Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sample introduction interface for on-chip nucleic acid-based analysis of Helicobacter pylori from stool samples†
O.
Mosley
a,
L.
Melling
a,
M. D.
Tarn
b,
C.
Kemp
b,
M. M. N.
Esfahani
c,
N.
Pamme
*b and
K. J.
Shaw
 *a
*a
aFaculty of Science and Engineering, Manchester Metropolitan University, Chester Street, Manchester, M1 5GD, UK. E-mail: k.shaw@mmu.ac.uk
bDepartment of Chemistry, University of Hull, Cottingham Road, Hull, HU6 7RX, UK. E-mail: n.pamme@hull.ac.uk
cSchool of Engineering, University of Hull, Cottingham Road, Hull, HU6 7RX, UK
First published on 10th May 2016
Abstract
Despite recent advances in microfluidic-based integrated diagnostic systems, the sample introduction interface, especially with regards to large volume samples, has often been neglected. We present a sample introduction interface that allows direct on-chip processing of crude stool samples for the detection of Helicobacter pylori (H. pylori). The principle of IFAST (immiscible filtration assisted by surface tension) was adapted to include a large volume sample chamber with a septum-based interface for stool sample introduction. Solid chaotropic salt and dry superparamagnetic particles (PMPs) could be stored on-chip and reconstituted upon sample addition, simplifying the process of release of DNA from H. pylori cells and its binding to the PMPs. Finally, the PMPs were pulled via a magnet through a washing chamber containing an immiscible oil solution and into an elution chamber where the DNA was released into aqueous media for subsequent analysis. The entire process required only 7 min while enabling a 40-fold reduction in working volume from crude biological samples. The combination of a real-world interface and rapid DNA extraction offers the potential for the methodology to be used in point-of-care (POC) devices.
Introduction
It is estimated that approximately two-thirds of the world's population harbours Helicobacter pylori (H. pylori), a Gram-negative microorganism that colonises the gastric mucosa in the human stomach.1H. pylori has been shown to have a significant role in the pathogenesis of chronic gastritis, peptic ulcers and more importantly gastric cancer.2 Different strains have varying abilities to cause inflammatory changes but the phenotype of H. pylori that expresses cytotoxin-associated protein (CagA) causes a higher degree of acute inflammation and is three times more likely to lead to gastric carcinogenesis.3 A wide variety of methods are available to detect H. pylori including immunochromatogenic assays, histology and culture. However, only polymerase chain reaction (PCR)-based assays have the ability to identify particular strains including those which are CagA+.4 Such methods which incorporate genotyping are therefore advantageous in identifying patients who are at higher risk of complications resulting from H. pylori infection.5Rapid and efficient diagnosis is thus important in eradicating the infection and reducing the risk of gastric cancer development. To this end, microfluidic and lab-on-a-chip (LOC) devices offer considerable advantages for use in point-of-care (POC) diagnostics6,7 due to increased analysis speed and sensitivity, reduced reagent usage and the possibility for full automation. Despite this great potential, the development of real-world sample introduction interfaces remains challenging. Currently, the majority of published integrated devices either use simulated samples or require excessive off-chip or on-chip sample pre-treatment to achieve the desired specimen volume reduction and target concentrations. Simulated samples include the use of a few microlitres of highly concentrated bacterial cell cultures8 or high virus titre matrices9 that rarely represent target concentrations and purities found in clinical samples. Low target concentrations, such as those present in urine and stool samples, therefore require the use of larger sample volumes to assure sensitivity of the assay. In particular, the analysis of stool samples results in the need for considerable off-chip sample pre-treatment, such as centrifugation and filtration steps10 and chemical lysis11 prior to addition to a microfluidic device, all of which can be somewhat time-consuming. Furthermore, research in this area has focussed on the detection of infectious agents, such as Clostridium difficile, which cause diarrhoea resulting in liquid stool samples that are easier to introduce into microfluidic systems for analysis.10,12,13 Thus, the development of a real-world interface for the direct manipulation of crude biological samples has largely been ignored, and represents a barrier between the research and clinical environments.
Superparamagnetic particles (PMPs) have become very popular as solid supports for nucleic acid purification in the detection of infectious diseases.14 With a suitable surface functionality, such as silica or chitosan, the particles will capture DNA in a sample and their magnetic properties allow them to be held in place by an external magnet while the sample is removed and washing steps are applied. However, these methods typically require a great deal of time and manual handling. As a consequence, magnetic particle-based procedures have been incorporated into microfluidic devices with great success,15–17 thanks to the reduction in diffusion distances, procedural times, and the ease of particle manipulation. Magnetic particle-based procedures integrated with microfluidic devices have proven particularly effective for the purification of nucleic acids prior to their amplification and analysis,18 but many techniques involve complex chip setups19,20 and laborious multi-step procedures involving the trapping of magnetic particles while solutions are pumped over them.20
A simple method of achieving DNA or RNA extraction involves the introduction of PMPs into a contained sample volume, before moving the particles via a magnet through multiple washing solutions, leaving behind any unwanted and unbound material. Early examples of this mechanism employed the use of droplets on open, superhydrophobic microfluidic platforms, in which magnetic particles would be moved between stationary sample and washing droplets separated via an immiscible phase such as oil21–24 or air.25–27 However, these techniques often require either mechanical25,26 or electromagnetic21,23,24 actuation, adding complexity to the system in terms of both fabrication and operation.
A recent development from the group of Beebe is that of immiscible filtration assisted by surface tension (IFAST), in which rather than having solutions contained in droplets they are instead added to interconnected microwells separated by small “gated” regions.28 The chambers are filled with alternating aqueous and oil phases to form “virtual walls” between each chamber, controlled by the surface tension, but allowing magnetic particles to be pulled through these walls and thus through each chamber in one smooth yet fast motion via a handheld magnet. This allows simple and rapid DNA extraction to be performed with minimal setup and materials, and by the “unskilled” end-user. The standard IFAST design features three chambers consisting of (i) aqueous sample solution to which PMPs are added, (ii) an immiscible oil phase for washing of particles, and (iii) elution buffer that can be collected for off-chip nucleic acid amplification and analysis. So far this method has been applied to the purification of RNA28–31 and DNA,31,32 as well as for cell isolation33–35 and immunoassays.36 Further developments have included automation of the devices,37,38 their fabrication from wax,30,38 and variants such as vertical IFAST (VerIFAST),34,35 and SNARE (selective nucleic acid removal via exclusion).31 Similar techniques have also been developed by other research groups, in which different immiscible phases have been employed including liquid wax,39 paraffin wax,40 and air.41–44 Furthermore, miscible phases have recently been employed for particle washing by using phaseguides to pattern interfaces between adjacent aqueous solutions.45,46
Here, we have exploited and considerably modified the IFAST principle to develop a sample introduction interface that enables direct processing of stool samples. Stool samples are particularly challenging for diagnostic analysis through molecular biology techniques as they exhibit high variability in terms of consistency of samples, the presence of PCR inhibitors and low target analyte concentrations, hence the requirement of the sample pre-treatment and pre-concentration steps described earlier. IFAST is usually conducted in chambers of 10 μL volumes, while in our high volume IFAST system the issue of low biomarker concentration is negated by the use of a large sample chamber, while the IFAST process itself allows rapid DNA purification, concentration, and elution, in a single device (Fig. 1). Initial experiments were performed using E. coli as a model Gram-negative pathogenic target before moving onto analysis of H. pylori (also Gram-negative) from clinical stool samples.
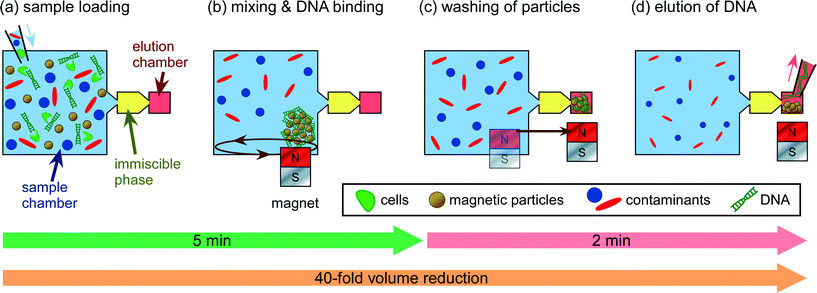 | ||
| Fig. 1 Schematic of the DNA extraction process, showing (a) sample loading and cell lysis, (b) mixing of PMPs with the sample for DNA binding, (c) transfer of PMPs through the immiscible phase for washing, and (d) elution of DNA from the PMPs followed by collection for off-chip analysis. The design was later amended to include two extra downstream chambers for additional washing (see Fig. 3 for further details). Schematics are not to scale. | ||
The novelty of the reported approach lies in (1) the design of the sample chamber which enables a 40-fold reduction in working volume, (2) the choice of detergent-free solid cell lysis and DNA binding agent that is reconstituted by the sample itself, (3) a unique PDMS/optical adhesive bonding approach which facilitated the formation of a stable but immiscible barrier, and (4) the real-world interface created using a septum-based sample introduction design.
Experimental
Chemicals, apparatus and samples
All aqueous solutions were prepared in filtered, purified water (18.2 MΩ cm at 25 °C). For IFAST devices, a Sylgard® 184 Silicone Elastomer Kit was purchased from Dow Corning, optical adhesive film (100 μm thickness, Adhesive PCR Film Seal) from Thermo Scientific, UK, and microscope cover slips (24 × 24 × 0.017 mm3) from Scientific Laboratory Supplies, UK. Guanidine hydrochloride (GuHCl) and MagneSil paramagnetic particles (2–14 μm diameter, ∼27 m2 g−1 surface area) were purchased from Promega, UK.47 Biomix™ for DNA amplification and the DNA size ladder, Hyperladder™ V, were obtained from Bioline Reagents Ltd, UK. Primers for UreC (H. pylori specific, forward: 5′ AAGCTTTTAGGGGTGTTAGGGGTTT 3′, reverse: 5′ AAGCTTACTTTCTAACACTAACGC 3′) and CagA (CagA strain specific, forward: 5′ AATACACCAACGCCTCCAAG 3′, reverse: 5′ TTGTTGCCGCTTTTGCTCTC 3′) were custom-made by Life Technologies, UK.48 Agarose and loading dye for electrophoresis were purchased from Sigma-Aldrich, UK. DNA quantification was performed on a Multiskan™ GO Microplate Spectrophotometer (Thermo Scientific, UK). DNA amplification was carried out using a Q-cycler 96 thermal cycler (Hain Lifesciences Ltd, UK). Escherichia coli (E. coli) cells (NCTC 9001) were obtained from the National Collection of Type Cultures, UK. Clinical stool samples were obtained from NHS Chesterfield Laboratories; samples were fully anonymised and were selected on the basis that they had previously been tested for H. pylori during routine clinical testing using a Proflow™ H. pylori test (ProLab, UK).Chip fabrication and setup
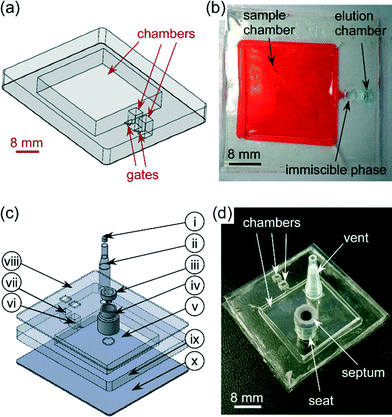
The integrated device consisted of either three (Fig. 2) or five (Fig. 3) chambers arranged in a linear configuration: a single sample chamber (26 × 26 × 4 mm3 [length × width × height]), and washing and elution chambers (each 3 × 3 × 4 mm3). These were interconnected by gated regions consisting of trapezoidal microfluidic conduits that narrowed from 3 mm to 500 μm in width, with a height of 500 μm. The devices themselves were fabricated in a novel manner. A mould was designed in SolidWorks 2011 (Dassault Systèmes SolidWorks Corp., France) and fabricated out of aluminium on a CNC milling machine (M7, Datron AG, Germany). Due to limitations in spatial resolution with the CNC machine and available tools, the mould was prepared featuring the final channel design. This was then used to fabricate a negative relief of the design in poly(methyl methacrylate) (PMMA) using an injection moulding machine (Babyplast 6/10P, Rambaldi+Co, Italy). The final device was prepared by pouring PDMS (consisting of a mixture of prepolymer and curing agent in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and degassed for 1 h) onto the PMMA mould and curing at 80 °C for 30 min, before peeling the PDMS substrate off. The process is shown in more detail in the ESI† (see Fig. S-1). The device was sealed with a double layer of optical adhesive film underneath the PDMS substrate to provide support and a microscope cover slip was adhered to the bottom of the sample chamber to overcome initial sample loading difficulties caused by the hydrophobic properties of the optical adhesive film. A single layer of optical adhesive film, featuring holes to allow access to the wash and elution chambers, was used to seal the top of the device.
1 ratio and degassed for 1 h) onto the PMMA mould and curing at 80 °C for 30 min, before peeling the PDMS substrate off. The process is shown in more detail in the ESI† (see Fig. S-1). The device was sealed with a double layer of optical adhesive film underneath the PDMS substrate to provide support and a microscope cover slip was adhered to the bottom of the sample chamber to overcome initial sample loading difficulties caused by the hydrophobic properties of the optical adhesive film. A single layer of optical adhesive film, featuring holes to allow access to the wash and elution chambers, was used to seal the top of the device.
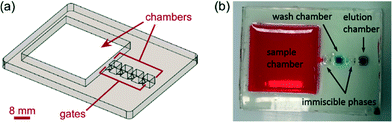 | ||
| Fig. 3 (a) Schematic of the 5-chamber DNA purification device. (b) Photograph of the PDMS chip filled with inks and oil. | ||
A real-world interface was constructed for sample introduction via the holes in the optical film lid above the sample chamber, consisting of a septum for sample introduction and an air vent (Fig. 2c and d). The septum was prepared by cutting a standard capillary gel electrophoresis septum to size and seating it in the top of a cut-to-size pipette tip (100 μL) that was attached to the optical film lid via double-sided tape. The vent was fabricated from a filter pipette tip (10 μL) that was also attached to the lid via double-sided tape. The vent allowed air to be expelled when the sample chamber was filled. The assembly and interfacing of the IFAST the device was the same for both the 3-chamber and 5-chamber chip designs.
DNA extraction by IFAST
The sample introduction setup was prepared as follows. Prior to attachment of the optical film lid, 1 μL silica-coated PMPs (80 mg mL−1) suspended in storage buffer were added to the sample chamber. The PMPs were held in place by a handheld neodymium–iron–boron (NdFeB) magnet (12 × 3 × 3 mm3, Magnet Sales, UK) while the buffer was removed via pipette. Solid GuHCl, a chaotropic salt that facilitates cell lysis, protein denaturation and the binding of DNA to silica surfaces, was then also added to the sample chamber, after which the optical film lid and interface was sealed onto the PDMS device. With the chip prepared, 400 μL of sample (either bacterial broth or liquid stool) was added to the chamber through the septum via a pipette. Bacterial broth samples were made up of E. coli cells cultured overnight in nutrient broth at 37 °C and 150 rpm. Stool samples were added to molecular biology grade water to a total volume of 400 μL. For the 3-chamber chip design; the final elution chamber was then filled with purified water (10 μL), followed by the immiscible phase being added to its chamber (10 μL). For the 5-chamber chip design the final elution chamber was filled with purified water (10 μL), followed by the central washing chamber being filled with 5 M GuHCl solution (10 μL) and then the immiscible phase was added to its two chambers (10 μL each).As described earlier, a number of immiscible phases have been used as the washing solution in IFAST, including liquid wax,39 paraffin wax,40 olive oil,28 and air.41–44 Here, mineral oil was chosen as the immiscible phase due to its purity and compatibility with downstream biochemical applications. Upon addition of the sample to the chamber, a handheld magnet was used to mix the PMPs with the sample for 5 min, reconstituting the GuHCl to a concentration of 5 M and allowing binding of the DNA to the particles. Finally, the PMPs were quickly transferred across the immiscible barrier by the handheld magnet and into purified water, where the DNA was allowed to elute from the particles for 2 min (Fig. 1). Unwanted components of the stool sample matrix were left behind in the sample chamber. The use of oil phases in both the 3- and 5-chamber chip designs acted to remove potential PCR inhibitors, such as complex polysaccharides, from the stool samples. In addition, the 5-chamber chip contained an additional wash step (5 M GuHCl) to ensure the DNA remained bound to the PMPs and could be separated from any remaining inhibitors, as previously described with other types of biological samples.28
Several parameters were tested using the described DNA extraction process, including: (i) lysis efficiency of Gram-negative bacterial cells (E. coli) using powdered GuHCl stored in the sample chamber (‘Evaluation of stored reagents parameters’ section), (ii) DNA extraction efficiency from cultured bacterial cells (E. coli) (‘DNA extraction efficiency’ section), (iii) evaluation of purity of DNA extracted from real clinical stool samples (‘Evaluation of clinical stool samples’ section), and (iv) amplification of H. pylori targets following IFAST-based extraction (‘Evaluation of clinical stool samples’ section).
Analysis of extracted DNA
Following the IFAST extraction process described above, the PMPs were held in the elution chamber via a magnet, and the elution solution removed for analysis. DNA concentration and purity were assessed by measuring the absorbance of 2 μL of elution solution, at 260 and 280 nm, using a Multiskan™ GO Microplate Spectrophotometer.DNA amplification was achieved using a 25 μL polymerase chain reaction (PCR) mixture prepared from the following: 5 μL of purified template DNA solution (taken directly from the IFAST device), 0.4 μM each primer, 1× Biomix™ containing 0.2 mM each dNTPs, reaction buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 2.5 mM MgCl2, and 0.01% (w/v) gelatin), and 2.5 U of Taq DNA polymerase. Samples were run on a Q-cycler 96 thermal cycler under the following conditions: an initial denaturation at 94 °C for 10 minutes followed by 40 cycles of 94 °C for 2 minutes, 55 °C for 2 minutes and 72 °C for 2 minutes, with a final extension of 72 °C for 10 minutes.
Following amplification, PCR products and a DNA size ladder (Hyperladder V) were run on a 2% (w/v) agarose gel until adequate separation had been achieved and visualised using a UV transilluminator.
Results and discussion
Device operation
IFAST and its comparative technologies have proven very successful for nucleic acid purification,28–32 but certain aspects of the method require consideration. IFAST relies on the formation of a stable interface between the aqueous and oil phases, yet it must also allow the particles to penetrate through the washing solution. This is not trivial and requires careful modulation of the interfacial energy at the sample/oil/elution interfaces. Conventional lysis and DNA binding buffers contain detergents that would lower the interfacial energy and can lead to the formation of an unstable interface. We therefore opted for the use of 5 M GuHCl as the binding agent. In accordance with the IFAST extraction methodology described in the experimental section ‘DNA extraction by IFAST’, 400 μL of either E. coli cell suspension or liquid stool sample was introduced into the sample chamber, followed by addition of elution, wash and immiscible phases into the relevant chambers. Addition of the biological sample allowed resuspension of the solid GuHCl and PMPs that were stored in the device. Manipulation of the PMPs via a handheld magnet enabled reconstitution of the GuHCl and allowed binding of the DNA to the PMPs, a process that took 5 min. The particles were then transferred from the sample chamber to the elution chamber, through the immiscible phase(s), by moving the magnet below each of the chambers in turn. Once in the final chamber, the DNA was allowed to elute from the PMPs for 2 min. Transfer of the PMPs enabled a 40-fold reduction in sample volume for analysis, from 400 μL of E. coli in nutrient broth or stool sample to 10 μL of elution buffer, in only 7 min.Evaluation of stored reagent parameters
The use of GuHCl as the lysis and binding reagent resulted in the following benefits: (1) the interfacial energy between the immiscible phases was increased due to the lack of added detergent, and (2) the increase in sample volume normally observed due to the addition of lysis and binding buffer is significantly reduced thanks to the fact that the GuHCl is reconstituted in the biological sample itself.49 Furthermore, it has been shown that GuHCl can be used to lyse cells and release DNA on-chip,49 and so this allows lysis of the bacterial cells to release and bind the DNA to the PMPs in a single step. A comparison was made between off-chip chemical lysis and on-chip chemical lysis, using direct addition of GuHCl powder to achieve a final concentration of 5 M in the sample solution (cultured E. coli cells as a model Gram negative specimen). Conventional off-chip thermal lysis was used as a control for comparison, whereby the biological sample was placed in a heat block for 5 min at 100 °C. Lysis was measured in terms of the total amount of DNA released from the cells once they had undergone treatment. Efficiency of the chemical lysis treatments is presented as a comparison to conventional thermal lysis. No significant difference was observed between efficiency of the on-chip and off-chip chemical lysis protocols (p = 0.928, ANOVA) but both protocols were more effective when dealing with a smaller number of target cells (p = 0.004, ANOVA) (Fig. 4). The lysis efficiency was also evaluated for a model Gram-positive specimen (Staphylococcus aureus) in order to evaluate whether the proposed system would be suitable for all types of bacteria. Successful lysis of Gram-positive bacteria was also demonstrated using the on-chip lysis method.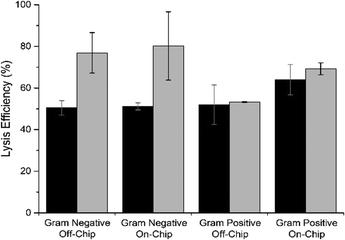 | ||
| Fig. 4 Lysis efficiency of the stored 5 M GuHCl reagent both on- and off-chip (n = 6) for 2.54 × 106 (black) and 2.54 × 104 cells (grey). | ||
The amount of particles that can be used in the IFAST device was restricted by the geometry of the microfluidic conduits. Previous studies demonstrated that 0.24 mg of MagneSil PMPs could be transported across the immiscible phase without particle loss or blocking of the device, and therefore this amount was chosen to achieve maximum DNA binding and transport.50
DNA extraction efficiency
The DNA extraction efficiency was measured by adding known amounts of DNA (using cultured E. coli cells as a model Gram-negative specimen) into the sample chamber on the 3-chamber IFAST device and comparing this to the amount of DNA recovered from the elution chamber. DNA extraction efficiency was shown to be greater when smaller amounts of DNA were present in the system, which is ideal for dealing with low levels of infection in clinical specimens (Fig. 5). The amount of PMPs used per reaction had the capacity to bind 320 ng of DNA, therefore the concentrations tested were below the saturation point of the particles.51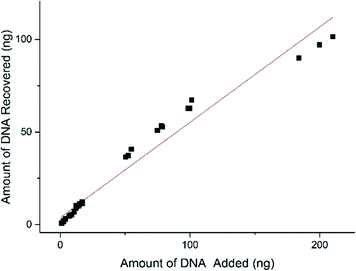 | ||
| Fig. 5 DNA extraction efficiency showing a strong linear correlation (Pearson's R, R2 = 0.98259) between the amount of DNA added to the system (ng) and the amount of DNA recovered from the system (ng). | ||
Negative controls were also included, in which samples containing no DNA were added to the IFAST device and underwent the DNA extraction process. No DNA was detected in the eluent of these samples (n = 3).
Evaluation of clinical stool samples
Known H. pylori positive stool samples from a local clinic were analysed for DNA concentration and purity after processing on the 3-chamber IFAST device. As expected, the DNA concentrations obtained varied from patient to patient due to varying levels of infection (Table 1). Some patients may have had a severe infection, increasing the DNA concentration, while others may have had a persisting infection after finishing antibiotics, thereby exhibiting a low DNA concentration. However, DNA purity values were consistently poor with a range of values between 1.1 and 1.3 (a value of between 1.8 and 2.0 indicates a ‘pure’ sample). Stool samples which had tested negative for H. pylori using the Proflow™ H. pylori test (ProLab, UK) were also analysed as control samples.| Sample | DNA concentration (ng μl−1) | Purity (260 nm/280 nm) |
|---|---|---|
| 1 | 31.8 | 1.3 |
| 2 | 63.5 | 1.1 |
| 3 | 19.1 | 1.3 |
| 4 | 295.0 | 1.1 |
| 5 | 93.8 | 1.2 |
| 6 | 154.0 | 1.3 |
| Average | 109.5 | 1.2 |
Following PCR, weak or no PCR products were observed, indicating that the samples were not sufficiently pure and free of inhibitors for successful amplification to be achieved. Therefore the chip design was modified to include an additional wash step, yielding the 5-chamber design shown in Fig. 3. An improvement was seen in the purity of the extracted samples using the modified chip design, with purity values up to 2.0 obtained (Table 2). In order to account for the wide variety in composition of stools, all subsequent samples analysed were assigned a value based on the Bristol Stool Chart which classifies samples on a 7-point scale from separate hard lumps (type 1) to entirely liquid (type 7), with an ideal stool being smooth and sausage-like (type 4).52 Comparison of the purity of the samples to their original appearance (based on values assigned from the Bristol Stool Chart) showed a strong correlation (R = 0.96 and P < 0.001 Pearson's R), with more liquid samples (e.g. types 6 and 7) producing higher extracted DNA purities. However, there was no correlation between the appearance of the stool and the amount of DNA that was obtained. In addition, successful amplification of the UreC target gene (PCR product size = 274 bp) was achieved on those samples which were extracted using the IFAST device (Fig. 6). As expected, no PCR products were observed for those stool samples which had previously tested negative for H. pylori. None of the clinical samples tested proved positive for CagA (expected PCR product size = 400 bp).
 | ||
| Fig. 6 Gel electrophoresis image showing: (L) DNA size ladder; (1–8) amplified faecal samples extracted using the IFAST device; (N) negative control. | ||
Conclusions
Microfluidic devices are of great potential for use in POC settings as they can be fully automated, allowing for minimum user intervention and reagent use, as well as reduced analysis times. However, the integration of real-world interfaces for sample introduction has often been neglected, especially with regards to large volumes of crude samples that contain only low concentrations of analyte. The aim of our work was therefore to develop a sample introduction interface which addresses these issues. The presented device bridges the gap between microfluidics and the requirements of real sample processing by enabling DNA purification and 40-fold pre-concentration within 7 min from crude stool samples.IFAST has been previously demonstrated for rapid nucleic acid and cell purification purposes, and here we have significantly adapted the procedure to enable analysis of stool samples via a real-world interface. Firstly, our design includes a large sample reservoir that accommodates 400 μL of crude sample without the need for sample pre-treatment. For clinical samples, the target analyte concentration may be very low and therefore the larger the sample volume that can be accommodated the more likely the chances of successful extraction of the target of interest. Secondly, on-chip cell lysis and DNA binding to the solid phase supports (PMPs) was achieved on-chip by preloading the chamber with the solid chaotrope49 and dried PMPs. Furthermore, sample loading was facilitated by an incorporated septum (Fig. 2b and c), keeping the sample sealed within the chamber. In a recent publication, cell lysis and DNA binding was performed on an IFAST device and proven to be as efficient as off-chip cell lysis prior to IFAST extraction,32 but required the addition of lysis buffer to the sample and incubation of 30 minutes in an oven. By comparison, the method described here requires only the addition of the crude sample to reconstitute the solid GuHCl and PMPs. This allows easy storage on the microfluidic device; increasing analysis speed and user friendliness. Detergent-free lysis is not a requirement for IFAST-based analysis as such systems have been shown to be compatible with common lysis and elution buffers containing detergents such as 1% Triton X-100, 1% LiDS or 2% SDS.28 The ability to reconstitute the reagent to a known concentration in the sample itself also makes it easier for the operator to use and reduces the number of manual steps required. Thirdly, instead of bonding the PDMS microfluidic layer to a glass substrate, we opted for an optical adhesive film as the bottom substrate of the chip. This very simple and rapid bonding approach has the added benefit that its hydrophobic surface properties allow the transfer of magnetic particles through the immiscible barrier without the addition of detergents to lower the interfacial energy. It also has advantages over plasma bonding in terms of ease of use and accessibility to equipment. Not every lab has access to a plasma oven for bonding but the optical adhesive is readily available from a number of suppliers and is more cost effective and easier to use. It is also specifically designed for PCR-based applications and has good optical properties which would be beneficial for future integration of real-time isothermal amplification to create a complete point-of-care system.
This miniaturised approach offers advantages over current commercially available stool DNA extraction kits, such as the QIAamp Stool DNA Mini Kit, as it offers a 7 fold reduction in the time taken for analysis, enables further pre-concentration of target DNA by eluting in a volume of 10 μL compared to 200 μL and is easy to use (e.g. multiple heating and centrifugation steps are not required, no proprietary chemicals are used).53
The simplicity and ease of use of the presented real world interface is perfectly suited for the requirements of a POC diagnostic device as results can be obtained whilst the patient is waiting, ensuring rapid identification of pathogenic strains of H. pylori from stool samples. Future work would look at evaluating the IFAST system with additional stool samples, allowing replicates of all possible sample types (based on the Bristol Stool Chart) to be performed, particularly with respect to the purity of the eluted DNA. This would also allow a more in depth study to be carried out on the number of H. pylori positive samples which express CagA. In addition, future work aims to integrate this work with real-time isothermal amplification of the pathogenic target to create a complete point-of-care system. This could lead to more immediate therapy and potentially a reduction in adverse conditions as a result of infection, such as gastric carcinogenesis. The microfluidic system could also be readily adapted to accommodate other pathogenic targets present in stool samples, such as Clostridium difficile or rotavirus.
Acknowledgements
The authors would like to thank Michael Collins, Lead Biomedical Scientist at NHS Chesterfield Laboratories, for provision of the stool samples and Marcus Hill for technical support.Notes and references
- D. o. B. Diseases, Helicobacter pylori and peptic ulcer disease, (accessed 2nd May, 2014).
- M. J. Blaser and J. Parsonnet, J. Clin. Invest., 1994, 94, 4–8 CrossRef CAS PubMed.
- J. Parsonnet, G. D. Friedman, N. Orentreich and H. Vogelman, Gut, 1997, 40, 297–301 CrossRef CAS PubMed.
- R. A. Wallace, P. J. Schluter, R. Forgan-Smith, R. Wood and P. M. Webb, J. Clin. Microbiol., 2003, 41, 4700–4704 CrossRef PubMed.
- C. Nogueira, C. Figueiredo, F. Carneiro, A. T. Gomes, R. Barreira, P. Figueira, C. Salgado, L. Belo, A. Peixoto, J. C. Bravo, L. E. Bravo, J. L. Realpe, A. P. Plaisier, W. G. V. Quint, B. Ruiz, P. Correa and L. J. Van Doorn, Am. J. Pathol., 2001, 158, 647–654 CrossRef CAS PubMed.
- V. Gubala, L. F. Harris, A. J. Ricco, M. X. Tan and D. E. Williams, Anal. Chem., 2012, 84, 487–515 CrossRef CAS PubMed.
- L. Gervais, N. de Rooij and E. Delamarche, Adv. Mater., 2011, 23, H151–H176 CrossRef CAS PubMed.
- X. Fang, H. Chen, L. Xu, X. Jiang, W. Wu and J. Kong, Lab Chip, 2012, 12, 1495–1499 RSC.
- G. V. Kaigala, R. J. Huskins, J. Preiksaitis, X.-L. Pang, L. M. Pilarski and C. J. Backhouse, Electrophoresis, 2006, 27, 3753–3763 CrossRef CAS PubMed.
- N. Bunyakul, C. Promptmas and A. J. Baeumner, Anal. Bioanal. Chem., 2015, 407, 727–736 CrossRef CAS PubMed.
- D. Verdoy, Z. Barrenetxea, J. Berganzo, M. Agirregabiria, J. M. Ruano-López, J. M. Marimón and G. Olabarría, Biosens. Bioelectron., 2012, 32, 259–265 CrossRef CAS PubMed.
- S. Huang, J. Do, M. Mahalanabis, A. Fan, L. Zhao, L. Jepeal, S. K. Singh and C. M. Klapperich, PLoS One, 2013, 8, e60059 CAS.
- S. Gillers, C. D. Atkinson, A. C. Bartoo, M. Mahalanabis, M. O. Boylan, J. H. Schwartz, C. Klapperich and S. K. Singh, J. Microbiol. Methods, 2009, 78, 203–207 CrossRef CAS PubMed.
- S. Berensmeier, Appl. Microbiol. Biotechnol., 2006, 73, 495–504 CrossRef CAS PubMed.
- N. Pamme, Lab Chip, 2006, 6, 24–38 RSC.
- M. A. M. Gijs, F. Lacharme and U. Lehmann, Chem. Rev., 2010, 110, 1518–1563 CrossRef CAS PubMed.
- A. van Reenen, A. M. de Jong, J. M. J. den Toonder and M. W. J. Prins, Lab Chip, 2014, 14, 1966–1986 RSC.
- N. Pamme, Curr. Opin. Chem. Biol., 2012, 16, 436–443 CrossRef CAS PubMed.
- T.-H. Kim, J. Park, C.-J. Kim and Y.-K. Cho, Anal. Chem., 2014, 86, 3841–3848 CrossRef CAS PubMed.
- K.-Y. Lien, C.-J. Liu, Y.-C. Lin, P.-L. Kuo and G.-B. Lee, Microfluid. Nanofluid., 2009, 6, 539–555 CrossRef CAS.
- U. Lehmann, C. Vandevyver, V. K. Parashar and M. A. M. Gijs, Angew. Chem., Int. Ed., 2006, 45, 3062–3067 CrossRef CAS PubMed.
- H. Tsuchiya, M. Okochi, N. Nagao, M. Shikida and H. Honda, Sens. Actuators, B, 2008, 130, 583–588 CrossRef CAS.
- R. Sista, Z. Hua, P. Thwar, A. Sudarsan, V. Srinivasan, A. Eckhardt, M. Pollack and V. Pamula, Lab Chip, 2008, 8, 2091–2104 RSC.
- C. H. Chiou, D. J. Shin, Y. Zhang and T. H. Wang, Biosens. Bioelectron., 2013, 50, 91–99 CrossRef CAS PubMed.
- J. Pipper, M. Inoue, L. F. P. Ng, P. Neuzil, Y. Zhang and L. Novak, Nat. Med., 2007, 13, 1259–1263 CrossRef CAS PubMed.
- J. Pipper, Y. Zhang, P. Neuzil and T.-M. Hsieh, Angew. Chem., Int. Ed., 2008, 47, 3900–3904 CrossRef CAS PubMed.
- Y. Zhang, S. Park, K. Liu, J. Tsuan, S. Yang and T.-H. Wang, Lab Chip, 2011, 11, 398–406 RSC.
- S. M. Berry, E. T. Alarid and D. J. Beebe, Lab Chip, 2011, 11, 1747–1753 RSC.
- S. M. Berry, C. Singh, J. D. Lang, L. N. Strotman, E. T. Alarid and D. J. Beebe, Integr. Biol., 2014, 6, 224–231 RSC.
- S. M. Berry, A. J. Lavanway, H. M. Pezzi, D. J. Guckenberger, M. A. Anderson, J. M. Loeb and D. J. Beebe, J. Mol. Diagn., 2014, 16, 297–304 CrossRef CAS PubMed.
- L. Strotman, R. O'Connell, B. P. Casavant, S. M. Berry, J. M. Sperger, J. M. Lang and D. J. Beebe, Anal. Chem., 2013, 85, 9764–9770 CrossRef CAS PubMed.
- L. N. Strotman, G. Lin, S. M. Berry, E. A. Johnson and D. J. Beebe, Analyst, 2012, 137, 4023–4028 RSC.
- S. M. Berry, L. N. Strotman, J. D. Kueck, E. T. Alarid and D. J. Beebe, Biomed. Microdevices, 2011, 13, 1033–1042 CrossRef PubMed.
- B. P. Casavant, D. J. Guckenberger, S. M. Berry, J. T. Tokar, J. M. Lang and D. J. Beebe, Lab Chip, 2013, 13, 391–396 RSC.
- B. P. Casavant, L. N. Strotman, J. J. Tokar, S. M. Thiede, A. M. Traynor, J. S. Ferguson, J. M. Lang and D. J. Beebe, Lab Chip, 2014, 14, 99–105 RSC.
- S. M. Berry, L. J. Maccoux and D. J. Beebe, Anal. Chem., 2012, 84, 5518–5523 CrossRef CAS PubMed.
- S. M. Berry, K. J. Regehr, B. P. Casavant and D. J. Beebe, J. Lab. Autom., 2013, 18, 206–211 CrossRef CAS PubMed.
- D. J. Guckenberger, P. C. Thomas, J. Rothbauer, A. J. LaVanway, M. Anderson, D. Gilson, K. Fawcett, T. Berto, K. Barrett, D. J. Beebe and S. M. Berry, J. Lab. Autom., 2014, 19, 267–274 CrossRef PubMed.
- K. Sur, S. M. McFall, E. T. Yeh, S. R. Jangam, M. A. Hayden, S. D. Stroupe and D. M. Kelso, J. Mol. Diagn., 2010, 12, 620–628 CrossRef CAS PubMed.
- R. C. den Dulk, K. A. Schmidt, G. Sabatte, S. Liebana and M. W. J. Prins, Lab Chip, 2013, 13, 106–118 RSC.
- H. Bordelon, N. M. Adams, A. S. Klemm, P. K. Russ, J. V. Williams, H. K. Talbot, D. W. Wright and F. R. Haselton, ACS Appl. Mater. Interfaces, 2011, 3, 2161–2168 CAS.
- K. M. Davis, J. D. Swartz, F. R. Haselton and D. W. Wright, Anal. Chem., 2012, 84, 6136–6142 CrossRef CAS PubMed.
- N. M. Adams, A. E. Creecy, C. E. Majors, B. A. Wariso, P. A. Short, D. W. Wright and F. R. Haselton, Biomicrofluidics, 2013, 7, 014104 Search PubMed.
- O. Strohmeier, A. Emperle, G. Roth, D. Mark, R. Zengerle and F. von Stetten, Lab Chip, 2013, 13, 146–155 RSC.
- R. Gottheil, N. Baur, H. Becker, G. Link, D. Maier, N. Schneiderhan-Marra and M. Stelzle, Biomed. Microdevices, 2014, 16, 163–172 CrossRef CAS PubMed.
- C. Phurimsak, E. Yildirim, M. D. Tarn, S. J. Trietsch, T. Hankemeier, N. Pamme and P. Vulto, Lab Chip, 2014, 14, 2334–2343 RSC.
- P. Otto, J. Assoc. Lab. Autom., 2002, 7, 34–37 CrossRef CAS.
- A. P. Lage, E. Godfroid, A. Fauconnier, A. Burette, J. P. Butzler, A. Bollen and Y. Glupczynski, J. Clin. Microbiol., 1995, 33, 2752–2756 CAS.
- C. Kemp, C. Birch, K. J. Shaw, G. J. Nixon, P. T. Docker, J. Greenman, J. F. Huggett, S. J. Haswell, C. A. Foy and C. E. Dyer, Anal. Methods, 2012, 4, 2141–2144 RSC.
- C. Kemp, J. M. Wojciechowska, M. N. Esfahani, G. Benazzi, K. J. Shaw, S. J. Haswell and N. Pamme, Proceedings for MicroTAS 2012, 2012, pp. 779–781 Search PubMed.
- P. Otto, J. Assoc. Lab. Autom., 2002, 7, 34–37 CrossRef CAS.
- NICE, Bristol Stool Chart, (accessed 29th May, 2015) Search PubMed.
- Qiagen, QIAamp DNA Stool Mini Kit Manual, 2012 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6lc00228e |
| This journal is © The Royal Society of Chemistry 2016 |