Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of an E. coli strain for one-pot biofuel production from ionic liquid pretreated cellulose and switchgrass†
Marijke
Frederix
ab,
Florence
Mingardon
c,
Matthew
Hu
ab,
Ning
Sun
bd,
Todd
Pray
bd,
Seema
Singh
abe,
Blake A.
Simmons
abe,
Jay D.
Keasling
abfg and
Aindrila
Mukhopadhyay
*ab
aJoint BioEnergy Institute, Emeryville, CA, USA. E-mail: amukhopadhyay@lbl.gov
bBiological Systems and Engineering Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA
cTotal New Energies Inc., Emeryville, CA, USA
dAdvanced Biofuels Process Demonstration Unit, Emeryville, CA, USA
eBiological and Engineering Sciences Center, Sandia National Laboratories, Livermore, CA, USA
fDepartment of Chemical & Biomolecular Engineering, University of California, Berkeley, CA 94720, USA
gDepartment of Bioengineering, University of California, Berkeley, CA 94720, USA
First published on 11th April 2016
Abstract
Biological production of chemicals and fuels using microbial transformation of sustainable carbon sources, such as pretreated and saccharified plant biomass, is a multi-step process. Typically, each segment of the workflow is optimized separately, often generating conditions that may not be suitable for integration or consolidation with the upstream or downstream steps. While significant effort has gone into developing solutions to incompatibilities at discrete steps, very few studies report the consolidation of the multi-step workflow into a single pot reactor system. Here we demonstrate a one-pot biofuel production process that uses the ionic liquid 1-ethyl-3-methylimidazolium acetate ([C2C1Im][OAc]) for pretreatment of switchgrass biomass. [C2C1Im][OAc] is highly effective in deconstructing lignocellulose, but nonetheless leaves behind residual reagents that are toxic to standard saccharification enzymes and the microbial production host. We report the discovery of an [C2C1Im]-tolerant E. coli strain, where [C2C1Im] tolerance is bestowed by a P7Q mutation in the transcriptional regulator encoded by rcdA. We establish that the causal impact of this mutation is the derepression of a hitherto uncharacterized major facilitator family transporter, YbjJ. To develop the strain for a one-pot process we engineered this [C2C1Im]-tolerant strain to express a recently reported D-limonene production pathway. We also screened previously reported [C2C1Im]-tolerant cellulases to select one that would function with the range of E. coli cultivation conditions and expressed it in the [C2C1Im]-tolerant E. coli strain so as to secrete this [C2C1Im]-tolerant cellulase. The final strain digests pretreated biomass, and uses the liberated sugars to produce the bio-jet fuel candidate precursor D-limonene in a one-pot process.
Introduction
Sustainably grown lignocellulosic biomass provides an attractive carbon source for the production of renewable biofuels.1 Prior to saccharification, pretreatment of the biomass is required to separate it from the lignin and to solubilize the available cellulose and hemicellulose to make it more accessible to enzymes that hydrolyze it into fermentable sugars. Pretreatment is traditionally done via techniques like exposure to dilute acids or bases at elevated temperatures or using ammonia fiber expansion.2 More recently however, hydrophilic ionic liquids (ILs) such as 1-ethyl-3-methylimidazolium acetate ([C2C1Im][OAc]) have emerged as highly effective pretreatment solvents that are more environmentally friendly, produce relatively low amounts of biomass-derived inhibitors and can be used at lower temperatures and pressures than conventional pretreatments that use acids or bases.3–5 One of the initial challenges in the use of ILs as pretreatment reagents was the cost of the reagents, and for an economical process the ILs had to be recovered and recycled, leaving less than 12 mM in a typical slurry.6 Significant improvements in IL synthesis have reduced their cost and current predictions estimate that in an economical industrial-scale production process, a residual level of 12–300 mM can be allowed to remain in the downstream sugar stream that will be used for cultivation of the biofuel-producing organism.6,7 Consequently, both the saccharification enzymes as well as the microbiological conversion have to be optimized to be able to accommodate the presence of higher levels of residual ILs. To this end, several studies have focused on both the discovery of IL-tolerant microbes8 as well as cellulases that can function in the presence of high levels of ILs.9,10 However, as with any process that requires purified proteins, the cost of the proteins remains a significant source of cost.Recent progress in consolidated bioprocessing addresses the cost of the saccharification enzymes and has led to the development of a variety of strains that express cellulase enzymes11,12 and in some examples are also coupled to a production pathway.13–15 However, these engineered strains will also be hampered by growth inhibitory compounds that are routinely present in pretreated hydrolysates,16 making the use of industrially relevant hydrolysates challenging. In the case of IL pretreated hydrolysates, the toxicity of residual ILs to the microbial production host17 is the major challenge to optimizing an integrated process, and has led to studies that focus on discovery of tolerance bestowing mechanisms18 and strain engineering to optimize the expression of such genes.19 A necessary next step in the area of biofuel production is to consolidate the findings from studies that have examined and found solutions to different segments of the bioconversion process, from cellulases that may allow efficient saccharification in IL-containing hydrolysate,9 strains that can withstand residual ILs8 and synthetic metabolic pathways that convert the biomass-derived sugars to desirable final products.20
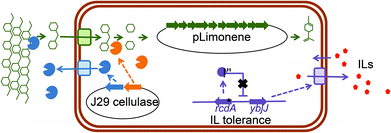
In this study, we characterized a spontaneous E. coli mutant that is tolerant to the IL 1-ethyl-3-methylimidazolium chloride ([C2C1Im]Cl), and identify the causal mutation to be located in the transcriptional regulator RcdA (also known as YbjK). We examined the genetic basis for the phenotype observed due to the mutant RcdA and demonstrated the ability of an rcdA mutant to produce fuels in IL containing medium and with IL pretreated biomass as the carbon source. Subsequently we screened a mini-library of secreted cellulases for their ability to confer growth on cellulose in the presence of ILs and integrated the best cellulase for our cultivation conditions into an E. coli rcdA mutant strain also engineered to contain a production pathway for the monoterpene bio-jet fuel precursor and commodity chemical, D-limonene (Fig. 1). Terpene-based compounds provide a range of candidates with energy content and combustion properties that make them suitable for gasoline, diesel as well as jet fuel needs.21,22 These advanced biofuels also have physicochemical properties that may align better with the existing infrastructure. Since E. coli DH1 has previously been shown to be an ideal host for monoterpene production,20 we first constructed an rcdA mutant in this strain background. This strain was able to produce significant levels of D-limonene in [C2C1Im][OAc] containing growth medium and with either phosphoric acid swollen cellulose (PASC) or [C2C1Im][OAc] pretreated switchgrass as a carbon source. This study demonstrates the one-pot biofuel production process for IL pretreated biomass, using an IL tolerant E. coli strain and takes into account the reduced requirement for IL recycling.
Experimental
Bacterial strains and plasmids
All strains and plasmids used in this work can be obtained from http://public-registry.jbei.org![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23 and are summarized in Table 1. E. coli DH10B (Invitrogen Carlsbad, CA) was used for cloning. All [C2C1Im]Cl and [C2C1Im][OAc] tolerance assays were done using E. coli BW25113-derived strains.24 The original FM002 strain was isolated from colonies of spontaneous mutants that appeared after plating FM001 on plates containing [C2mim+] Cl−. The kanamycin resistance cassette of the KEIO collection E. coli strains, BW25113 rcdA and BW25113 ompA, was removed using the plasmid pCP20, per methods described in Cherepanov et al.25 to generate the strains JBEI-12574 and JBEI-13315. An E. coli DH1 (ATCC33849) derived rcdA mutant strain (JBEI-13314) was generated using the lambda red methodology26 and was used as a host strain for D-limonene production. All plasmids constructed in this study were designed using the j5 DNA assembly design software27 and were assembled using the method described by Gibson et al.28
23 and are summarized in Table 1. E. coli DH10B (Invitrogen Carlsbad, CA) was used for cloning. All [C2C1Im]Cl and [C2C1Im][OAc] tolerance assays were done using E. coli BW25113-derived strains.24 The original FM002 strain was isolated from colonies of spontaneous mutants that appeared after plating FM001 on plates containing [C2mim+] Cl−. The kanamycin resistance cassette of the KEIO collection E. coli strains, BW25113 rcdA and BW25113 ompA, was removed using the plasmid pCP20, per methods described in Cherepanov et al.25 to generate the strains JBEI-12574 and JBEI-13315. An E. coli DH1 (ATCC33849) derived rcdA mutant strain (JBEI-13314) was generated using the lambda red methodology26 and was used as a host strain for D-limonene production. All plasmids constructed in this study were designed using the j5 DNA assembly design software27 and were assembled using the method described by Gibson et al.28
| Description | Ref. | |
|---|---|---|
| Strains | ||
| JBEI-12929 | FM001, E. coli BW25113 ΔdmsD + pBbA8c-RFP | This work |
| JBEI-7851 | FM002, spontaneous mutant tolerant to 200 mM [C2C1Im]Cl derived from E coli BW25113 ΔdmsD + pBbA8c-RFP, P7Q mutation | This work |
| JW5114 | E. coli BW25113 rcdA (b0846) mutant | 24 |
| JW0829 | E. coli BW25113 ybjJ (b0845) mutant | 24 |
| JW0940 | E. coli BW25113 ompA (b0957) mutant | 24 |
| JW1010 | E. coli BW25113 pgaA (b1024) mutant | 24 |
| JW1023 | E. coli BW25113 csgD (b1040) mutant | |
| JW1529 | E. coli BW25113 ydeI (b1536) mutant | 24 |
| JW5778 | E. coli BW25113 nanC (b4311) mutant | |
| JW1151 | E. coli BW25113 ycgZ (b1164) mutant | 24 |
| JW5826 | E. coli BW25113 asr (b1597) mutant | 24 |
| JW2095 | E. coli BW25113 yehA (b2108) mutant | 24 |
| JW1022 | E. coli BW25113 csgE (b1039) mutant | 24 |
| JBEI 13314 | E. coli DH1 rcdA mutant | This work |
| JBEI-13315 | E. coli BW25113 ompA (b0957) mutant, Km cassette removed using pCP20 | This work |
| JBEI-12574 | E. coli BW25113 rcdA (b0846) mutant, Km cassette removed using pCP20 | This work |
| Plasmids | ||
| JBEI-6409 | pLimonene, limonene production plasmid | 20 |
| JBEI-7585 | pCP20, plasmid to remove Km resistance cassette from KEIO collection | 26 |
| JBEI-2558 | pBbS8k (low copy vector with arabinose inducible promoter) | 56 |
| JBEI-4338 | pCellulose | 15 |
| JBEI-12905 | pCellulose/J24 | This work |
| JBEI-12906 | pCellulose/J29 | This work |
| JBEI-12934 | pCellulose/J20 | This work |
| JBEI-12940 | pCellulose/Csac | This work |
| JBEI-12935 | pBbS8k-ybjJ | This work |
| JBEI-12927 | pBbS8k-ompA | This work |
| JBEI-12925 | pBbS8k-ybjJ-ompA | This work |
Growth conditions and media
All E. coli strains expressing cellulases or the limonene production pathway were grown at 30 °C, while all other strains were grown at 37 °C. The medium used was Luria-Bertani (LB) or EZ rich minimal medium (Teknova, Hollister, CA). When PASC29 was used as carbon source it was added to EZ rich to obtain a final concentration of 10 g per L cellulose (equivalent to 11 g glucose per L). For hydrolysate-derived medium, [C2C1Im][OAc]-treated biomass (prepared at the ABPDU per methods described below) was added to obtain a final glucose concentration of 10 g per L and 2.3 g per L xylose. Cultures with pCellulose and pCellulose-derived plasmids (Table 1) were always grown in the presence of 10 g per L glucose at 30 °C to repress transcriptional activity from the cspD promoter, unless PASC was used as the carbon source. All chemicals were purchased from Sigma-Aldrich (St Louis, MO). Antibiotics were added as required to maintain plasmid selection. [C2mim+]Cl− (98%) was purchased from Sigma-Aldrich (St Louis, MO) and [C2mim+][OAc] (90%) was purchased from BASF (Ludwigshafen, Germany).Growth assays
To measure growth, 5 μl of preculture was inoculated into 100 μl of growth medium in 96-well plates and grown at 30 °C in a Tecan F200 microtiterplate reader (Maennedorf, Switzerland), measuring the optical density at a wavelength of 590 nm (OD590) at 20 min intervals. When PASC and IL-pretreated biomass were used as a carbon source, because of the turbidity of the growth medium, growth was measured by counting colony forming units (CFU). 4 mL of culture was inoculated with 100 μl of fresh preculture, grown shaking at 30 °C, and CFU was determined by counting the number of colonies after plating 100 μl of a 10−6 dilution on LB-agar plates.Whole-genome resequencing
DNA from FM001 and FM002 was randomly sheared into ∼400 bp fragments that were used to create Illumina libraries. These libraries were sequenced on Illumina sequencers, generating 100 bp paired-end reads that were aligned to the reference genome (GenBank accession number NC_000913) using the Burrows–Wheeler alignment tool30 and down sampled to generate an average read depth of 250 times. A total of 55 putative variants were identified using the samtools and mpileup programs.31 Variants that failed our filtering criteria (minimum quality = 10, minimum P value for strand bias = 0.0001, minimum P value for end bias = 0.0001, single-nucleotide polymorphisms within 10 bp around a gap) were removed from the analysis. There was only one single base pair that was different between FM001 and FM002, a C20A mutation in the gene rcdA leading to a P7Q mutation in the amino acid sequence.Limonene production assays
The E. coli DH1 rcdA mutant (JBEI 13314) was used for all limonene production assays. Strains harboring the limonene production plasmid (JBEI-6409) were grown in EZ-Rich medium with the appropriate antibiotics to maintain plasmid selection, with glucose, PASC, or IL-pretreated biomass hydrolysate as a carbon source at a final concentration of 1% (10 g L−1) (Teknova, Hollister, CA). Precultures (from fresh transformation plates) were grown shaking overnight at 30 °C and used to inoculate production cultures at a starting optical density (OD600) of 0.1. Cultures containing glucose or IL-pretreated biomass hydrolysate as carbon source were then grown at 30 °C until OD600 between 0.6–0.8 and induced with 25 μM isopropyl β-D-1-thiogalactopyranoside (IPTG), at which point a dodecane overlay (10 mL for 50 mL cultures) was also added to capture the produced limonene. Limonene production was sampled after 48 h and analyzed using gas-chromatography mass-spectrometry (GC-MS) according to methods described previously.20 Cultures containing PASC as carbon source were grown for 8 hours with no [C2mim+][OAc] present and then for 10 hours with 100 mM [C2mim+][OAc], after which they were induced with 25 μM IPTG, overlaid with dodecane (10 mL for 50 mL cultures) and allowed to grow for 48 h before sampling and GC-MS analysis for limonene production.Preparation of the hydrolysate using IL pretreated biomass
The hydrolysate was prepared using pilot-scale reactors at the DOE Advanced Biofuels Process Demonstration Unit (ABPDU), Lawrence Berkeley National Laboratory.32 Switchgrass was pretreated in a 210 L customized Andritz reactor at 160 ± 3 °C for 2 h using [C2C1Im][OAc] as the solvent. The total mass of the reaction was 40 kg with 15% (w/w) solid loading. After 2 h incubation, the reactor was cooled to 60 °C and 40 L ethanol was pumped into the mixture as the anti-solvent. The slurry was released from the reactor and homogenized using a laboratory blender to break down large chunks. The solid was then separated from the IL-rich liquid by filtration through cheese cloth. The recovered solid was further washed with warm water (60 °C) to remove the IL residual. The washed solids were collected to determine moisture content and solid recovery from IL pretreatment. Enzymatic saccharification was performed in 50L IKA SPP50 reactor with 15% (w/w) solid loading. The total batch mass was 20 kg with cellulase (CTec2) 54 mg enzyme per g glucan and xylanase (HTec2) 6 mg enzyme per g glucan (total protein loading: 9.2 mg protein per g glucan). After 160 h incubation, the reaction was stopped and the slurry was transferred out. The lignin rich solid was separated from the hydrolysate using a basket centrifuge with a polypropylene filter bag (pore size 25–30 micron) as the liner. The hydrolysate was stored at 4 °C for future use and analysis.Results and discussion
Isolation of a spontaneous IL tolerant mutant
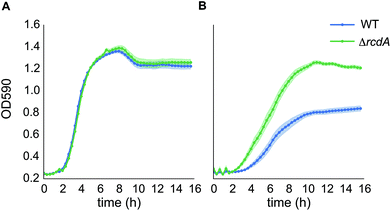
Selected mutants from the E. coli KEIO collection were screened for strains with increased tolerance to [C2C1Im]Cl. While none of the KEIO mutants in this screen were significantly more or less tolerant than wild type, we did observe the appearance of spontaneous mutants with the desired phenotype. One of these mutants (FM002), found in the ΔdmsD mutant (FM001), had a very reproducible IL tolerant phenotype (Fig. S1A and B†). Further study showed that in addition to increased tolerance to 200 mM [C2C1Im]Cl it also had increased tolerance to 150 mM [C2C1Im][OAc], a related IL. This suggests that the tolerance is related to the [C2C1Im]+ cation (Fig. S1C and D†). The genetic basis of the increased tolerance phenotype of FM002 was identified via whole genome resequencing and indicated one original mutation in basepair 20 (C20A) of the transcriptional regulator rcdA (b0846), which resulted in a P7Q amino acid change. Bioinformatics analysis of the RcdA protein sequence showed that this mutation is at the start of the helix-turn-helix DNA binding domain of the protein. Considering the biochemical nature of proline (hydrophobic with a strong conformational rigidity) to glutamine (polar) change, we hypothesized that the identified mutation renders RcdA non-functional. To verify this, the rcdA mutant from the KEIO collection was tested for tolerance to [C2C1Im]Cl and indeed found to be more tolerant than wild type BW25113 (Fig. 2). As was the case for FM002, the BW25113 rcdA mutant was also found to be tolerant to [C2C1Im][OAc] (Fig. S1E and F†).Phenotypic characterization of RcdA regulatory targets
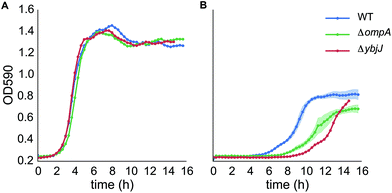
Since RcdA is a transcriptional regulator, the increased tolerance phenotype of the rcdA mutant was hypothesized to be due to its regulatory targets. The RcdA binding box and its potential targets had already been determined via a genomic SELEX screening procedure in E. coli BW25113 by Shimada et al.33 The presence of 20 RcdA binding sites in the genome had led to the prediction of 27 putative gene regulatory targets. A subset of these was selected for further analysis, based on their predicted function. Amongst these were two transcriptional regulators appY and ycgF, which have been shown to be involved in stress response,34,35 two cytoplasmatic stress response genes ydeI and asr,36,37 several genes involved in biofilm formation (csgD, csgB, yehA, fimB),38–40 an inner membrane protein ybjJ, which is located next to rcdA and is predicted to be a transporter of the major facilitator superfamily (MFS), and three pore-forming outer membrane proteins (ompA, pgaA and nanC).41–43 The mutants for these genes were obtained from the KEIO collection24 and screened for their potential involvement in [C2C1Im]+ tolerance by a liquid growth assay in 96 well plates in the presence of 200 mM [C2C1Im]Cl (Fig. S2A–D†). This led to the identification of two mutants with reduced tolerance to [C2C1Im]Cl−, ΔompA and ΔybjJ, both of which encode proteins involved in transport across the inner and outer membrane (Fig. 3).Overexpression of YbjJ and OmpA
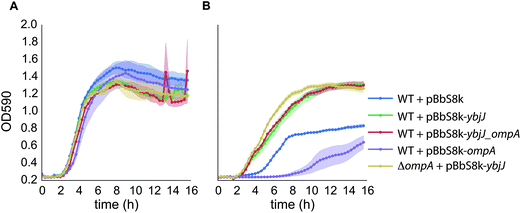
YbjJ is an inner membrane transporter and OmpA an outer membrane porin and both may impact [C2C1Im]+ tolerance individually, or together as a complex. To evaluate these hypotheses, both genes were overexpressed separately (JBEI-12935 and JBEI-12927 respectively) and together in an operon (JBEI-12925), on a low copy plasmid transcribed from the arabinose-inducible araBAD promoter (PBAD). They were subsequently introduced into wild type BW25113 and their growth in the presence of 200 mM [C2C1Im]Cl was evaluated in a liquid growth assay in 96 well plates (Fig. 4A and B). Overexpression of OmpA and YbjJ in an operon increased tolerance significantly, as did overexpression of YbjJ on its own. However, overexpression of OmpA did not increase tolerance, but rather led to a growth burden that was further increased in the presence of [C2C1Im]Cl. However, in strains overexpressing YbjJ alone, native levels of OmpA are still present in wild type E. coli BW25113 and may still have synergistic benefit in the function. To fully eliminate the possibility that YbjJ requires OmpA for function, YbjJ was overexpressed in a ΔompA mutant. The resulting strain was found to have increased tolerance to [C2C1Im]Cl equivalent to the overexpression of YbjJ in the wild type E. coli BW25113, showing that the YbjJ on its own is sufficient for tolerance.Production of fuels in the presence of [C2C1Im][OAc]
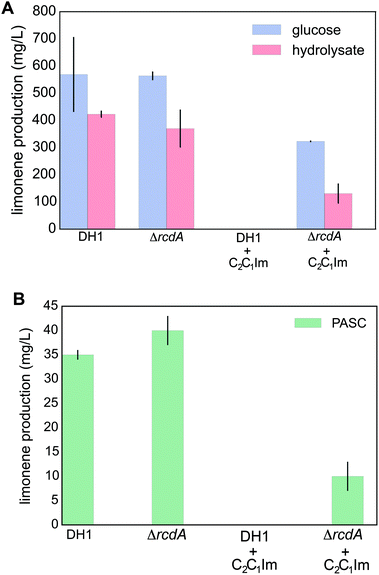
To evaluate if the rcdA mutant could be used for biofuel production from IL-pretreated biomass, we generated an rcdA deletion mutant in E. coli DH1, since this is the preferred production host for terpenes.44 The main goal of this study was to use IL-pretreated biomass for fuel production, therefore we chose to conduct all fuel production assays with [C2C1Im][OAc], one of the most effective ILs established for biomass pretreatment.45First we confirmed tolerance of the E. coli DH1 rcdA mutant to [C2C1Im][OAc] using plate growth assays (Fig. S3†). Then we developed the strain to produce the terpene jet-fuel precursor candidate D-limonene and determined the production using either glucose or switch grass hydrolysate as the carbon source (Fig. 5A). In optimal conditions using glucose as a carbon source and without toxicity from [C2C1Im][OAc], an E. coli DH1 D-limonene producer strain carrying the plasmid pLimonene20 yields about ∼550 mg L−1. In the presence of 100 mM [C2C1Im][OAc] this strain shows limited growth and no D-limonene could be detected. In contrast, the E. coli DH1 rcdA mutant D-limonene producer strain produced ∼300 mg L−1 in the presence of [C2C1Im][OAc] (Fig. 5A).
We also evaluated the ability of both strains to produce limonene in EZ rich media containing IL pretreated and saccharified switchgrass as a carbon source. IL pretreated switchgrass was generated at the ABPDU, based on a standard process as described in the Methods section, and involved extensive washing leading to a final residual [C2C1Im][OAc] concentration of 12 mM. No toxicity was observed when the biofuel production strains were grown in hydrolysate-derived growth medium compared to glucose-derived growth medium. Using this hydrolysate-containing media, a yield of ∼400 mg per L D-limonene was obtained for both wild type and the rcdA mutant.
To test if the strains could produce limonene in IL pretreated biomass that contains toxic levels of [C2C1Im][OAc], the IL was added as described in the Methods section. In this test, no growth or production was observed in the wild type producer strain (Fig. 5A). However the rcdA mutant producer produced approximately 150 mg per L D-limonene, which is comparable to the production with glucose and no [C2C1Im][OAc] in the media. To our knowledge, this is the first report of producing an advanced biofuel directly from a solution containing IL pretreated biomass.
One-pot biofuel production process in [C2C1Im]+ containing medium
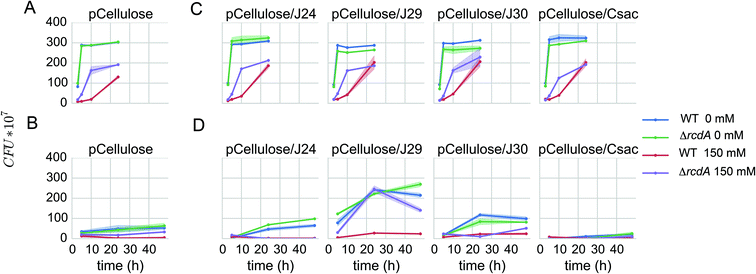
In the cellulosic biofuel production workflow, after pretreatment of biomass, the solubilized cellulose needs to be converted into glucose by cellulases. Traditionally this is done in a separate step using purified enzymes, but it can also be done through a consolidated process in which the required cellulases have been engineered into the biofuel production host. In previous work15 a cellulose degrading plasmid, pCellulose, had been developed for the expression of a secreted cellulase (a translational fusion between the N-terminus of the E. coli protein OsmY and Cel from Bacillus D04) and an intracellular β-glucosidase (Cel3A from Cellvibrio japonicas) by expressing them from native E. coli promoters (pcspD and pwrbA respectively) (Fig. 1 and Table 1). These enzymes had not been tested for activity in the presence of [C2C1Im][OAc]. We transformed pCellulose into BW25113 and the BW25113 rcdA mutant, and tested the ability to grow on medium containing PASC as a carbon source. While pCellulose did allow growth in culture medium without [C2C1Im][OAc], the growth rates were significantly lower than previously reported. In the presence of [C2C1Im][OAc], the rcdA mutant grew slightly better than the WT, but never to the same level as the control without [C2C1Im][OAc], indicating that the enzymes in pCellulose might not perform optimally under these conditions (Fig. 6A, B), and that the Cel enzyme may be sensitive to [C2C1Im][OAc].To overcome the potential toxicity of [C2C1Im][OAc] to cellulases, we took advantage of the collection of IL-tolerant cellulases that had been assembled by Gladden et al.9 We evaluated four [C2C1Im][OAc] tolerant cellulases for their ability to confer growth on PASC in the presence of [C2C1Im][OAc]: J24 (Paenibacillus provencensis), J29 (Paenibacillus sp. KSM-N546), J30 (Thermobacillus composti KWC4) and Csac (Caldicellulosiruptor saccharolyticus).9,46 To evaluate their potential consolidated with a production strain, variants of pCellulose plasmid were constructed in which cel was replaced by the genes encoding the IL tolerant cellulases (Table 1). These plasmids were subsequently transformed to BW25113 and the BW25113 rcdA mutant and the growth on PASC derived medium was assayed by CFU counting assays. All strains showed equivalent growth on glucose as a carbon source (Fig. 6C).
When expressed in the rcdA mutant, one pCellulose derivative, pCellulose-J29 outperformed the others. No significant growth difference was observed in the presence and absence of [C2C1Im][OAc], indicating that J29 is adequately produced, secreted and active in our cultivation conditions (Fig. 6D). Next, D-limonene production using PASC as an unsaccharified carbon source was evaluated by transforming the E. coli DH1 pLimonene control strain and the E. coli DH1 rcdA pLimonene strain with the pCellulose-J29 plasmid. In the absence of [C2C1Im][OAc], both control and the rcdA mutant produce approximately 35 mg per L D-limonene. In the presence of 150 mM [C2C1Im][OAc], no growth or fuel production was observed for the control, but a yield of 10 mg L−1 was obtained for the rcdA mutant production strain (Fig. 5B).
Conclusions
The major contribution of this study is the development of a one-pot biofuel production process that can directly convert both saccharified IL-pretreated hydrolysate and unsaccharified cellullose into a next-generation bio-jet fuel precursor in the presence of ILs. In contrast to previous studies that used highly washed pretreated biomass,15 significant IL was included in the growth medium, as would be expected in a typical industrial process that uses IL based pretreatment and provides an important baseline for production under industrially relevant conditions.6,7 For a successful process, two aspects of IL toxicity needed to be overcome: both the biofuel production strain and the secreted cellulases that convert the solubilized cellulose into fermentable sugars were required to be IL-tolerant.To overcome the first challenge, a spontaneous [C2C1Im] tolerant mutant strain of E. coli BW25113 was used. Analysis of the isolated mutant identified a mutation in the transcriptional regulator RcdA. We used the E. coli DH1 rcdA mutant as our background for growth in medium containing saccharified [C2C1Im] pretreated biomass as carbon source in the presence of the [C2C1Im] reagent, and demonstrated the production of the bio-jet fuel precursor D-limonene, at final titer of 150 mg L−1. These levels are lower than the best reported yields obtained using glucose as a carbon source, however the control E. coli production strain is unable to produce any D-limonene in [C2C1Im][OAc] containing medium. To our knowledge this is the first demonstration of one-pot microbial D-limonene production from IL-containing saccharified hydrolysate as the substrate.
We also established the genetic basis for the mutant rcdA phenotype. RcdA has mainly been studied for its involvement in the regulation of biofilm formation via CsgD. However, previous work by Shimada et al.33 report a comprehensive list of its regulatory targets. In our study, two of these targets, ybjJ and ompA, were found to have a role in tolerance to imidazolium-based ILs, as a deletion of these genes led to increased sensitivity. OmpA is an abundant protein in the E. coli outer membrane and forms a non-specific channel.41 An E. coli ΔompA mutant has been found to be more sensitive to compounds like sodium dodecyl sulfate, cholate, acid and to high osmolarity,47 which correlate with sensitivity measured in our study to [C2C1Im]Cl. ybjJ encodes an inner membrane protein belonging to the major facilitator superfamily (MFS) of pumps and is transcribed divergently from rcdA. YbjJ does not show any significant similarity to another MFS IL efflux pump, EilA, identified previously.18 It has been shown that RcdA can function both as an activator and repressor of gene expression and our results indicate that RcdA functions as a transcriptional repressor for the expression of ybjJ and ompA and is consistent with previous reports by Shimada et al.33 that also show RcdA to be a repressor for ybjJ expression. While the deletion of both ybjJ and ompA increased the sensitivity to [C2C1Im]Cl, when overexpressed in trans, only YbjJ displayed the tolerance phenotype demonstrated by the rcdA mutant. These results indicate that the primary mechanism for enhanced IL tolerance in the RcdA(P7Q) mutant is due to the derepression of ybjJ.
The second challenge for developing a one-pot biofuel production is in the direct use of pretreated, but unsaccharified biomass. This is possible by engineering the microbial host to also produce cellulases for the saccharification of pretreated biomass as has been shown before.15 However, the recent improvements7 in processes using ILs allow greater residual IL levels to be retained in the final substrate necessitating that the cellulase also be functional in the presence of ILs. To achieve this, we screened a mini-library of previously reported IL tolerant cellulases for their ability to grow on PASC, in the presence or absence of IL. One enzyme, J29, emerged as a highly active and IL-tolerant cellulase in the conditions used to cultivate E. coli for bio-fuel production. J29 has been shown to be 100% active in 294 mM [C2C1Im][OAc] and retains >50% activity in 882 mM [C2C1Im][OAc].9 While its optimal temperature is reported to be 65 °C, we found its activity to be sufficient at 30 °C, the optimal temperature for D-limonene production by E. coli DH1. A future study focused on the full biochemical characterization of the cellulases in our library will be very valuable in determining the underlying mechanism for the superior performance of J29 relative to the other enzymes and may use methods that were developed to examine cellulases after exposure to reagents such as [C2C1Im][OAc].48
We consolidated an E. coli DHI rcdA mutant background with a plasmid borne D-limonene production pathway and the plasmid pCellulose-J29, and demonstrated the production of 10 mg per L D-limonene on growth medium containing PASC as carbon source in the presence of 100 mM [C2C1Im][OAc]. This yield is significantly lower than the best D-limonene production reported using idealized glucose based growth conditions. However the demonstrated one-pot biofuel production from non-saccharified cellulose provides the necessary next step in the development of a strain that can function and produce a biofuel product in a highly industrially relevant condition.
Several challenges remain to be overcome as we continue to build and test strains and production processes with greater industrial value. The growth benefit provided by the rcdA mutant, via the derepression of ybjJ, can be further optimized by fine-tuning the levels of this MFS pump as well as by combining additional IL tolerance genes. In this study we also do not address the presence of other potentially inhibiting compounds in the growth medium (for example, lignin derived aromatic compounds)49 and the toxicity of the final product itself,50–52 both of which are factors that likely negatively impact fuel production. Tolerance mechanisms for many of these inhibitory compounds have been studied and reported and often involve export pumps,53,54 and pose their own metabolic burden on the cell when overexpressed.55 Our group and others have shown in previous studies that the overexpression of pumps needs to be tightly controlled.19,50 One foreseeable challenge is to fully understand how different tolerance mechanisms can be combined in a synergistic manner. Other areas for the successful development of a consolidated biofuel process include the improved secretion of cellulases and continued metabolic engineering of the fuel pathways, that when incorporated into the microbial chromosome will generate the final industrial hosts.
Acknowledgements
We thank Noppadon Sathitsuksanoh (JBEI) for providing PASC, Ling Liang and Tina Trang Luong (ABPDU) for their effort in hydrolysate preparation, and Wendy Schackwitz (US DOE Joint Genome Institute) for the genome re-sequencing and data analysis support for the mutant E. coli strain FM0002. This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.References
- D. Tilman, R. Socolow, J. A. Foley, J. Hill, E. Larson, L. Lynd, S. Pacala, J. Reilly, T. Searchinger, C. Somerville and R. Williams, Science, 2009, 325, 270–271 CrossRef CAS PubMed.
- V. Chaturvedi and P. Verma, 3 Biotech., 2013, 3, 415–431 CrossRef.
- M. Mora-Pale, L. Meli, T. V. Doherty, R. J. Linhardt and J. S. Dordick, Biotechnol. Bioeng., 2011, 108, 1229–1245 CrossRef CAS PubMed.
- C. Li, B. Knierim, C. Manisseri, R. Arora, H. V. Scheller, M. Auer, K. P. Vogel, B. A. Simmons and S. Singh, Bioresour. Technol., 2010, 101, 4900–4906 CrossRef CAS PubMed.
- C.-Z. Liu, F. Wang, A. R. Stiles and C. Guo, Appl. Energy, 2012, 92, 406–414 CrossRef CAS.
- D. Klein-Marcuschamer, B. A. Simmons and H. W. Blanch, Biofuels, Bioprod. Biorefin., 2011, 5, 562–569 CrossRef CAS.
- N. M. Konda, J. Shi, S. Singh, H. W. Blanch, B. A. Simmons and D. Klein-Marcuschamer, Biotechnol. Biofuels, 2014, 7, 86 CrossRef PubMed.
- J. I. Khudyakov, P. D'haeseleer, S. E. Borglin, K. M. Deangelis, H. Woo, E. A. Lindquist, T. C. Hazen, B. A. Simmons and M. P. Thelen, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E2173–E2182 CrossRef CAS PubMed.
- J. M. Gladden, J. I. Park, J. Bergmann, V. Reyes-Ortiz, P. D'haeseleer, B. F. Quirino, K. L. Sale, B. A. Simmons and S. W. Singer, Biotechnol. Biofuels, 2014, 7, 15 CrossRef PubMed.
- J. I. Park, E. J. Steen, H. Burd, S. S. Evans, A. M. Redding-Johnson, T. Batth, P. I. Benke, P. D'haeseleer, N. Sun, K. L. Sale, J. D. Keasling, T. S. Lee, C. J. Petzold, A. Mukhopadhyay, S. W. Singer, B. A. Simmons and J. M. Gladden, PLoS One, 2012, 7, e37010 CAS.
- D. Gao, Y. Luan, Q. Wang, Q. Liang and Q. Qi, Microb. Cell Fact., 2015, 14, 159 CrossRef PubMed.
- S. M. Gaida, A. Liedtke, A. H. W. Jentges, B. Engels and S. Jennewein, Microb. Cell Fact., 2016, 15, 6 CrossRef PubMed.
- L. R. Lynd, W. H. van Zyl, J. E. McBride and M. Laser, Curr. Opin. Biotechnol., 2005, 16, 577–583 CrossRef CAS PubMed.
- D. La Grange, R. Den Haan and W. Van Zyl, Appl. Microbiol., 2010, 87, 1195–1208 Search PubMed.
- G. Bokinsky, P. P. Peralta-Yahya, A. George, B. M. Holmes, E. J. Steen, J. Dietrich, T. Soon Lee, D. Tullman-Ercek, C. A. Voigt, B. A. Simmons and J. D. Keasling, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 19949–19954 CrossRef CAS PubMed.
- A. Mukhopadhyay, N. J. Hillson and J. D. Keasling, Microbial Stress Tolerance for Biofuels: Systems Biology, Springer Science & Business Media, 2011 Search PubMed.
- M. Ouellet, S. Datta, D. C. Dibble, P. R. Tamrakar, P. I. Benke, C. Li, S. Singh, K. L. Sale, P. D. Adams, J. D. Keasling, B. A. Simmons, B. M. Holmes and A. Mukhopadhyay, Green Chem., 2011, 13, 2743 RSC.
- T. L. Ruegg, E.-M. Kim, B. A. Simmons, J. D. Keasling, S. W. Singer, T. S. Lee and M. P. Thelen, Nat. Commun., 2014, 5, 3490 Search PubMed.
- M. Frederix, K. Hütter, J. Leu, T. S. Batth, W. J. Turner, T. L. Rüegg, H. W. Blanch, B. A. Simmons, P. D. Adams, J. D. Keasling, M. P. Thelen, M. J. Dunlop, C. J. Petzold and A. Mukhopadhyay, PLoS One, 2014, 9, e101115 Search PubMed.
- J. Alonso-Gutierrez, R. Chan, T. S. Batth, P. D. Adams, J. D. Keasling, C. J. Petzold and T. S. Lee, Metab. Eng., 2013, 19, 33–41 CrossRef CAS PubMed.
- P. P. Peralta-Yahya, F. Zhang, S. B. del Cardayre and J. D. Keasling, Nature, 2012, 488, 320–328 CrossRef CAS PubMed.
- B. G. Harvey, M. E. Wright and R. L. Quintana, Energy Fuels, 2010, 24, 267–273 CrossRef CAS.
- T. S. Ham, Z. Dmytriv, H. Plahar, J. Chen, N. J. Hillson and J. D. Keasling, Nucleic Acids Res., 2012, 40, e141 CrossRef PubMed.
- T. Baba, T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner and H. Mori, Mol. Syst. Biol., 2006, 2, 2006.0008 CrossRef PubMed.
- P. P. Cherepanov and W. Wackernagel, Gene, 1995, 158, 9–14 CrossRef CAS PubMed.
- K. A. Datsenko and B. L. Wanner, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6640–6645 CrossRef CAS PubMed.
- N. J. Hillson, R. D. Rosengarten and J. D. Keasling, ACS Synth. Biol., 2012, 1, 14–21 CrossRef CAS PubMed.
- D. G. Gibson, L. Young, R.-Y. Chuang, J. C. Venter, C. A. Hutchison and H. O. Smith, Nat. Methods, 2009, 6, 343–345 CrossRef CAS PubMed.
- K. Deng, J. M. Guenther, J. Gao, B. P. Bowen, H. Tran, V. Reyes-Ortiz, X. Cheng, N. Sathitsuksanoh, R. Heins, T. E. Takasuka, L. F. Bergeman, H. Geertz-Hansen, S. Deutsch, D. Loqué, K. L. Sale, B. A. Simmons, P. D. Adams, A. K. Singh, B. G. Fox and T. R. Northen, Front. Bioeng. Biotechnol., 2015, 3, 153 Search PubMed.
- H. Li and R. Durbin, Bioinformatics, 2009, 25, 1754–1760 CrossRef CAS PubMed.
- H. Li, B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis and R. Durbin, Bioinformatics, 2009, 25, 2078–2079 CrossRef CAS PubMed.
- C. Li, D. Tanjore, W. He, J. Wong, J. L. Gardner, K. L. Sale, B. A. Simmons and S. Singh, Biotechnol. Biofuels, 2013, 6, 1–14 CrossRef PubMed.
- T. Shimada, Y. Katayama, S. Kawakita, H. Ogasawara, M. Nakano, K. Yamamoto and A. Ishihama, MicrobiologyOpen, 2012, 1, 381–394 CrossRef CAS PubMed.
- H. Yang, E. Wolff, M. Kim, A. Diep and J. H. Miller, Mol. Microbiol., 2004, 53, 283–295 CrossRef CAS PubMed.
- Y. Nakasone, T. Ono, A. Ishii, S. Masuda and M. Terazima, Biochemistry, 2010, 49, 2288–2296 CrossRef CAS PubMed.
- J. Lee, S. R. Hiibel, K. F. Reardon and T. K. Wood, J. Appl. Microbiol., 2010, 108, 2088–2102 CrossRef CAS PubMed.
- V. Seputiene, D. Motiejūnas, K. Suziedelis, H. Tomenius, S. Normark, O. Melefors and E. Suziedeliene, J. Bacteriol., 2003, 185, 2475–2484 CrossRef CAS PubMed.
- M. Hammar, A. Arnqvist, Z. Bian, A. Olsén and S. Normark, Mol. Microbiol., 1995, 18, 661–670 CAS.
- C.-G. Korea, R. Badouraly, M.-C. Prevost, J.-M. Ghigo and C. Beloin, Environ. Microbiol., 2010, 12, 1957–1977 CrossRef CAS PubMed.
- C. J. Dorman and C. F. Higgins, J. Bacteriol., 1987, 169, 3840–3843 CAS.
- E. Sugawara and H. Nikaido, J. Biol. Chem., 1992, 267, 2507–2511 CAS.
- Y. Itoh, J. D. Rice, C. Goller, A. Pannuri, J. Taylor, J. Meisner, T. J. Beveridge, J. F. Preston and T. Romeo, J. Bacteriol., 2008, 190, 3670–3680 CrossRef CAS PubMed.
- C. Wirth, G. Condemine, C. Boiteux, S. Bernèche, T. Schirmer and C. M. Peneff, J. Mol. Biol., 2009, 394, 718–731 CrossRef CAS PubMed.
- A. M. Redding-Johanson, T. S. Batth, R. Chan, R. Krupa, H. L. Szmidt, P. D. Adams, J. D. Keasling, T. S. Lee, A. Mukhopadhyay and C. J. Petzold, Metab. Eng., 2011, 13, 194–203 CrossRef CAS PubMed.
- N. Sun, H. Liu, N. Sathitsuksanoh, V. Stavila, M. Sawant, A. Bonito, K. Tran, A. George, K. Sale and S. Singh, Biotechnol. Biofuels, 2013, 6(1), 39 CrossRef CAS PubMed.
- J. I. Park, M. S. Kent, S. Datta, B. M. Holmes, Z. Huang, B. A. Simmons, K. L. Sale and R. Sapra, Bioresour. Technol., 2011, 102, 5988–5994 CrossRef CAS PubMed.
- Y. Wang, Biochem. Biophys. Res. Commun., 2002, 292, 396–401 CrossRef CAS PubMed.
- E. Husson, S. Buchoux, C. Avondo, D. Cailleu, K. Djellab, I. Gosselin, O. Wattraint and C. Sarazin, Bioresour. Technol., 2011, 102, 7335–7342 CrossRef CAS PubMed.
- L. J. Jönsson, B. Alriksson and N.-O. Nilvebrant, Biotechnol. Biofuels, 2013, 6, 16 CrossRef PubMed.
- M. J. Dunlop, J. D. Keasling and A. Mukhopadhyay, Syst. Synth. Biol., 2010, 4, 95–104 CrossRef PubMed.
- M. J. Dunlop, Z. Y. Dossani, H. L. Szmidt, H. C. Chu, T. S. Lee, J. D. Keasling, M. Z. Hadi and A. Mukhopadhyay, Mol. Syst. Biol., 2011, 7, 487 CrossRef PubMed.
- J. L. Foo, H. M. Jensen, R. H. Dahl, K. George, J. D. Keasling, T. S. Lee, S. Leong and A. Mukhopadhyay, mBio, 2014, 5, e01932 CrossRef CAS PubMed.
- P. Fernandes, B. S. Ferreira and J. M. S. Cabral, Int. J. Antimicrob. Agents, 2003, 22, 211–216 CrossRef CAS PubMed.
- A. Mukhopadhyay, Trends Microbiol., 2015, 23, 498–508 CrossRef CAS PubMed.
- S. Wagner, L. Baars, A. J. Ytterberg, A. Klussmeier, C. S. Wagner, O. Nord, P.-A. Nygren, K. J. van Wijk and J.-W. de Gier, Mol. Cell. Proteomics, 2007, 6, 1527–1550 CAS.
- T. S. Lee, R. A. Krupa, F. Zhang, M. Hajimorad, W. J. Holtz, N. Prasad, S. K. Lee and J. D. Keasling, J. Biol. Eng., 2011, 5, 12 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6gc00642f |
| This journal is © The Royal Society of Chemistry 2016 |