aspx?id=2&cid=1http://www.sigmaaldrich.com/life-science/molecular-biology/molecular-biology-products.html?TablePage=15447365. (Accessed: 12th July 2016).
- BART – the new robotic sample changer for MX beamlines at Diamond – Diamond Light Source, Available at: http://www.diamond.ac.uk/Home/Corporate-Literature/Annual-Review/Review2015/Villages/Macromolecular-Crystallography-Village/Macromolecular-Crystallography-Village-Developments/BART---the-new-robotic-sample-changer-for-MX-beamlines-at-Diamond.html. (Accessed: 12th July 2016) Search PubMed.
- MASSIF-1: Rise of the robots transforms MX, Available at: http://www.esrf.eu/home/news/general/content-news/general/massif-1-rise-of-the-robots-transforms-mx.html. (Accessed: 12th July 2016) Search PubMed.
- J. Melin and S. R. Quake, Annu. Rev. Biophys. Biomol. Struct., 2007, 36, 213–231 CrossRef CAS PubMed.
- Microfluidics Market worth $7.5 Billion by 2020, Available at: http://www.marketsandmarkets.com/PressReleases/microfluidics.asp. (Accessed: 8th July 2016) Search PubMed.
Footnotes |
| † Contributed equally. |
| ‡ Present address: National Institute of Standards and Technology, 100 Bureau Dr, Gaithersburg, MD 20899, USA. |
| This journal is © The Royal Society of Chemistry 2016 |